Abstract
BWI-1 (buckwheat trypsin inhibitor), a member of the potato inhibitor I family, suppresses the growth of T-acute lymphoblastic leukemia cells and induces apoptosis in human solid tumor cell lines. Here, we report the crystal structure of rBTI (recombinant buckwheat trypsin inhibitor), a recombinant protein of BWI-1, at 1.84 Å resolution and the structure of rBTI in complex with bovine trypsin at 2.26 Å resolution. A conformational change of Trp53 at the P8′ position in rBTI was observed upon its binding to trypsin, which is not seen in other members of the potato inhibitor I family reported previously. The role of the P8′ residue in the potato inhibitor I family was examined by measuring the association and dissociation rates of four rBTI mutants with different substitutions at the P2 and P8′ positions when binding to trypsin. One of the mutants, P44T, was found to be a much stronger inhibitor than wild-type rBTI, with a picomolar (pM) dissociation constant. Our results could provide valuable insights for designing a new rBTI-based antitumor drug in the future.
Introduction
Canonical inhibitors of serine protease function according to the standard mechanism of protease inhibition in which they bind tightly in the active site of a cognate protease in a substrate-like manner (substrate residues of protease inhibitors surrounding the cleavage site are designated by the nomenclature of Schechter and Berger [1]. The scissile bond is the starting point. In the direction of the N terminus, substrate residues are numbered P1, P2, P3 and so on, and in the direction of the C terminus, residues are numbered P1′, P2′, P3′ and so on.) [2]. However, unlike substrates, canonical inhibitors cannot be easily hydrolyzed by proteases, which is attributed to the rigidity of their convex binding loop [3]. The protein core of a canonical inhibitor serves as a scaffold for the binding loop and is responsible for maintaining the binding loop stability. A previous study revealed that an inhibitor could quickly form an acyl-enzyme intermediate with a protease but was hydrolyzed very slowly. Thus, a clogged gutter mechanism was proposed to underscore two key factors in protease inhibition: the intramolecular hydrogen-bonding network and the correct orientation of the religating amide [4].
The potato inhibitor I family belongs to the canonical inhibitors, and their P2, P1 ′, P6′, and P8′ residues are highly conserved due to their importance in the formation of the internal hydrogen-bonding network between the binding loop and protein core. Mutations of either P2 Thr or P1′ Glu in CI-2 (chymotrypsin inhibitor 2) result in a dramatic increase of the dissociation constant between CI-2 and chymotrypsin [5]. P6′ and P8′ mutants of CMTI-V (cucurbita maxima trypsin inhibitor V) have been proven to be very unstable. The P6′ mutant, in particular, can be easily hydrolyzed by trypsin [6].
Recently, attentions have been drawn to another member of the potato inhibitor I family from buckwheat seeds, BWI-1 (Buckwheat Inhibitor 1). BWI-1 was sequenced and characterized in buckwheat seeds soon after its discovery [7], [8], [9]. A previous cytobiology study revealed that BWI exhibits suppression activity against human T-Acute lymphoblastic leukemia cell lines [10]. In the past few years, Wang and her colleagues has focused on the antitumor activity of the BWI-1 recombinant protein rBTI (recombinant buckwheat trypsin inhibitor) [11] and has investigated its effects on the induction of apoptosis in several human solid tumor cell lines (EC907, HepG2 and HeLa) [12]. Additionally, the resistance of tobacco and potatoes to biotic stress can be improved by introducing the BWI-1 encoding gene [13].
Interestingly, BWI-1 has an uncommon binding loop sequence with a Pro at the P2 position and Trp at the P8′ position, suggesting a unique mode of intramolecular interactions between the binding loop and the protein core. Because the inhibition activity of certain canonical inhibitors is strongly affected by their intramolecular hydrogen-bonding network [4], it is logical to propose that BWI-1 inhibits proteases in an unusual way.
Here, we report the crystal structure of rBTI at 1.84 Å resolution and the structure of rBTI-trypsin complex at 2.26 Å resolution. Curiously, structural superposition revealed a significant conformational change of P8′ Trp in rBTI upon binding to trypsin. Several rBTI mutants were constructed to mimic different binding loop conformations of potato inhibitor I family members. Their association and dissociation rates upon binding to bovine trypsin were determined, allowing us to correlate several binding loop conformations with their inhibition abilities in the potato inhibitor I family. Out of our expectations, one of the mutants, P44T, was found to be a much stronger inhibitor compared to the wild-type with a picomolar (pM) dissociation constant. These results allow us to propose a detailed model for the structural basis of protease inhibition of the potato Inhibitor I family.
Results and Discussion
Overall structure of native rBTI and its complex with bovine trypsin
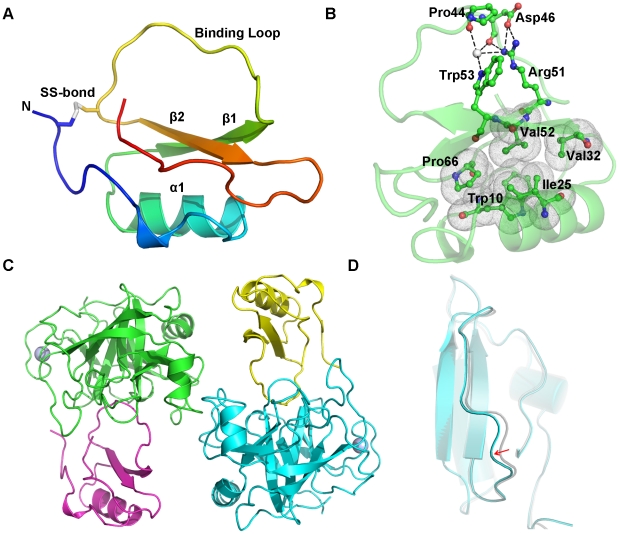
The structure of rBTI is composed of 69 amino acid residues. Its main structural elements comprise a single α-helix (α1, residues 18 to 28), a central parallel β-sheet consisting of two strands (β1, residues 30 to 38; β2, residues 51 to 56), a binding loop (residues 39 to 50) and two irregular structures at the N-terminus (residues 3 to 15) and C-terminus (residues 61 to 69)(Figure 1A). A hydrophobic core is formed in rBTI among α1, β1, β2 and two short loops (side-chains of Trp10, Ile25, Val32, Val52 and Pro66), as shown in Figure 1B. The Cys4-Cys49 disulfide bond stabilizes the binding loop by connecting it with the N-terminus. The binding loop of rBTI is a convex loop sandwiched between β1 and β2. Within the loop, the P1 residue, Arg45, is an ideal substrate of trypsin, which ensures the inhibitor's tight binding to trypsin. As in other canonical inhibitors, the binding loop of rBTI forms a hydrogen-bonding network with the protein core [6], [14], [15](Figure 1B). This hydrogen-bonding network is one of the key factors that causes inhibitors to be hydrolyzed at a very slow rate [3].
Figure 1.
(A) Cartoon representation of the overall structure of rBTI. Different structural elements are shown in different colors, and the disulfide bridge is indicated. (B) A view of the hydrogen-bonding network and the hydrophobic core in rBTI. rBTI is shown in a cartoon presentation in green. Residues involved in hydrogen-bonding network and hydrophobic core are shown as ball-and-stick models. The grey sphere indicates a water molecule. Hydrogen bonds are indicated by black dashes and hydrophobic interactions are indicated by dotted clouds. (C) Overview of the structure of rBTI-trypsin complexes within an asymmetric unit. rBTIs are shown in yellow and magenta; trypsins are shown in green and cyan. The calcium ions in trypsin are shown as light-blue spheres. (D) Superposition of trypsin-bound rBTI and free rBTI. The binding loop of trypsin-bound rBTI (cyan) is shifted by a small distance from that of free rBTI (grey). The RMSD value calculated by superposition of trypsin-bound rBTI's and free rBTI's binding loops is 0.26 Å.
In the crystal structure of the rBTI-trypsin complex, one crystallographic asymmetric unit contains two rBTI-trypsin complexes, that is, two rBTIs and two trypsins, as shown in Figure 1C. The structure of bovine trypsin in complex with rBTI aligns well with other trypsin structures deposited in the PDB database. Trypsin-bound rBTI has an overall structure similar to that of free rBTI, with both consisting of one α-helix and a central parallel β-sheet. By superposing trypsin-bound rBTI over free rBTI, we found that the Arg45 at the P1 position was buried deeply into the binding pocket of trypsin, leading to a small but noticeable shift of the binding loop towards trypsin (RMSD 0.26 Å, Figure 1D). This movement disrupts several hydrogen bonds between the binding loop and protein core, indicating that a significant conformational change of the binding loop occurs upon binding to trypsin.
Comparison of the binding loops between rBTI and LUTI
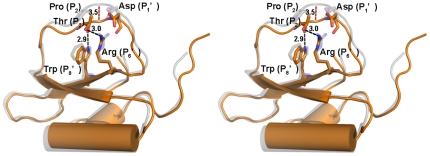
As noted, the P6′ and P8′ residues play important roles in maintaining the stability of the binding loops of inhibitors in the potato inhibitor I family. Mutations of the P6′ and P8′ residues would destabilize the binding loop structure, resulting in a significant decrease in the inhibitor's activity [6], [15]. Therefore, the P6′ and P8′ residues are highly conserved among members of the potato inhibitor I family. Almost all reported structures in the potato inhibitor I family exhibit an Arg at the P8′ position, with the exception of rBTI and LUTI (linum usitatissimum trypsin inhibitor), in which the P8′ residue is Trp (Figure S1). It is noteworthy that although the sequence homology of LUTI and rBTI is very high (58% sequence identity), their binding loops have completely different conformations.
In the structure of LUTI, Thr44 at the P2 position forms hydrogen bonds with Arg51 at the P6′ position and Trp53 at the P8′ position (Figure 2). Cierpicki et al. proposed that because Trp has a shorter side-chain than Arg, the binding loop of LUTI would be much closer to the protein core than other members of the potato inhibitor I family [14].
Figure 2. Stereo view of the superposition of LUTI and rBTI showing the different conformations of their binding loops.
LUTI is shown in brown, and rBTI is shown in grey. Red dashes indicate the distance between the Cα atoms of the P1 residues in rBTI and LUTI.
However, in the case of rBTI, there is a Pro at the P2 position instead of Thr. Therefore, no hydrogen bond can be formed between the P2 and P8′ residues or between the P2 and P6′ residues, resulting in an extended binding loop distant from the protein core. After superposing LUTI over rBTI, we observed that the binding loop of rBTI was approximately 3.5 Å more distant from the protein core than that of LUTI. Moreover, the P2 Pro appears to be responsible for the conformational change of the binding loop after rBTI binds to trypsin.
Conformational change of the binding loop of rBTI
Given that rBTI presents an uncommon residues (Trp) at the P8′ position and that its binding loop is different from that of LUTI, which also has a Trp at the P8′ position, further investigation of the P8′ Trp of rBTI appears to be a promising means through which to shed some light on the inhibition mechanism of rBTI.
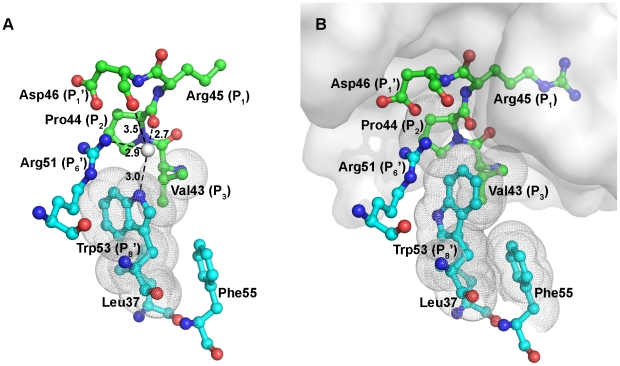
It is generally believed that the P8′ residue, as well as other residues that form the intramolecular hydrogen-bonding network in the potato inhibitor I family, maintains a relatively stable conformation upon binding to proteases [3]. However, superposition of free rBTI and trypsin-bound rBTI suggests that a significant conformational change occurs when rBTI binds to trypsin. As shown in Figure 3, in free rBTI, a water molecule forms hydrogen bonds with Trp53 at the P8′ position, Arg51 at the P6′ position, the α carbonyl oxygen of Pro44 at the P2 position and the α carbonyl oxygen of Asp46 at the P1′ position. This water molecule mediates the interactions between the binding loop and the protein core, stabilizing the structure of the binding loop. When rBTI binds to trypsin, its P1 residue fits into trypsin's binding pocket and forms hydrogen bonds with trypsin residues. Because of this tight binding, the binding loop of rBTI is pulled towards trypsin slightly (Figure 1D), which shifts the P2 and P1′ residues further from the protein core and causes a rupture of the hydrogen bonds around the water molecule, leading to the dissociation of this water molecule. As a consequence, the binding loop of rBTI cannot maintain a stable conformation and risks being hydrolyzed by trypsin, similarly to a regular substrate. To counteract with this situation, rBTI employs a local conformational change mechanism: the Trp53 at the P8′ position undergoes a rotameric switch, with its indole ring flipping upwards and occupying the original position of the water molecule (Figure S3). In this new conformation, Trp53 forms a stable cation-π interaction (examined by the CaPTURE program [16]) with Arg51 at the P6′ site and interacts with hydrophobic residues from the binding loop, thereby stabilizing the binding loop.
Figure 3. Structural differences between free rBTI and trypsin-bound rBTI at the local region around the P8′ position.
(A) Interactions between the P2 and P8′ residues in free rBTI. (B) Interactions between the P2 and P8′ residues in trypsin-bound rBTI. Side-chains of residues in the binding loop are shown in green, while those in the protein core are shown in cyan. The grey sphere indicates a water molecule. Trypsin is shown as surface. Hydrophobic interactions are indicated by dotted clouds. Part of the side chain of Arg45 in free rBTI is missing due to poor electron densities.
Binding loop conformations of the potato inhibitor I family and their inhibition activities
Following the discovery of the rotameric switch of the P8′ Trp in rBTI, two questions arose: why does rBTI exhibit such a rare inhibition mechanism; and what roles do the P8′ and P2 residues play in its inhibition activity? To address these questions, we referred to the structure of several classical members of the potato inhibitor I family and designed four rBTI mutants. These mutants each include substitutions at the P2 and P8′ positions to mimic different binding loop conformations of the potato inhibitor I family members. Interactions of wild-type rBTI and rBTI mutants with bovine trypsin were investigated by means of an optical biosensor using the surface plasmon resonance (SPR) effect. Their association rate (ka) and dissociation rate (kd) and the dissociation constant (KD) were determined (Table 1, Figure S2).
Table 1. Association rates (ka), dissociation rates (kd) and dissociation constants (KD) for the interactions of wild-type rBTI and its mutants with bovine trypsin.
| Inhibitor | P8′ residue | P2 residue | Speculated interaction | Reference Model | ka, M−1s−1 | kd, s−1 | KD(kd/ka), M |
| WT rBTI | Trp53 | Pro44 | Hydrophobic force | rBTI | 4.62×105 | 1.25×10−3 | 2.69×10−9 |
| P44T | Trp53 | Thr44 | Hydrogen bond | LUTI | 3.46×105 | 5.24×10−7 | 1.52×10−12 |
| W53R/P44T | Arg53 | Thr44 | Hydrogen bond | CI-2 | 6.79×105 | 5.54×10−4 | 8.15×10−10 |
| W53F | Phe53 | Pro44 | Hydrophobic force | - | 3.99×105 | 2.46×10−3 | 6.16×10−9 |
| W53R | Arg53 | Pro44 | - | - | 6.59×105 | 9.97×10−3 | 1.51×10−8 |
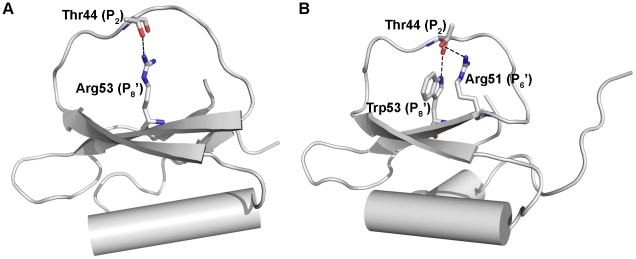
Both rBTI mutants with an Arg substitution at the P8′ position (W53R, W53R/P44T) show elevated association rates compared to the wild-type rBTI for tryspin. As proposed previously, the closer packing of the binding loop of LUTI against the protein core compared to CI-2 and eglin C is attributed to the shorter side-chain of Trp compared to Arg [14]. We speculated that it is the same in rBTI: because Arg has a longer side-chain than Trp or Phe, mutants with Arg at the P8′ position (W53R, W53R/P44T) exhibit more extended binding loops than other forms of rBTI (wild-type rBTI, P44T, W53F) (Figure 4A). Thus, the binding loops of W53R and W53R/P44T are more accessible to the substrate pocket of trypsin, resulting in elevated association rates. As noted earlier, for wild-type rBTI, a water molecule mediates the interaction between the binding loop and protein core, suggesting that the binding loop of wild-type rBTI is also extended. This could explain why rBTI exhibits a third higher association rate among the wild-type rBTI and all rBTI mutants.
Figure 4. Putative rBTI mutant structures based on inhibitors homologous to rBTI.
(A) Possible structure of W53R/P44T based on CI-2. (B) Possible structure of P44T based on LUTI.
We also found that mutants that are able to form hydrogen bonds between the P2 and P8′ residues (P44T, W53R/P44T) present significantly lower dissociation rates compared to wild-type for trypsin. The most surprising outcome from our kinetic analyses is that the dissociation rate of P44T is three orders of magnitude lower than that of W53R/P44T, indicating that P44T is a very strong inhibitor to trypsin. We speculated that the much lower dissociation rate of P44T might be attributed to two factors: one is that a second hydrogen bond formed between P2 and P6′ (prediction based on the structure of LUTI, Figure 4B); the other is that the side-chain of Trp is more rigid than that of Arg. A combination of these two factors stabilizes the binding loop conformation, resulting in a more than 103-fold decrease in the dissociation constant when it binds trypsin.
Among all investigated forms of rBTI, W53R exhibited the highest dissociation rate. In BIAcore binding assays, W53R was the only mutant that did not require the regeneration of immobilized trypsin, suggesting that it had lost the normal function of an inhibitor. The reason might be the lack of stable interactions formed between its P2 Pro and P8′ Arg. Therefore, the binding loop of W53R is very unstable and is vulnerable to hydrolysis by trypsin. This result underpins the important role of the P8′ residue in the inhibitory functioning of the potato inhibitor I family. However, our speculation needs further experimental proof.
Of all the inhibitors that cannot form a hydrogen bond between P2 and P8′, wild-type rBTI showed the lowest dissociation rate. We assumed that this was related to the conformational change within rBTI: upon binding to trypsin, Trp53 flips up and forms a relatively more stable conformation. In comparison with W53F, we found that wild-type rBTI had a slightly higher association rate and a lower dissociation rate, suggesting that the rotameric switch of Trp53 might improve the inhibition activity of wild-type rBTI.
Based on these results, we inferred that in the potato inhibitor I family, at least, the association rate and dissociation rate of an inhibitor to a protease are not necessarily related, which is consistent with the results of a previous study [17]. In our study, we found that the association rate was determined by the extended shape of the binding loop, as well as its amino acid composition [18]. However, the dissociation rate is impacted by the overall structure of the inhibitor, in particular, the internal hydrogen-bonding network between the binding loop and the protein core. Based on the analysis of our data, we further speculated that in the potato inhibitor I family, inhibitors with more extended binding loops bind to protease at a higher association rate; interactions between the P8′ and P2 residues of an inhibitor can significantly affect the dissociation rate to its cognate protease, which can be summarized as: kd-hydrogen<kd-nonhydrogen<kd-non in which kd-hydrogen is the dissociation rate of inhibitors that have a hydrogen bond between the P8′ and P2 residues, while kd-nonhydrogen denotes the dissociation rate of inhibitors presenting interactions other than hydrogen-bonding between the P8′ and P2 residues, and kd-non denotes inhibitors exhibiting no interaction between the P8′ and P2 residues. Among inhibitors that form hydrogen bonds between P8′ and P2, those that can form extra hydrogen bonds or present a more rigid P8′ side-chain exhibit the lowest dissociation rate. Our conclusion might also apply to other types of canonical inhibitors that form hydrogen bonds at different positions.
Research on the potato inhibitor I family began in the mid-19th century. However, conformational changes of the P8′ residues of these inhibitors upon binding to proteases in a similar way to what is seen for rBTI have not been reported before. In the present study, we compared several different binding loop conformations in the potato inhibitor I family on the same protein core. Such comparisons allowed us to better understand the relationship between the conformation of the binding loops and their inhibition activities.
This comparison also provided us a clearer picture of the inhibition mechanism of rBTI: the rotameric switch of the P8′ Trp results in rBTI being a relatively weak inhibitor but does not compromise its proteolytic stability, which is a reflection of biological diversity. In this case, rBTI could be suitable for certain physiological processes that require a weak, but stable, inhibitor.
It is also worth mentioning that the P44T mutant we constructed presents a dissociation constant three orders of magnitude lower than that of wild-type rBTI. This mutant may represent a good example of improving an inhibitor's binding affinity to its cognate protease. Therefore, it could provide useful information for improving the binding affinity of inhibitor drugs to their target proteins.
As noted earlier, although rBTI can suppress tumor cell growth and induce apoptosis in several tumor cell lines [10], [11], [12], the mechanism underlying its antitumor activity is currently unknown. This structural study of rBTI represents a first step towards understanding its antitumor mechanism. Moreover, The rBTI mutants with an improved inhibition activity for trypsin investigated in the present study may facilitate designing inhibitors with higher antitumor activities and are of potential therapeutic value.
Materials and Methods
Expression, purification and crystallization of rBTI and the rBTI-trypsin complex
rBTI was prepared as described previously [11]. The rBTI crude sample was then applied to Superdex75 (GE Healthcare). The elution buffer used was 25 mM Tris-HCl (pH 8.0) and 50 mM NaCl. The fractions containing rBTI were collected and concentrated to 20 mg/ml. rBTI was crystallized by vapor diffusion. Crystals were grown at 18°C in hanging drops over a reservoir of 24% (w/v) PEG MME2000, 220 mM (NH4)2SO4, 100 mM NaAc (pH 4.4) and 100 mM NaI. Drops were prepared by mixing equal volumes of protein and reservoir solutions. After two weeks, thin and rod-like crystals were harvested, soaked in a cryoprotectant mixture (paraffin oil and NVH oil in a ratio of 7∶3) and flash-frozen in liquid nitrogen.
rBTI and bovine trypsin (AppliChem) were mixed in a 1∶1.3 stoichiometric molar ratio and incubated at room temperature for a half an hour to form complexes. The incubation buffer contained 50 mM Tris-HCl (pH 8.0) and 200 mM NaCl. After incubation, the incubation sample was applied to Superdex75 to remove excessive rBTI. The elution buffer used was 50 mM Tris pH 8.0 and 20 mM NaCl. The complex of rBTI with bovine trypsin was also crystallized by vapor diffusion. Crystals were grown at 18°C in hanging drops over a reservoir of 15% (w/v) PEG3350, 200 mM MgCl2 and 100 mM Tris-HCl (pH 9.0). Drops were prepared by mixing equal volumes of protein and reservoir solutions. Rod-like crystals grew over the course of two weeks. They were then harvested, soaked in a cryoprotectant solution (100 mM Tris-HCl pH 9.0, 20% (w/v) PEG3350, 20% (v/v) glycerol and 200 mM MgCl2) and flash-frozen in liquid nitrogen.
X-ray data collection and processing
For rBTI, synchrotron X-ray data were collected from a single crystal at 100 K using a MAR555 CCD detector at beamline 1W2B, BSRF (Beijing Synchrotron Radiation Facility). For rBTI in complex with bovine trypsin, X-ray data were collected from a single crystal at 100 K using a Raxis4 IP detector at the Institute of Microbiology, Chinese Academy of Science. All data were processed and scaled with the HKL2000 software suite [19].
Phasing
The rBTI-trypsin complex was the first to yield diffraction quality crystals. An incomplete structure containing only trypsin was solved by molecular replacement using the program Phaser [20]. The search model was derived from a previous structure of trypsin (PDB entry 2CMY). After failed attempts at molecular replacement using two search models (LUTI, linum usitatissimum trypsin inhibitor, PDB entry 1DWM; CMTI-V, cucurbita maxima trypsin inhibitor-V, PDB entry 1 HYM) that share the highest sequence homology with rBTI, we used MrBump [21] to perform a search for homologous structures. Then, one of the two rBTIs in the asymmetric unit was solved using a search model of BGTI (Bitter Gourd Trypsin Inhibitor, PDB entry 1VBW). The other rBTI was solved by superposing one rBTI-trypsin complex over the other trypsin using trypsin as the reference structure in the asymmetric unit.
The rBTI structure was solved by molecular replacement using the program Phaser [20] with the solved trypsin-bound rBTI structure as a search model.
Model Refinement
Models were rebuilt using the model-building module of the PHENIX software suite [22]. Cycles of manual rebuilding in COOT [23] were alternated with automated refinement using the refinement module of PHENIX. Test sets comprised 5% of the total reflections were excluded from refinement to allow the calculation of the free R-factors. Composite omit maps generated by the CNS software suite [24] and prime and switch maps generated by Resolve [25] were used as reference maps in manual rebuilding. Model validations were carried out using PROCHECK [26]. Superpositions were performed using COOT, andall figures representing structures were created using the graphics software PyMOL. A summary of the data collection and refinement statistics is presented in Table 2. The coordinates and structure factors of rBTI were deposited into RCSB Protein Data Bank with accession code 3RDY. The coordinates and structure factors of rBTI-trypsin complex were deposited with accession code 3RDZ.
Table 2. Summary of Data Collection and Refinement Statistics.
| rBTI-trypsin complex | rBTI | |
| Wavelength (Å) | 1.5418 | 1.00 |
| Space group | P21 | P43212 |
| Resolution rangea (Å) | 12.0−2.26(2.34−2.26) | 15.0−1.84(1.91−1.84) |
| Unique Reflections | 26279 | 8344 |
| Unit Cell (a,b,c) (Å) | 66.7, 50.2, 84.5 | 62.7,62.7,45.9 |
| Completenessa (%) | 99.8(97.7) | 99.8(100) |
| Redundencya | 3.7(3.6) | 24.1(23.2) |
| Averagea b I/σ | 22.6(4.9) | 46.4(4.7) |
| Rmerge a (%) | 5.5(25.3) | 8.0(50) |
| a.s.u content | ||
| No. trypsin | 2 | - |
| No. rBTI | 2 | 1 |
| No. Non-hydrogen atoms | 4247 | 494 |
| No. Ca2+ | 2 | - |
| No. water molecules | 300 | 94 |
| R factor and Rfree (%) | 18.2/22.6 | 19.1/21.6 |
| r.m.s deviations: | ||
| Bond length (Å) | 0.0075 | 0.012 |
| Bond angles (deg) | 1.116 | 1.390 |
| B-factors (Å2): | ||
| Protein | 30.9 | 28.8 |
| Main-chain | 29.5 | 25.7 |
| Side-chain and water | 32.4 | 31.3 |
Outer shell values are given in parentheses.
I is the intensity; σ is the standard deviation.
Expression and purification of the mutants
The rBTI expression construct was mutagenized using the PCR-based QuickChange method (Stratagene). We designed four mutants based on the structure of the homologous inhibitors of rBTI. They were P44T, W53F, W53R and W53R/P44T double mutant. The mutants of rBTI were expressed and purified in the same way as wild-type rBTI.
BIAcore Binding Assays
Interactions of wild-type rBTI and its mutants with bovine trypsin (AppliChem) were measured using the optical biosensor BIAcore 3000 and CM5 optical chips. Carboxymethlated dextran on the chip surface was activated with the mixture 0.2 M EDC/0.05 M NHS. The subsequent immobilization of bovine trypsin was carried out by injecting a trypsin solution (20 µg/ml in 10 mM acetate buffer, pH 5.5) at a flow rate of 5 µl/min over the activated sensor surface. The residual active groups of dextran were blocked by 1 M ethanolamine.
Interactions of different inhibitors with immobilized trypsin were studied using concentrations of 7.4 nM, 22.2 nM, 66.7 nM and 200 nM (at a flow rate of 30 µl/min for 1 min) in running buffer containing 100 mM NaCl, 10 mM Na2HPO4∶NaH2PO4 (pH 8.0) (Figure S2). After the injection of each inhibitor sample, except W53R, the chip was regenerated by the injection of 10 mM Glycine-HCl (pH 3.0). A channel without immobilized protein was used as a reference. Running buffers without inhibitors were used to generate the baseline. Kinetic parameters were calculated using the program BIAevaluation, and the mathematical model was 1∶1 Langmuir binding.
Supporting Information
Sequence alignment of several members of the potato inhibitor I family. The binding loop are marked with grey alpha boxes.
(TIF)
Sensograms of the interaction of wild-type rBTI and rBTI mutants with immobilized bovine trypsin. (A) Wild-type rBTI. (B) W53R/P44T double mutant. (C) W53F. (D) P44T. (E) W53R.
(TIF)
(A and B) P8′ Trp omit maps of free rBTI (A) and trypsin-bound rBTI (B). The 2Fo-Fc map is shown in blue, and the Fo-Fc map is shown in green and red. The positive electron density is shown in green, and the negative density is shown in red. (C and D) 2Fo-Fc (blue) and Fo-Fc (red and green) maps of P8′ Trp with incorrect conformations in free rBTI (C) and trypsin-bound rBTI (D). The positive electron density is shown in green, and the negative density is shown in red.
(TIF)
Acknowledgments
We thank the Beijing Synchrotron Radiation Facility for their help with data collection. We also acknowledge Prof. Zhenfeng Liu and Prof. Tao Jiang for their helpful suggestions and Yuanyuan Chen for her kind help with the BIAcore assays.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the 973 Project (Grant No. 2011CBA00902), the National Natural Science Foundation of China (Grant No. 31021062) and the Knowledge Innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-YW-R-123 and KSCX2-EW-J-3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schechte I, Berger A. On Size of Active Site in Proteases .I. Papain. Biochemical and Biophysical Research Communications. 1967;27:157. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 2.Laskowski M, Kato I. Protein Inhibitors of Proteinases. Annual Review of Biochemistry. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 3.Krowarsch D, Cierpicki T, Jelen F, Otlewski J. Canonical protein inhibitors of serine proteases. Cell Mol Life Sci. 2003;60:2427–2444. doi: 10.1007/s00018-003-3120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radisky ES, Koshland DE., Jr A clogged gutter mechanism for protease inhibitors. Proc Natl Acad Sci U S A. 2002;99:10316–10321. doi: 10.1073/pnas.112332899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson SE, Fersht AR. Contribution of Residues in the Reactive-Site Loop of Chymotrypsin Inhibitor-2 to Protein Stability and Activity. Biochemistry. 1994;33:13880–13887. doi: 10.1021/bi00250a042. [DOI] [PubMed] [Google Scholar]
- 6.Cai M, Huang Y, Prakash O, Wen L, Dunkelbarger SP, et al. Differential modulation of binding loop flexibility and stability by Arg50 and Arg52 in Cucurbita maxima trypsin inhibitor-V deduced by trypsin-catalyzed hydrolysis and NMR spectroscopy. Biochemistry. 1996;35:4784–4794. doi: 10.1021/bi953038a. [DOI] [PubMed] [Google Scholar]
- 7.Belozersky MA, Dunaevsky YE, Musolyamov AX, Egorov TA. Complete amino acid sequence of the protease inhibitor from buckwheat seeds. FEBS Lett. 1995;371:264–266. doi: 10.1016/0014-5793(95)00899-k. [DOI] [PubMed] [Google Scholar]
- 8.Dunaevsky YE, Pavlukova EB, Belozersky MA. Isolation and properties of anionic protease inhibitors from buckwheat seeds. Biochem Mol Biol Int. 1996;40:199–208. doi: 10.1080/15216549600201692. [DOI] [PubMed] [Google Scholar]
- 9.Dunaevsky YE, Gladysheva IP, Pavlukova EB, Beliakova GA, Gladyshev DP, et al. The anionic protease inhibitor BWI-1 from buckwheat seeds. Kinetic properties and possible biological role. Physiologia Plantarum. 1997;101:483–488. doi: 10.1111/j.1399-3054.1997.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 10.Park SS, Ohba H. Suppressive activity of protease inhibitors from buckwheat seeds against human T-Acute lymphoblastic leukemia cell lines. Applied Biochemistry and Biotechnology. 2004;117:65–74. doi: 10.1385/abab:117:2:065. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Li Y, Li C, Yuan J, Wang Z. Expression of a buckwheat trypsin inhibitor gene in Escherichia coli and its effect on multiple myeloma IM-9 cell proliferation. Acta Biochim Biophys Sin (Shanghai) 2007;39:701–707. doi: 10.1111/j.1745-7270.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- 12.Li YY, Zhang Z, Wang ZH, Wang HW, Zhang L, et al. rBTI induces apoptosis in human solid tumor cell lines by loss in mitochondrial transmembrane potential and caspase activation. Toxicology Letters. 2009;189:166–175. doi: 10.1016/j.toxlet.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Khadeeva NV, Kochieva EZ, Tcherednitchenko MY, Yakovleva EY, Sydoruk KV, et al. Use of buckwheat seed protease inhibitor gene for improvement of tobacco and potato plant resistance to biotic stress. Biochemistry-Moscow. 2009;74:260–267. doi: 10.1134/s0006297909030031. [DOI] [PubMed] [Google Scholar]
- 14.Cierpicki T, Otlewski J. Determination of a high precision structure of a novel protein, Linum usitatissimum trypsin inhibitor (LUTI), using computer-aided assignment of NOESY cross-peaks. J Mol Biol. 2000;302:1179–1192. doi: 10.1006/jmbi.2000.4116. [DOI] [PubMed] [Google Scholar]
- 15.Cai M, Gong YX, Wen L, Krishnamoorthi R. Correlation of binding-loop internal dynamics with stability and function in potato I inhibitor family: relative contributions of Arg(50) and Arg(52) in Cucurbita maxima trypsin inhibitor-V as studied by site-directed mutagenesis and NMR spectroscopy. Biochemistry. 2002;41:9572–9579. doi: 10.1021/bi0258952. [DOI] [PubMed] [Google Scholar]
- 16.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salameh MA, Soares AS, Navaneetham D, Sinha D, Walsh PN, et al. Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin. J Biol Chem. 2010;285:36884–36896. doi: 10.1074/jbc.M110.171348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grzesiak A, Helland R, Smalas AO, Krowarsch D, Dadlez M, et al. Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases. J Mol Biol. 2000;301:205–217. doi: 10.1006/jmbi.2000.3935. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keegan RM, Winn MD. Automated search-model discovery and preparation for structure solution by molecular replacement. Acta Crystallogr D Biol Crystallogr. 2007;63:447–457. doi: 10.1107/S0907444907002661. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of several members of the potato inhibitor I family. The binding loop are marked with grey alpha boxes.
(TIF)
Sensograms of the interaction of wild-type rBTI and rBTI mutants with immobilized bovine trypsin. (A) Wild-type rBTI. (B) W53R/P44T double mutant. (C) W53F. (D) P44T. (E) W53R.
(TIF)
(A and B) P8′ Trp omit maps of free rBTI (A) and trypsin-bound rBTI (B). The 2Fo-Fc map is shown in blue, and the Fo-Fc map is shown in green and red. The positive electron density is shown in green, and the negative density is shown in red. (C and D) 2Fo-Fc (blue) and Fo-Fc (red and green) maps of P8′ Trp with incorrect conformations in free rBTI (C) and trypsin-bound rBTI (D). The positive electron density is shown in green, and the negative density is shown in red.
(TIF)