Abstract
A model for the secondary structure of mouse beta Maj globin messenger RNA is presented based on enzymatic digestion data, comparative sequence and computer analysis. Using 5'-32P-end-labeled beta globin mRNA as a substrate, single-stranded regions were determined with S1 and T1 nucleases and double-stranded regions with V1 ribonuclease from cobra venom. The structure data obtained for ca. 75% of the molecule was introduced into a computer algorithm which predicts secondary structures of minimum free energy consistent with the enzymatic data. Two prominent base paired regions independently derived by phylogenetic analysis were also present in the computer generated structure lending support for the model. An interesting feature of the model is the presence of long-range base pairing interactions which permit the beta globin mRNA to fold back on itself, thereby bringing the 5'- and 3'-noncoding regions within close proximity. This feature is consistent with data from other laboratories suggesting an interaction of the 5'- and 3'-domains in the mammalian globin mRNAs.
Full text
PDF


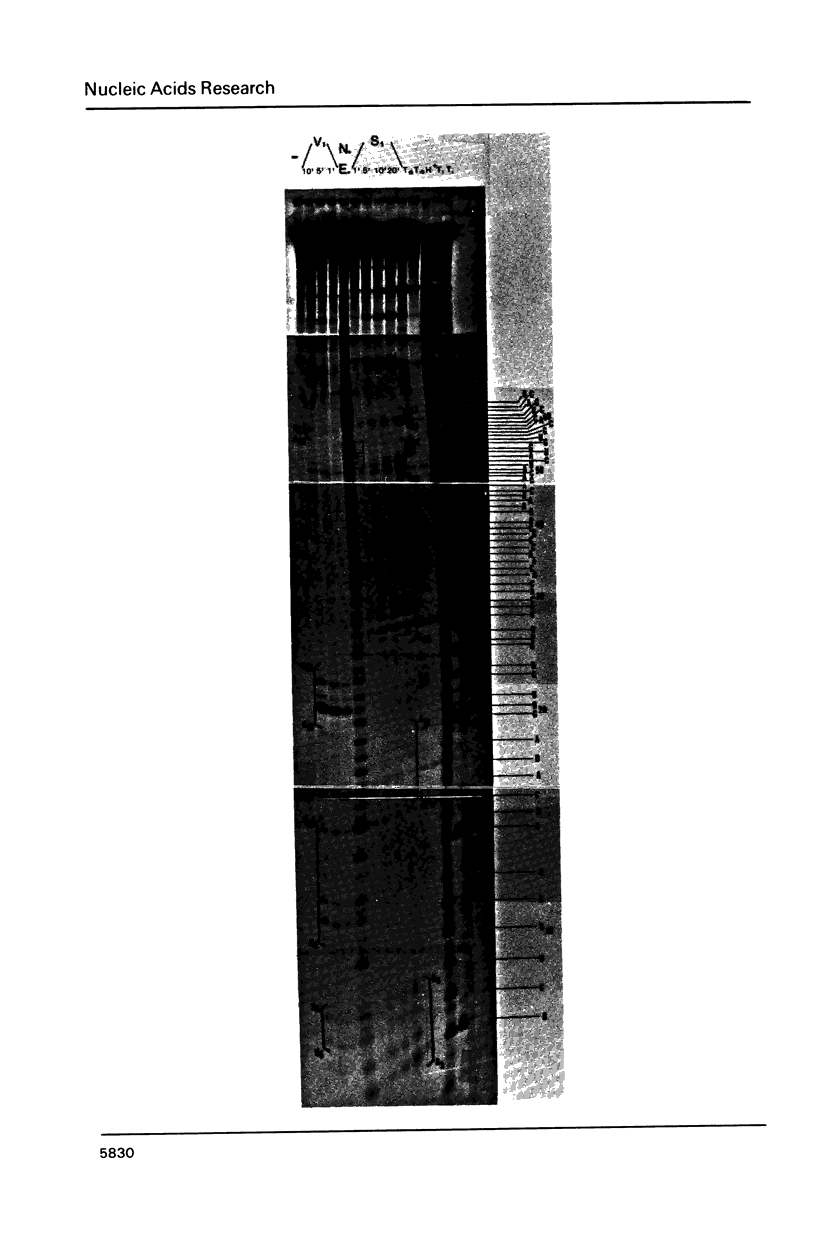

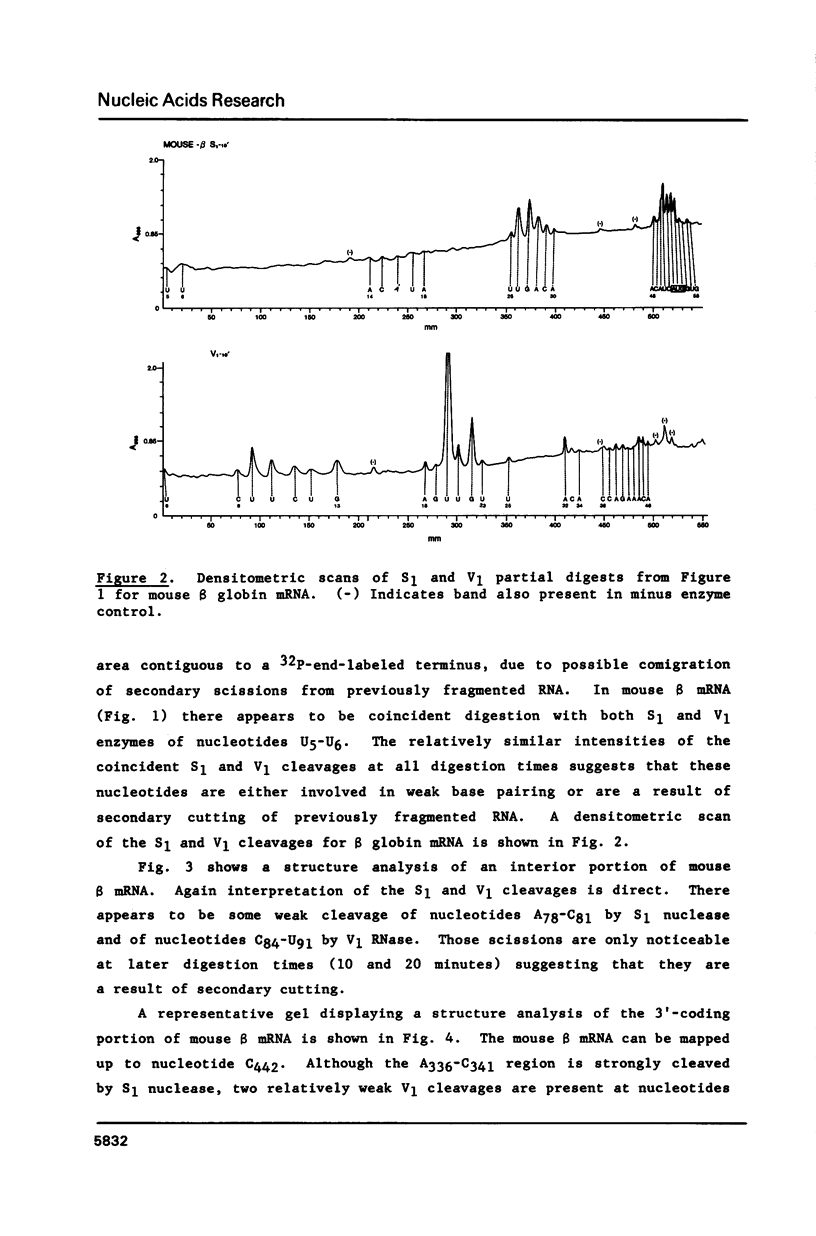
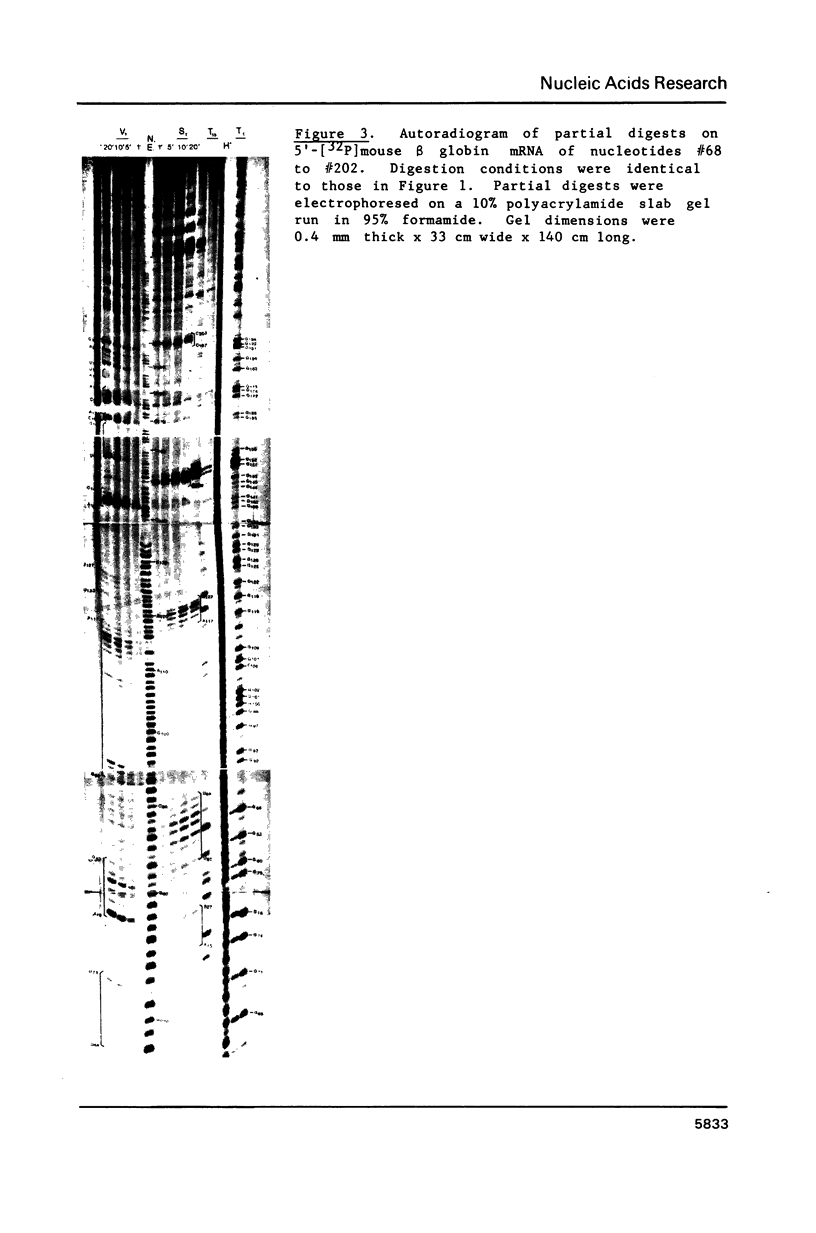


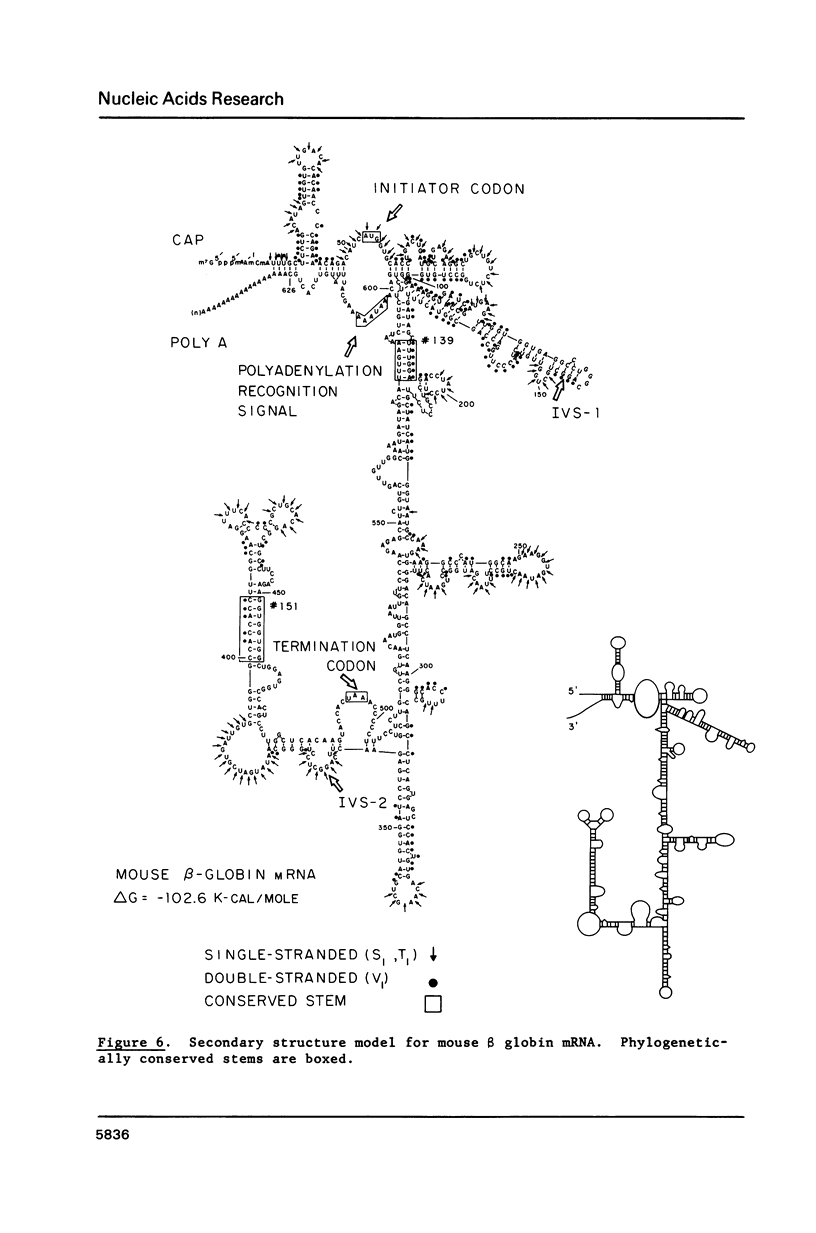





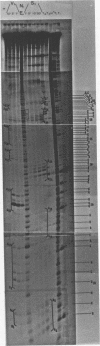
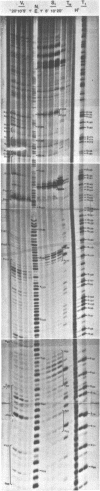
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auron P. E., Rindone W. P., Vary C. P., Celentano J. J., Vournakis J. N. Computer-aided prediction of RNA secondary structures. Nucleic Acids Res. 1982 Jan 11;10(1):403–419. doi: 10.1093/nar/10.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browner M. F., Lawrence C. B. Comparative sequence analysis as a tool for studying the secondary structure of mRNAs. Nucleic Acids Res. 1985 Dec 9;13(23):8645–8660. doi: 10.1093/nar/13.23.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connaughton J. F., Rairkar A., Lockard R. E., Kumar A. Primary structure of rabbit 18S ribosomal RNA determined by direct RNA sequence analysis. Nucleic Acids Res. 1984 Jun 11;12(11):4731–4745. doi: 10.1093/nar/12.11.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J., Morel C., Lebleu B., Herzberg M. Structure of globin mRNA and mRNA-protein particles. Use of dark-field electron microscopy. Eur J Biochem. 1973 Jul 16;36(2):465–472. doi: 10.1111/j.1432-1033.1973.tb02931.x. [DOI] [PubMed] [Google Scholar]
- Favorova O. O., Fasiolo F., Keith G., Vassilenko S. K., Ebel J. P. Partial digestion of tRNA--aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry. 1981 Feb 17;20(4):1006–1011. doi: 10.1021/bi00507a055. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gehrke L., Auron P. E., Quigley G. J., Rich A., Sonenberg N. 5'-Conformation of capped alfalfa mosaic virus ribonucleic acid 4 may reflect its independence of the cap structure or of cap-binding protein for efficient translation. Biochemistry. 1983 Oct 25;22(22):5157–5164. doi: 10.1021/bi00291a015. [DOI] [PubMed] [Google Scholar]
- Holder J. W., Lingrel J. B. Determination of secondary structure in rabbit globin messenger RNA by thermal denaturation. Biochemistry. 1975 Sep 23;14(19):4209–4215. doi: 10.1021/bi00690a009. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979 Jul 26;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel D. A., Tilghman S. M., Leder P. The sequence of the chromosomal mouse beta-globin major gene: homologies in capping, splicing and poly(A) sites. Cell. 1978 Dec;15(4):1125–1132. doi: 10.1016/0092-8674(78)90040-5. [DOI] [PubMed] [Google Scholar]
- Ladhoff A. M., Uerlings I., Rosenthal S. Electron microscopic evidence of circular molecules in 9-S globin mRNA from rabbit reticulocytes. Mol Biol Rep. 1981 May 22;7(1-3):101–106. doi: 10.1007/BF00778739. [DOI] [PubMed] [Google Scholar]
- Liebhaber S. A., Kan Y. W. Different rates of mRNA translation balance the expression of the two human alpha-globin loci. J Biol Chem. 1982 Oct 25;257(20):11852–11855. [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Lingrel J. B. Mouse hemoglobin messenger ribonucleic acid. Translational capacities of rabbit and duck reticulocyte cell-free systems programmed with mouse 9 S ribonucleic acid. J Biol Chem. 1972 Jul 10;247(13):4174–4179. [PubMed] [Google Scholar]
- Lockhard R. E., Rajbhandary U. L. Nucleotide sequences at the 5'termini of rabbit alpha and beta globin mRNA. Cell. 1976 Dec;9(4 Pt 2):747–760. doi: 10.1016/0092-8674(76)90138-0. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Shelness G. S., Williams D. L. Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. Evaluation of primer extension for high resolution structure mapping of mRNA. J Biol Chem. 1985 Jul 15;260(14):8637–8646. [PubMed] [Google Scholar]
- Spena A., Krause E., Dobberstein B. Translation efficiency of zein mRNA is reduced by hybrid formation between the 5'- and 3'-untranslated region. EMBO J. 1985 Sep;4(9):2153–2158. doi: 10.1002/j.1460-2075.1985.tb03909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Toots I., Misselwitz R., Böhm S., Welfle H., Villems R., Saarma M. Two distinct conformations of rat liver ribosomal 5S RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3381–3389. doi: 10.1093/nar/10.11.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNase H-catalyzed site-specific deadenylylation of rabbit alpha- and beta- globin mRNAs. Secondary structure of 3'-noncoding regions. J Biol Chem. 1984 Mar 10;259(5):3299–3307. [PubMed] [Google Scholar]
- Warner J. R., Rich A., Hall C. E. Electron Microscope Studies of Ribosomal Clusters Synthesizing Hemoglobin. Science. 1962 Dec 28;138(3548):1399–1403. doi: 10.1126/science.138.3548.1399. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]