Abstract
Background
Although Campylobacter jejuni-infections have a high prevalence worldwide and represent a significant socioeconomic burden, it is still not well understood how C. jejuni causes intestinal inflammation. Detailed investigation of C. jejuni-mediated intestinal immunopathology is hampered by the lack of appropriate vertebrate models. In particular, mice display colonization resistance against this pathogen.
Methodology/Principal Findings
To overcome these limitations we developed a novel C. jejuni-infection model using gnotobiotic mice in which the intestinal flora was eradicated by antibiotic treatment. These animals could then be permanently associated with a complete human (hfa) or murine (mfa) microbiota. After peroral infection C. jejuni colonized the gastrointestinal tract of gnotobiotic and hfa mice for six weeks, whereas mfa mice cleared the pathogen within two days. Strikingly, stable C. jejuni colonization was accompanied by a pro-inflammatory immune response indicated by increased numbers of T- and B-lymphocytes, regulatory T-cells, neutrophils and apoptotic cells, as well as increased concentrations of TNF-α, IL-6, and MCP-1 in the colon mucosa of hfa mice. Analysis of MyD88−/−, TRIF−/−, TLR4−/−, and TLR9−/− mice revealed that TLR4- and TLR9-signaling was essential for immunopathology following C. jejuni-infection. Interestingly, C. jejuni-mutant strains deficient in formic acid metabolism and perception induced less intestinal immunopathology compared to the parental strain infection. In summary, the murine gut flora is essential for colonization resistance against C. jejuni and can be overcome by reconstitution of gnotobiotic mice with human flora. Detection of C. jejuni-LPS and -CpG-DNA by host TLR4 and TLR9, respectively, plays a key role in immunopathology. Finally, the host immune response is tightly coupled to bacterial formic acid metabolism and invasion fitness.
Conclusion/Significance
We conclude that gnotobiotic and “humanized” mice represent excellent novel C. jejuni-infection and -inflammation models and provide deep insights into the immunological and molecular interplays between C. jejuni, microbiota and innate immunity in human campylobacteriosis.
Introduction
In industrialized countries Campylobacter jejuni is among the most frequent causative agents of bacterial enteritis [1]. The Gram-negative pathogen is transmitted via the food chain from farm animals to humans by the ingestion of undercooked meat, non-pasteurized milk, and water [2], [3]. After infection C. jejuni colonizes the human distal small intestine and colon. Clinical symptoms include abdominal pain, fever, myalgia, and watery or bloody diarrhea. After active invasion of colonic epithelial cells, C. jejuni induces mucosal inflammation characterized by neutrophil infiltration, crypt abscesses, focal ulcerations, and plasma cell proliferation [4]–[6]. Although the vast majority of infections are self-limited, in some cases infection might result in chronic sequelae such as Reiter's syndrome, reactive polyarthropathy, and Guillain-Barré syndrome [2], [7].
Whereas colonization factors of C. jejuni are well known [8], the molecular mechanisms underlying immunopathology of C. jejuni-infection in the host are still poorly understood. Insights into C. jejuni - host interactions are limited due to the scarcity of suitable experimental in vivo models [9]. While chickens are well suited to investigate colonization, vertebrate models using newborn pigs, weanling ferrets, gnotobiotic canine pups, and primates have numerous limitations including high costs, handling issues, and a lack of reproducibility [9], [10]. Mice are highly convenient for the study of bacterial pathogenicity and can help to overcome several of these limitations. Murine models of C. jejuni-infection, however, have the disadvantage of sporadic colonization and/ or absence of clinical disease manifestations [1]. This is in part due to the colonization resistance against C. jejuni displayed by conventional mice with a normal commensal microflora. The use of germfree animals or mice with a limited gut flora has alleviated these shortcomings with varying degrees of success [10], [11]. It was demonstrated earlier that C. jejuni colonized the entire gastrointestinal (GI) tract of isolator-raised germfree mice and induced clinical signs of disease including granulocyte infiltrates, bloody diarrhea, and humoral immune responses, reproducibly occurring after infection [12]–[16]. However, germfree mice have an abnormal development of gut-associated lymphoid tissue [17], [18]. Therefore, germfree mice might not represent a suitable experimental model of C. jejuni-infections in humans due to the lack of an intact innate immune system. Furthermore, mice deficient in the central innate immunity adaptor protein MyD88 essential in Toll-like receptor (TLR) signaling were stably colonized by C. jejuni but did not develop intestinal inflammation [19]. Interaction of C. jejuni with individual TLRs, namely TLR2, TLR4, and TLR9 recognizing bacterial lipoproteins (LP), lipopolysaccharide (LPS), or CpG-DNA, respectively, still await detailed investigation particularly in vivo and in the intestines in situ. In many in vitro studies, however, TLR signaling could be demonstrated to be involved in C. jejuni-induced immune responses. Whereas C. jejuni bacteria were not recognized by human TLR5 [20], live and lysed C. jejuni cells activated TLR2, TLR4 and TLR9 via MyD88 in different human, murine or avian cell lines [21]–[25]. The role of TLR9 signaling in human C. jejuni enteritis is not clear yet. C. jejuni activated avian, but not human TLR9 transiently expressed in a transfected cancer cell line in vitro [21]. However, the role of TLR9 in C. jejuni enteritis has not been investigated in vivo so far. For proper LPS-mediated TLR4 activation, the adapter proteins MyD88 or TRIF are essentially required [26]. In recent studies, IL-10 deficient mice harboring a conventional gut flora developed significant signs of intestinal immunopathology following C. jejuni-infection [27]–[29]. In addition, enteritis and bacterial translocation were also observed in TLR4−/− IL-10−/− double deficient mice [29]. Taken together, these results point towards pivotal functions of the host-specific gut microflora and the innate immune system in both, infection control and inflammation.
In order to optimize murine models for C. jejuni enteritis mimicking human immunopathology, we used our gnotobiotic murine model in which the intestinal flora was completely eradicated by quintuple antibiotic treatment [30]–[32]. Then, we reconstituted these germfree mice with a complete human gut flora to display the human intestinal environment in mice with a fully developed immune system. The results obtained with C. jejuni-infected gnotobiotic or “humanized” mice presented here prove for the first time that the host specific gut flora plays a crucial role in colonization resistance against C. jejuni. Furthermore, we demonstrate that TLR4- and, strikingly, TLR9-mediated signaling are essentially involved in C. jejuni-induced immunopathology. Lastly, we show that the host responses to C. jejuni infection are tighly coupled to the pathogen's formic acid metabolism and perception essential for cell invasion fitness by analyzing the respective deletion mutants in our novel in vivo models. Thus, our results presented here indicate for the first time that gnotobiotic mice reconstituted with human gut flora provide deep insights into the “ménage à trois” of C. jejuni, human commensal gut flora and the host innate immune system.
Results
Gnotobiotic mice as a novel model for studying C. jejuni - host interactions
Given that the commensal murine gut microbiota is essential for the resistance against C. jejuni colonization and thus suitable mouse models of C. jejuni-infection are scarce, we established a novel murine experimental system to investigate the interplay (“ménage à trois”) of the gut microbiota and innate immune system on the host side with the pathogen after oral C. jejuni-infection and permanent intestinal colonization. For this purpose, we generated gnotobiotic mice by quintuple antibiotic treatment for six weeks (refer to [30]). It was essential to handle these animals under strictly sterile conditions throughout the entire antibiotic treatment and experimental procedures in order to prevent from secondary contaminations. Results from cultural and molecular analyses confirmed that these gnotobiotic mice were completely free of any culturable and non-culturable bacteria (Fig. S1). Similar to isolator-raised germfree animals, the antibiotics-treated mice developed a mega-cecum that is a characteristic morphologic indicator for a sterile gut.
We determined the colonization capacity of C. jejuni in gnotobiotic mice after antibiotic treatment as compared to mice harboring a conventional gut flora following peroral infection with the C. jejuni reference strains ATCC 43431 [33], [34], 81–176 [35], [36] and B2 [37]. These strains originate from humans with severe enteritis, are invasive in vitro and were extensively investigated in earlier studies. C. jejuni 81–176 carries a virulence plasmid that encodes components of a type IV protein secretion machinery [38].
The results confirmed that conventional mice bred in our SPF facilities display a strong colonization resistance, as at the time of necropsy at day (d) 12 post infection (p.i.) C. jejuni irrespective of the strain used could not be detected in fecal samples in the majority of the animals (Fig. S2). Kinetic studies revealed that C. jejuni was expelled from the GI tract already within 48 hours after infection (data not shown). Strikingly, colonization resistance was completely abrogated in gnotobiotic mice generated by quintuple antibiotic treatment as all three C. jejuni strains colonized at high concentrations of 109–1010 colony forming units (CFU) per gram fecal sample at day 12 p.i. (Fig. S2).
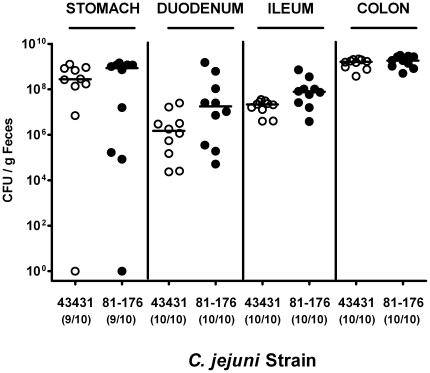
In addition, C. jejuni densities within the GI tract of gnotobiotic mice were analyzed in more detail ( Fig. 1 ). Following peroral infection, C. jejuni ATCC 43431 and 81–176 readily colonized the stomach, duodenum, ileum and colon at comparable levels with the highest bacterial loads in the large intestine reaching >109 CFU per gram luminal content at d12 p.i.. Kinetic experiments revealed that in gnotobiotic animals, C. jejuni colonization was stable throughout a time period of more than 100 days p.i. (data not shown), indicating that this model is well suited to examine long-term evolution and host-adaptation of the pathogen in vivo.
Figure 1. C. jejuni colonization along the gastrointestinal tract of gnotobiotic mice.
Gnotobiotic mice generated by antibiotic gut decontamination were orally infected with C. jejuni strains ATCC 43431 (open circles) or 81–176 (closed circles) as described in methods. The pathogen densities in distinct compartments of the gastrointestinal tract were determined by quantification of live C. jejuni in luminal samples taken from stomach, duodenum, ileum, and colon at day 12 p.i. by cultural analysis (CFU, colony forming units). Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) are indicated. Data shown were pooled from three independent experiments.
TLR4 and TLR9 play an important role in C. jejuni immunopathology
In order to investigate the role of TLR4 and TLR9 signaling in the development of immunopathology after stable C. jejuni colonization in vivo, we generated gnotobiotic mice deficient in TLR4, TLR9, MyD88 or TRIF by antibiotic treatment.
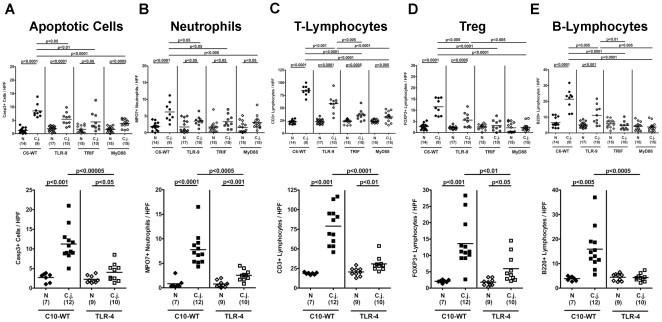
At day 12 following peroral infection with C. jejuni ATCC 43431, significant histopathological changes in colon sections of wildtype (WT) mice such as loss of goblet cells, crypt elongation, and immune cell infiltration could be observed which were less pronounced in infected TLR4-deficient mice (Figure S3). In order to more specifically and precisely analyze distinct immune cell responses in the inflamed colon in situ and to quantitate the degree of histopathology we performed immunohistochemical stainings of colon section. Given that apoptosis is commonly used as a diagnostic marker in the histopathological evaluation and grading of intestinal diseases, we quantitatively determined apoptotic cells and, in addition, the influx of neutrophils acting as inflammatory effector cells within the colon mucosa following C. jejuni-infection. Twelve days p.i., C. jejuni ATCC 43431 colonization densities were comparable within the respective segments of the GI tract (stomach, duodenum, ileum, and colon) irrespective of the genotype of mice under investigation with highest bacterial counts in the colon (approximately 109 CFU/ g luminal colon content; data not shown). Gnotobiotic mice deficient in TLR4, TLR9, TRIF or MyD88, however, displayed significantly less inflammatory responses compared to respective wildtype controls as indicated by lower numbers of Caspase3+ apoptotic and MPO7+ neutrophilic granulocytes in the colon in situ ( Fig. 2 A, B ). Furthermore, we observed significantly lower numbers of CD3+ T-lymphocytes, FOXP3+ regulatory T-cells (Tregs), and B220+ B-lymphocytes within the colonic mucosa of TLR4−/−, TLR9−/−, TRIF−/−, and MyD88−/− infected mice compared to WT mice at d12 p.i. ( Fig. 2 C–E ). Interestingly, Treg and B-lymphocyte counts of infected TRIF−/− and MyD88−/− animals did not differ when compared to non-infected controls (Fig. 2 D, E). In addition, B220+ cells within the colon mucosa of TLR4−/− mice did not increase within 12 days following C. jejuni-infection ( Fig. 2 E ). These results hightlight the gnotobiotic mouse model following antibiotic treatment as a well-suited vertebrate model for the study of C. jejuni colonization and pathogen-host interaction mounting in immunopathological responses similar to those seen in humans with Campylobacter enteritis. Strikingly, we demonstrate for the first time in vivo that MyD88- and TRIF-dependent sensing of Campylobacter-LPS and -CpG-DNA via TLR4- and TLR9-signaling is essentially involved in C. jejuni-mediated immunopathology.
Figure 2. Impact of host bacterial sensing on immunopathology in the colon following C. jejuni-infection.
Gnotobiotic wildtype (WT; C57BL/10 (C10) or C57BL/6 (C6) as indicated), TLR4−/− (lower panel), TLR9−/−, TRIF−/−, and MyD88−/− (upper panel) mice generated by antibiotic gut decontamination were orally infected with C. jejuni strain ATCC 43431. The average numbers of apoptotic cells (positive for caspase-3, panel A), neutrophilic granulocytes (neutrophils, positive for MPO-7, panel B), T-lymphocytes (positive for CD3, panel C), regulatory T-cells (Tregs, positive for FOXP3, panel D) and B-lymphocytes (positive for B220, panel E) from at least six high power fields (HPF, 400× magnification) per animal were determined microscopically in immunohistochemically stained colon sections. Numbers of animals of the respective genotype analyzed are given in parentheses. Means (black bars) and levels of significance (P-values) as compared to the respective infected WT control group (determined by the Student's t-test) are indicated. Data shown were pooled from three independent experiments.
Generation of mice reconstituted with a complete human gut flora
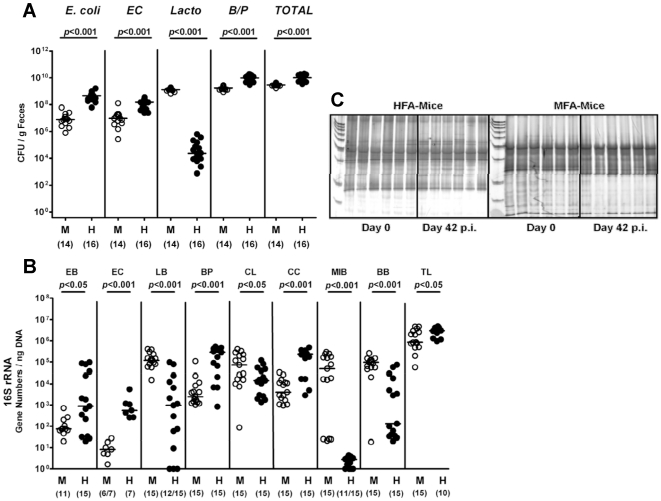
Using our gnotobiotic mouse model we were able to study the dual interaction between C. jejuni and the murine host. In order to get deeper insights into the triangle relationship (“ménage à trois”) of the pathogen, the host innate immune system and the human versus the murine microbiota, we replenished gnotobiotic mice with a human or murine gut flora by gavage. Quantitative analyses of the main bacterial communities by culture ( Fig. 3 A ) and molecular techniques ( Fig. 3 B–C , Fig. S4) revealed that the human or murine microbiota was effectively and permanently established within the gut for more than six weeks as independently confirmed by PCR-based genetic fingerprinting of fecal samples ( Fig. 3 C ). According to the typical differences in the microbiota composition of the two host species, lactobacilli were predominantly found in the murine flora associated (mfa) mice whereas human flora associated (hfa) animals harbored higher total bacterial and higher individual species loads of enterobacteria (E. coli), enterococci, Bacteroides/ Prevotella spp., and clostridia as determined by cultural ( Fig. 3 A ) and molecular ( Fig. 3 B ) analyses of feces at d0 (right before C. jejuni-infection). A quantitative molecular six-weeks follow-up revealed that C. jejuni-infected “humanized” mice displayed higher enterobacteria, Bacteroides/ Prevotella spp. and Clostridium coccoides loads in their feces as compared to mfa animals (Fig. S4 A).
Figure 3. Microflora analysis of gnotobiotic mice with human or murine gut flora.
Gnotobiotic mice generated by antibiotic gut decontamination were recolonized with a human (HFA, H) or murine (MFA, M) microflora as described (see methods). Main gut bacterial groups were quantified by culture and molecular analysis of fecal samples before and after C. jejuni-infection (d42 p.i.). (A) Culture. Total bacterial counts (TOTAL) and numbers of E. coli, enterococci (EC), Lactobacilli (Lacto) and Bacteroides/ Prevotella spp. (B/P) were determined in feces samples of MFA (M) and HFA (H) mice right before C. jejuni-infection (day 0) by detection of colony forming units (CFU) per gram feces on appropriate culture media (see methods). Bacterial species were identified by biochemical analysis and reconfirmed by comparative sequence analyses of 16S rRNA genes. Numbers of animals harboring the respective bacterial species are given in parentheses. Medians and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments. (B) RT-PCR analysis of the murine and human flora. Quantitative Real-Time-PCR amplifying bacterial 16S rRNA variable regions. 16S rRNA gene numbers/ ng DNA from luminal colon content from hfa (H) or mfa (M) mice after stable re-colonization before C. jejuni-infection (day 0) of the following bacterial groups were determined: Enterobacteriaceae (EB), Enterococci (EC), Lactic acid bacteria (LB). Bacteroides/Prevotella spp. (BP), Clostridium leptum group (CL), Clostridium coccoides group (CC), Mouse intestinal bacteroidetes (MIB), Bifidobacteria (BB), and total eubacterial load (TL). Numbers of animals harboring the respective bacterial rRNA are given in parentheses. Medians and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments. (C) Genetic fingerprinting of the murine and human flora. Molecular fingerprints were generated by PCR-based DGGE analysis of total DNA isolated from luminal colon contents as described (see methods) of six individual hfa (left panel) and mfa mice (right panel). Samples were taken after stable recolonization, but before C. jejuni-infection (day 0) and 42 days after infection (day 42 p.i.).
C. jejuni colonization in “humanized” mice
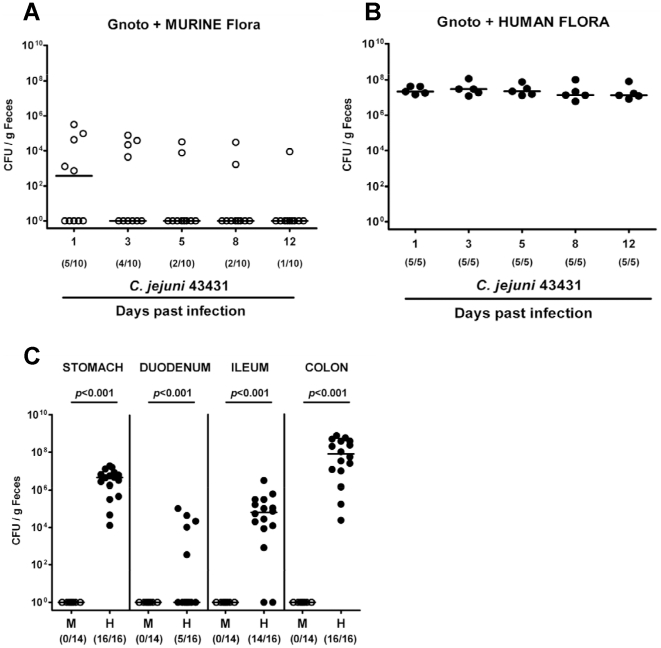
Two weeks following reconstitution of gnotobiotic mice with human or murine flora (to assure proper establishment of the microbiota along the GI tract), hfa and mfa mice were perorally infected with C. jejuni ATCC 43431 (d0). Kinetic analyses of the pathogen loads were performed in fecal samples and in distinct compartments of the entire GI tract after necropsy (d12 p.i.). Results revealed that mfa mice were protected from intestinal colonization as indicated by effective clearance of C. jejuni from the GI tract during the first two to three days p.i. ( Fig. 4 A ). In “humanized” mice, however, C. jejuni ATCC 43431 did stably colonize ( Fig. 4 B ) with bacterial loads of up to 108 CFU per g feces. Long-term kinetic experiments in hfa mice revealed that C. jejuni ATCC 43431 colonization of stomach, ileum, and colon was stable for more than six weeks p.i. with the highest bacterial loads in the colon ( Fig. 4 C ).
Figure 4. C. jejuni colonization in mice with a murine or human gut microbiota.
Following oral infection with C. jejuni ATCC 43431, kinetic analyses of the pathogen loads in feces samples of gnotobiotic mice reconstituted with a murine (A) or human (B) gut flora until d12 p.i. were performed by culture (as described in methods; CFU, colony forming units). In addition, C. jejuni burdens in distinct compartments of the gastrointestinal tract (stomach, duodenum, ileum, colon; C) of mfa (M, open circles) and hfa (H, filled circles) mice at d12 p.i. were determined by quantification of live C. jejuni by culture. Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) and significance levels (P-values, hfa as compared to mfa animals) determined by Mann-Whitney-U test are indicated. Data shown are representative for three independent experiments.
C. jejuni-induced intestinal immunopathology in “humanized” mice
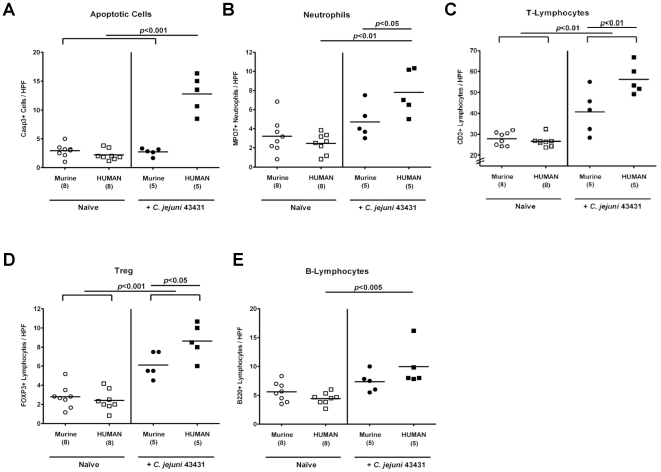
In humans, C. jejuni induces the recruitment of pro-inflammatory immune cell populations to sites of inflammation in the colon [4], [5]. Therefore, we investigated apoptotic cell, neutrophil, T- and B-lymphocyte as well as Treg recruitment in the colonic mucosa during C. jejuni-infection in both, hfa and mfa mice by immunohistochemical staining of Caspase-3, MPO7, CD3, B220, and FOXP3 in colon sections of infected mice, respectively ( Fig. 5 ). Remarkably, after reconstitution of gnotobiotic mice with a human or murine gut microbiota no differences were observed in the numbers of apoptotic cells, neutrophilic granulocytes, T- and B-lymphocytes or Tregs in the colon mucosa in situ. However, quantitative analyses of the respective immune cell populations after C. jejuni ATCC 43431-infection revealed that C. jejuni-infection induced a pronounced inflammatory response in the colon of hfa mice ( Fig. 5 ). Twelve days p.i., a five-fold increase in apoptotic cells within the colon mucosa was observed in hfa mice as compared to infected mfa and uninfected control animals ( Fig. 5 A ). This increase of apoptotic cells was accompanied by a three-fold increase in neutrophil numbers in the colon mucosa of C. jejuni-infected hfa mice ( Fig. 5 B ). Furthermore, higher T- and B-lymphocyte as well as Treg counts were found in the colon mucosa at d12 following C. jejuni-infection with significantly higher T-lymphocyte and Treg numbers in hfa as compared to mfa mice ( Fig. 5 C–E ).
Figure 5. Immunopathological responses in the colon of “humanized” mice in situ following C. jejuni-infection.
Mfa and hfa mice were generated and orally infected with C. jejuni strain ATCC 43431 by gavage as described in methods section. The average numbers of apoptotic cells (postive for caspase-3, panel A), neutrophilic granulocytes (neutrophils, positive for MPO-7, panel B), T-lymphocytes (positive for CD3, panel C), Tregs (positive for FOXP3, panel D), and B-lymphocytes (positive for B220, panel E) from at least six high power fields (HPF, 400× magnification) per animal were determined microscopically in immunohistochemically stained colon sections of naïve (Naïve, open symbols) and infected (+C. jejuni ATCC 43431, filled symbols) mfa (circles) and hfa (squares) mice at day 12 p.i.. Numbers of analyzed animals are given in parentheses. Medians (black bars) and levels of significance (P-values) as compared to the indicated groups (determined by Student's t-test) are indicated. Data shown are representative for three independent experiments.
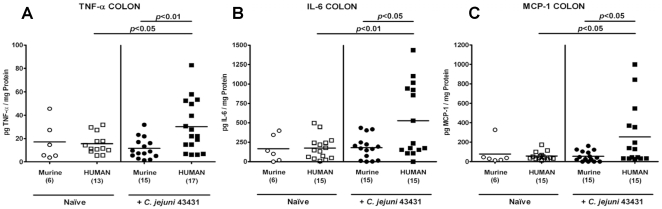
Next, we determined secretion of pro-inflammatory cytokines in the colon of C. jejuni-infected mice. At d12 p.i., hfa mice displayed higher TNF-α, IL-6 and MCP-1 concentrations in their large intestine as compared to mfa and uninfected control animals ( Fig. 6 A–C ). It is noteworthy that following C. jejuni-infection the respective means of TNF-α, IL-6, and MCP-1 levels increased by factors 2, 3, and 5, respectively, as compared to naïve hfa controls. The multi-fold increases of secreted pro-inflammatory cytokines, in the context of significant increases in immune cell numbers in colons sections of infected hfa mice (Fig. 5), point towards a biological relevant effect despite the high standard deviations within the infected hfa groups.
Figure 6. Pro-inflammatory cytokine responses in the colon of C. jejuni-infected “humanized” mice.
Mfa and hfa mice were generated and orally infected with C. jejuni strain ATCC 43431 as described (see methods). Secretion of (A) TNF-α, (B) IL-6 and (C) MCP-1 was determined in supernatants of ex vivo colon cultures from naïve animals (Naïve, open symbols) or infected (+C. jejuni ATCC 43431, filled symbols) mfa (circles) or hfa (squares) mice at day 12 p.i.. Numbers of analyzed animals are given in parentheses. Means (black bars) and levels of significance (P-values) as compared to the indicated groups (determined by Student's t-test) are indicated. Data shown were pooled from two (for naïve mice) or three (for infected animals) independent experiments.
Taken together, following C. jejuni-infection the “humanized” mice are not only susceptible to colonization but also display typical pro-inflammatory features of human campylobacteriosis.
The role of C. jejuni formic acid metabolism and perception in inducing immunopathology
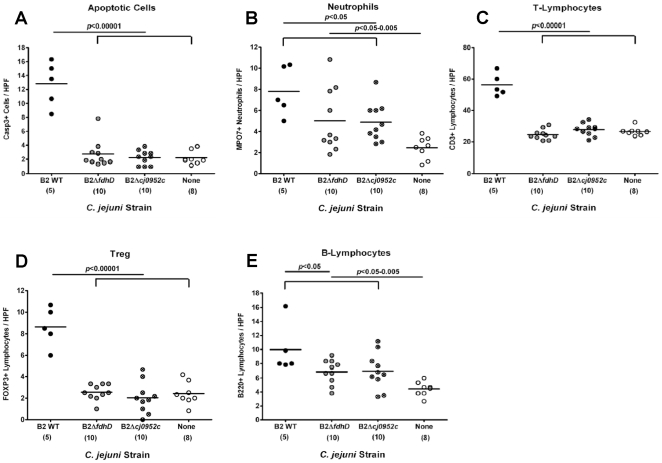
We further investigated gnotobiotic and hfa mice as suitable experimental models for the study of C. jejuni virulence factors in colonization and immunopathology. For this purpose we performed infection experiments with two different C. jejuni mutant strains deficient in factors essential for cellular invasion. Both mutant strains were recently generated by transposon mutagenesis and displayed a reduced invasion capacity into epithelial cell lines in vitro [37]. The mutant strains carrying transposon (TnKan) insertions in the genes for formate dehydrogenase subunit D fdhD (strain B2ΔfdhD) or the formic acid receptor Tlp7 (strain B2Δcj0952c) were generated in the C. jejuni B2 background as described earlier [37]. The formic acid receptor was shown to be associated with the cell invasion capacity of C. jejuni [37]. Briefly, if compared to the parental strain, the B2Δcj0952c mutant displayed a five times reduced invasion capacity (as determined in gentamicin protection assays with Caco-2 cells). To investigate the role of formate dehydrogenase and the formic acid receptor in immunopathology we infected hfa mice with the B2ΔfdhD or B2Δcj0952c mutant strain (Fig. S5). Control mice were infected with the parental C. jejuni strain B2. Results indicate that the invasion capacity and the formic acid metabolism are not required for intestinal colonization, but play essential roles in C. jejuni-mediated immunopathology. Both mutant strains colonized hfa mice similarly at high loads (up to 107–108 CFU/ g feces) as compared to the parental B2 strain (Fig. S5 A–C). Immunopathology, however, was exclusively induced by the parental strain, but not by either mutant ( Fig. 7 ). Numbers of apoptotic cells, T-lymphocytes and Tregs within the colon mucosa 12 days after infection with either mutant remained comparable to non-infected mice, but highly significantly increased multi-fold after oral infection with the parental strain ( Fig. 7 A, C, D ). In addition, the numbers of neutrophils and B-lymphocytes increased in the colon of hfa mice infected with either mutant strain compared to non-infected controls ( Fig. 7 B, E ). However, the MPO7+ or B220+ cell counts were higher in animals infected with the parental strain as compared to mice infected with either mutant.
Figure 7. Impact of C. jejuni formic acid metabolism and perception on immunopathology in “humanized” mice.
Hfa mice were generated as described (see methods) and orally infected with the C. jejuni wildtype strain (B2 WT), or with isogenic mutants deficient in the formate dehydrogenase subunit D (B2ΔfdhD) or the formic acid receptor (B2Δcj0952c). The average numbers of apoptotic cells (positive for Caspase-3, panel A), neutrophilic granulocytes (neutrophils, positive for MPO-7, panel B), T-lymphocytes (positive for CD3, panel C), regulatory T-cells (Treg, positive for FOXP3, panel D) and B-lymphocytes (positive for B220, panel E) from at least six high power fields (HPF, 400× magnification) per animal were determined microscopically in immunohistochemically stained colon sections of naïve, non-infected (None, open circles) and infected (B2 WT: filled circles; B2ΔfdhD: grey circles; B2Δcj0952c: crossed circles) mice at d12 post infection. Numbers of analyzed animals are given in parentheses. Means (black bars) and levels of significance (P-values) as compared to the respective groups (determined by Student's t-test) are indicated. Data shown are representative for three independent experiments.
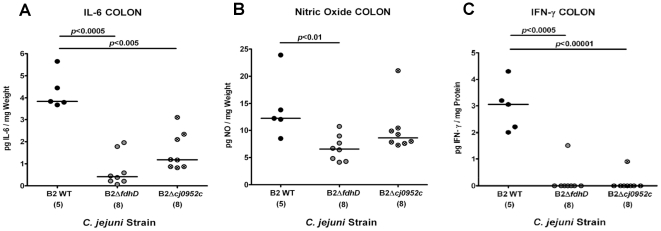
Furthermore, lower numbers of in situ immune cell counts were accompanied by significantly lower IL-6 and IFN-γ protein concentrations in ex vivo colon cultures obtained from mice infected with the respective mutant as compared to the parental strain infected controls (at d12 p.i.). In addition, mice infected with the C. jejuni mutant B2ΔfdhD displayed lower nitric oxide secretion in the colon as compared to mice infected with the parental strain ( Fig. 8 A–C ).
Figure 8. Impact of C. jejuni formic acid metabolism and perception on cytokine responses in “humanized” mice.
Hfa mice were generated as described (see methods) and orally infected with the C. jejuni wildtype strain (B2 WT), or with isogenic mutants deficient in the formate dehydrogenase subunit D (B2ΔfdhD) or the formic acid receptor (B2Δcj0952c). Secretion of (A) IL-6, (B) NO and (C) IFN-γ was determined in supernatants of ex vivo colon cultures obtained from infected (B2 WT: filled circles; B2ΔfdhD: grey circles; B2Δcj0952c: crossed circles) mice at d12 p.i.. Numbers of analyzed animals are given in parentheses. Means (black bars) and levels of significance (P-values) as compared to the respective groups (determined by Student's t-test) are indicated. Data shown are representative for three independent experiments.
C. jejuni translocation in gnotobiotic and hfa, but not mfa mice
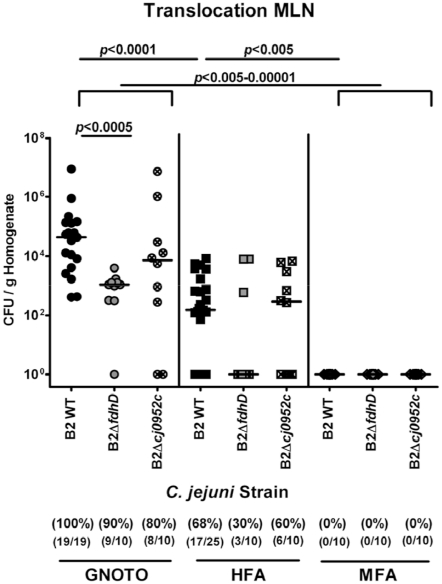
Studies have shown that live C. jejuni is able to translocate to the mesenteric lymph nodes (MLNs) in infected isolator-raised germfree animals [12]–[14]. Therefore, we determined the translocation capacity of the parental strain and both mutants, B2ΔfdhD and B2Δcj0952c in vivo. Quantitative cultural analysis of live bacteria in MLNs, spleen, liver, kidney, and cardiac blood of infected gnotobiotic and hfa mice revealed that at d12 p.i. C. jejuni translocates to MLNs ( Fig. 9 ) but not to other organs or blood (data not shown) in gnotobiotic or hfa mice. Mfa mice displaying a strong colonization resistance against C. jejuni served as negative controls. Interestingly, the translocation frequencies (100% vs. 68%) as well as the bacterial loads (medians of approximately 105 vs. 102 CFU/ g organ homogenate) of the C. jejuni B2 parental strain in MLNs were higher in infected gnotobiotic versus “humanized” animals at day 12 p.i., with no differences as compared to other C. jejuni strains such as ATCC 43431 and 81–176 (not shown). In gnotobiotic and hfa mice, both, the C. jejuni B2ΔfdhD and B2Δcj0952c mutant strain, translocated less frequently into MLNs as compared to the parental strain with lower counts of approximately two orders of magnitude following infection with the B2ΔfdhD mutant versus the parental strain ( Fig. 9 ).
Figure 9. Impact of C. jejuni formic acid metabolism and perception on bacterial translocation to mesenteric lymphnodes.
Gnotobiotic (GNOTO; circles) and with human flora associated (HFA; squares) mice were generated as described (see methods). Conventionally colonized (MFA; diamonds) animals served as negative controls. Mice were orally infected with the C. jejuni wildtype strain (B2 WT; solid symbols), or with isogenic mutants deficient in the formate dehydrogenase subunit D (B2ΔfdhD; grey symbols) or the formic acid receptor (B2Δcj0952c; crossed symbols). At d12 post infection, live C. jejuni were quantified in homogenized MLNs by culture (as described in methods; CFU, colony forming units). Medians (black bars) and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Absolute and relative numbers of animals harboring C. jejuni are given in parentheses. Data shown are pooled from three independent experiments.
Discussion
The commensal intestinal flora constitutes a barrier to effectively prevent the host from colonization and infection by gut pathogens. The earlier observation that C. jejuni readily colonized isolator-raised germfree mice [13]–[15] pointed towards an essential role of the conventional gut microbiota in the colonization resistance against C. jejuni. The results presented here do further underline the impact of the murine microbiota in colonization resistance as was independently confirmed by effective C. jejuni-infection of gnotobiotic, but not mfa mice. In comparison with isolator-raised germfree mice, we favor our animal model for the following reasons: First, gnotobiotic mice generated by antibiotic treatment exhibit a physiologically developed immune system. Second, these mice can be generated under standardized breeding conditions in a sterile environment within a common animal facility and without the need for special equipment (such as isolators for instance) and, thus, costs for generating and housing are significantly lower.
An experimental model of a human pathogen in mice previously depleted of their intestinal gut microbiota and subsequently replenished with human gut flora allows the study of host-pathogen interactions in the natural host environment. Given that these “humanized” mice, but not mice with a murine flora, are susceptible to C. jejuni-infection indicates for the first time, that the host-specific microflora composition is essential for susceptibility or resistance to Campylobacter-infections resembling the natural host conditions. However, individual bacterial species or the complex microflora composition responsible for colonization resistance have not been identified so far and await further detailed investigation.
The susceptibility of gnotobiotic and hfa mice to C. jejuni-infection renders the novel models presented here excellently suited for standardized and reproducible analyses of C. jejuni colonization and immunopathology in a higher vertebrate experimental system. Most importantly, the inflammatory responses in the colon of C. jejuni-infected “humanized” mice in situ mimicked key features of immunopathological responses in human campylobacteriosis [4], [5]: First, tissue damage was most pronounced in the lower intestinal tract as indicated by increased numbers of apoptotic cells in the colon mucosa. Second, C. jejuni-induced mucosal injury was accompanied by an increased recruitment of innate immune and effector cells (e.g. T- and B-lymphocytes, Tregs and neutrophils) in the colon mucosa of infected mice. Lastly, an increased secretion of pro-inflammatory cytokines such as TNF-α, IL-6, and MCP-1 were found in ex vivo colon cultures taken from infected hfa mice. Interestingly, Treg numbers increased in the colons of hfa mice during the course of C. jejuni-infection suggesting that production of IL-10 and other anti-inflammatory mediators might be involved in limiting C. jejuni-induced immunopathology. This was further supported by Bell and coworkers (2009) who showed that C. jejuni is able to induce a strong inflammatory response in IL-10 deficient mice [27].
Using our gnotobiotic infection model we could further dissect the interplay between C. jejuni and the host innate responses. Infection of gnotobiotic mice deficient in MyD88, TRIF or TLR4 revealed for the first time in vivo that MyD88- and TRIF-dependent TLR4-recognition of bacterial LPS is essentially involved in the development of immunopathology during C. jejuni enteritis as indicated by lower numbers of apoptotic cells, neutrophils, T- and B-lymphocytes as well as Tregs found in the colon mucosa of the respective gene-deficient mice at day 12 p.i.. Similarly, other in vitro studies have shown the important role of TLR4-signaling in C. jejuni LPS infected cells [22] and activation of human TLR4 by C. jejuni [21].
Strikingly, our results obtained from infected TLR9-deficient mice for the first time point towards a pivotal role of bacterial CpG-DNA in mediating C. jejuni-induced immunopathology. The activation of innate immune cells via TLR9, however, depends on the cell type, and on both, the methylation status and sequence of the DNA ligand under study [39]. Furthermore, a selected set of Campylobacter strains did not activate human TLR9 expressed in a transfected human cancer cell line [21]. However, this does not rule out a role for TLR9 in human campylobacteriosis. The C. jejuni population is genetically highly variable and could present DNA of diverging sequence or methylation status to host immune cells in a strain dependent manner. The C. jejuni strains used for analysis of campylobacteriosis in mice presented here were neither studied for TLR9 activation in human nor murine cell lines before. Thus, the fact that C. jejuni-infected gnotobiotic mice lacking TLR9 displayed less severe immunopathology points towards an important role of TLR9 in the murine model. The role of TLR9 in human C. jejuni-infection awaits further confirmation by additional studies with cells of the adaptive and innate immune system.
Also noteworthy is the contribution of bacterial DNA and TLR9 signaling in augmenting murine intestinal inflammation supported by our previous studies: In murine intestinal graft-versus-host disease we observed recently that TLR9 is essentially involved in aggravation of colitis [40].
Finally, the murine C. jejuni-infection models presented here are valuable tools to investigate C. jejuni virulence factors in vivo. Hfa mice infected with C. jejuni mutant strains deficient in formic acid metabolism or perception involved in cell invasion fitness harbored comparable bacterial loads throughout the GI tract. However, both mutant strains significantly displayed less immunopathological responses as indicated by lower numbers of apoptotic cells, neutrophils, T-lymphocytes, and Tregs in the colon mucosa as compared to mice infected with the parental strain. This diminished cellular response was paralleled by lower expression of pro-inflammatory cytokines (IL-6, nitric oxide, and IFN-γ) and less translocation of living C. jejuni into MLNs in animals infected with the B2ΔfdhD or B2Δcj0952c strain as compared to mice infected with the wildtype strain. Given that these results underline the suitability of the animal model presented here for in vivo analyses of C. jejuni gene mutant strains, there is a possibility of polar effects on downstream genes which cannot be ruled out. Thus, the relevance of the fdhD or the cj0952c gene mutation for amelioration of immunopathology and translocation following oral infection cannot unequivocally be attributed to the gene products encoded by the deleted genes. Nevertheless, since formic acid is a metabolite produced by many commensal gut bacteria and thus part of the intestinal intraluminal milieu, these findings point towards an essential role of formic acid metabolism and perception in the intestinal “lifestyle” of C. jejuni and its cell invasiveness in vivo.
Taken together, we conclude that our novel murine models for colonization and inflammation following C. jejuni-infection shown in the present study, mimic many important aspects of human C. jejuni-infection and thus offer promising novel tools to better understand the arousing “ménage à trois” between the pathogen, commensal gut bacteria, and host innate immune responses.
Materials and Methods
Ethics Statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin, Germany; registration number TVV G0173/07). Animal welfare was monitored twice daily by assessment of clinical conditions. Fresh human fecal samples for recolonizing gnotobiotic mice (hfa) were collected from healthy volunteers. Before sample collection written informed consent was obtained from all volunteers. Since fecal samples were obtained from co-workers of our laboratory and thus outside a clinical environment and used for re-colonization of mice only, experiments were exempted from approval by the Charité - Universitätsmedizin ethical committee according to German legacy (§15, Legal Basis for Clinical Trials).
Mice
All animals were maintained in the facilities of the “Forschungsinstitut für Experimentelle Medizin” (FEM, Charité - Universitätsmedizin, Berlin, Germany), under specific pathogen-free (SPF) conditions. Mice deficient in TLR4 (C57BL/10ScSn (C10) background), TLR9, TRIF or MyD88 (C57BL/6 (C6) background), used in the experiments are described in detail elsewhere [40]. Age and sex matched mice between 10 and 12 weeks of age were used.
Generation of gnotobiotic mice
To eradicate the commensal gut flora, mice were transferred to sterile cages and treated by adding ampicillin (1 g/L; Ratiopharm), vancomycin (500 mg/L; Cell Pharm), ciprofloxacin (200 mg/L; Bayer Vital), imipenem (250 mg/L; MSD), and metronidazole (1 g/L; Fresenius) to the drinking water ad libitum for 6–8 weeks as described earlier [30].
Generation of gnotobiotic mice with a human or murine gut flora
Fresh human and murine fecal samples free of enteropathogenic bacteria, parasites, and viruses were collected from five individual healthy volunteers and animals, respectively, pooled and dissolved in an equal volume of sterile PBS, aliquoted and stored at −80°C until use. For reconstitution experiments, aliquots were thawed and bacterial communities quantified by cultural and molecular methods (refer to [30]) before gavage of mice with 0.3 mL of the respective suspension. Between independent experiments bacterial counts of groups varied of less than 0.5 log orders of magnitude.
C. jejuni-infection of mice
Conventional, gnotobiotic or mice reconstituted with human or murine gut flora were infected with 109 viable CFU of C. jejuni strains ATCC 43431, 81–176, B2 or mutant strains B2ΔfdhD and B2Δcj0952c deficient in the formate dehydrogenase subunit D or the formic acid receptor gene, respectively [37], by gavage in a total volume of 0.3 mL PBS on three consecutive days.
Sampling procedures and histopathology
Mice were sacrificed by isofluran treatment (Abbott, Germany). Cardiac blood and tissue samples from liver, spleen, kidneys, MLNs, and gastrointestinal (GI) tract (stomach, duodenum, ileum, colon) were removed under sterile conditions. GI samples from each mouse were collected in parallel for histological, microbiological, immunological, and molecular analyses. Histopathological changes were determined in colon samples immediately fixed in 5% formalin and embedded in paraffin. Sections (5 µm) were stained with hematoxylin and eosion (HE) and examined by light microscopy (magnification ×200).
Immunohistochemistry
In situ immunohistochemical analysis of colon paraffin sections was performed as described previously [40]. Primary antibodies against CD3 (#N1580, Dako, Denmark, dilution 1∶10), myeloperoxidase-7 (MPO-7, # A0398, Dako, 1∶10000), FOXP-3 (FJK-16s, eBioscience, 1∶100), B220 (eBioscience, San Diego, CA, USA, 1∶200), and cleaved caspase-3 (Asp175, Cell Signaling, USA, 1∶200) were used. For each animal, the average number of positively stained cells within at least six high power fields (HPF, 400× magnification) were determined microscopically by three independent investigators (MMH, RP, AAK).
Quantitative analysis of C. jejuni translocation
Live C. jejuni were detected in MLNs, spleen, liver, kidneys, and cardiac blood by culture. Tissues were homogenized in sterile PBS and analyzed in dilution series on karmali agar (Oxoid, Wesel, Germany) in a microaerophilic atmosphere at 37°C for at least 48 hours. Cardiac blood (0.2 mL) was immediately streaked out on karmali agar plates.
Analysis of the intestinal microflora
Cultural analyses, biochemical identification, and molecular detection of luminal bacterial communities from stomach, duodenum, ileum, and colon were performed as previously described [30], [31], [41]. Genetic fingerprints of the intestinal microflora were generated by PCR-based denaturing-gradient-gel-electrophoresis (PCR-DGGE) amplifying the 16S rRNA variable region V6–8 [30]. Quantitative real-time PCR (qRT-PCR) was performed on a StepOnePlus RealTime-PCR System (Applied Biosystems) using the Maxima SYBR Green/ROX qPCR Master Mix (Fermentas). The reaction volume was 20 µl using 2 µl template DNA at following cycling conditions (40 cycles): Initial denaturation / enzyme activation at 95°C for 10 min, initial denaturation at 95°C for 15 s followed by annealing / elongation at 60°C for 1 min. Fluorescence detection was performed at 80°C (75°C for BP/MIB; 70°C for EC) after each cycle. A melting curve for each run was performed to check for the correct amplicon. Total bacteria (TL), gamma-Proteobacteria / Enterobacteriaceae (EB), Lactobacillus group (LB), Clostridium leptum group (CL), Clostridium coccoides group (CC), Bacteroides group including Prevotella and Porphyromonas (BP), Mouse Intestinal Bacteroidetes (MIB), Enterococci (EC) and Bifidobacteria (BB) were separately detected and quantified using 16S primer sets described earlier [40]. The C. jejuni loads of feces samples and luminal GI contents were determined with a flagellin A based qRT-PCR (flaA AW 50 fw, 5′-ATGGGATTTCGTATTAACAC-3′, flaA AW 5 re 5′-GATATAGCTTGACCTAAAGTA-3′) resulting in a 197bp product (primers taken from: Dr. K. Pietsch, Freiburg, personal communication). The Campylobacter-flaA-PCR cycling conditions were different to the above mentioned: Annealing was at 55°C for 20 s, elongation at 60°C for 30 s and fluorescence detection at 70°C. In general, the 16S rRNA or flaA gene numbers / ng DNA of each sample were determined, not the actual bacterial numbers.
Cytokine detection in colon culture supernatants
Colon biopsies were cut longitudinally, washed in PBS, and strips of 1 cm2 were placed in 24-flat-bottom well culture plates (Nunc, Wiesbaden, Germany) containing 500 µl serum-free RPMI 1640 medium supplemented with penicillin (100 U/ ml) and streptomycin (100 ug/ ml; PAA Laboratories). After 18 h at 37°C, culture supernatants were tested for TNF-α, IL-6, MCP-1, and IFN-γ by the Mouse Inflammation Cytometric Bead Assay (CBA; BD Biosciences) on a BD FACSCanto II flow cytometer (BD Biosciences). Nitric oxide (NO) was determined by Griess-reaction as described earlier [30].
Statistical analysis
Mean values, medians, standard deviations, and levels of significance were determined using appropriate tests as indicated (two-tailed Student's t-Test, Mann-Whitney-U Test). Two-sided probability (P) values≤0.05 were considered significant. All experiments were repeated at least twice.
Supporting Information
PCR-based detection of gut bacteria in feces from conventionally colonized, but not gnotobiotic mice. PCR analysis with eubacterial primer amplifying 16S V6–8 region of DNA from fecal samples obtained from nine conventionally raised mice (conventional) and gnotobiotic mice (gnotobiotic) were performed and run on 1% agarose gels stained with ethidiumbromide.
(TIF)
C. jejuni colonization in gnotobiotic and conventional mice. Conventional wildtype mice (SPF, open circles), and gnotobiotic mice (GNOTO, filled circles, generated by antibiotic gut decontamination) were orally infected with C. jejuni strains 81–176, ATCC 43431 or B2 (as indicated on the x-axis) as described (see methods). The colonization capacity was determined by quantification of live C. jejuni in feces samples at day 12 p.i. by cultural analysis (CFU, colony forming units). Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) and significance levels (P-values, as compared to SPF animals) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments.
(TIF)
Histopathology in colon sections following C. jejuni infection. Paraffin sections of colon samples were HE-stained as described (see methods). (A) In naïve, uninfected wildtype mice many goblet cells (dashed arrows), normal crypt architecture, and no immune cell infiltration were observed. (B) TLR4-deficient mice displayed mild immune cell infiltration (solid arrow), whereas in wildtype animals (C) loss of goblet cells (dashed arrows), crypt elongation, and moderate immune cell infiltration into the lamina propria (solid arrow) could be detected at day 12 following C. jejuni ATCC 43431-infection. Representative photomicrographs (magnification ×200) from three independent experiments are shown.
(TIF)
Quantitative molecular analysis of fecel samples obtained from mfa and hfa mice. (A) Quantitative Real-Time-PCR amplifying bacterial 16S rRNA variable regions. 16S rRNA gene numbers / ng DNA from luminal colon content of mfa (M, open squares) or hfa (H, filled squares) recolonized mice after C. jejuni-infection (day 42 p.i.) of the following bacterial groups were determined: Enterobacteriaceae (EB), enterococci (EC), lactic acid Bacteria (LB), Bacteroides/ Prevotella spp. (BP), Clostridium leptum group (CL), Clostridium coccoides group (CC), mouse intestinal bacteroidetes (MIB), Bifidobacteria (BB), and total eubacterial load (TL). Medians (black bars) and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Quantitative loads of the respective bacterial groups at day 0 (after stable re-colonization; circles) and at day 42 (d42, squares) after C. jejuni-infection obtained from MFA (B; open symbols) and HFA (C; filled symbols) mice were analyzed by qRT-PCR. Numbers of animals harboring the respective bacterial rRNA are given in parentheses. Medians are indicated as black bars. Data shown were pooled from three independent experiments.
(TIF)
Impact of C. jejuni formic acid metabolism and perception on colonization in “humanized” mice. Human flora associated mice (hfa) were generated as described (see methods) and orally infected with the C. jejuni wildtype (WT; A) strain B2, or with isogenic mutants deficient in the formate dehydrogenase subunit D (B2ΔfdhD; B) or the formic acid receptor (B2Δcj0952c; C). The kinetic analysis of colonization capacity was determined by quantification of live C. jejuni in luminal colon samples until day 12 post infection by culture (CFU, colony forming units). Medians (black bars) and days post infection (on x-axis) are indicated. Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Data shown were pooled from three independent experiments.
(TIF)
Acknowledgments
We thank Michaela Wattrodt, Ursula Rüschendorf, Ines Puschendorf, Gernot Reifenberger, Uwe Lohmann, Angelina Meier, and the staff of the animal research facility for excellent technical assistance, animal breeding, and genotyping of mice. We are grateful to Simone Spieckermann for immunohistochemistry staining of colon sections.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the German Research Foundation (DFG) to UG (GR906/13-1, CampyGerm), UBG (GO363/12-1, CampyGerm; SFB633, TP A7), AK and CL (SFB633, TP Z1), MM and MMH (SFB633, TP B6), LMH and BO (SFB633, Immuco), and from the German Federal Ministry of Education and Research (BMBF) to SB (TP1.1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Friedman CR, Neimann J, Wegener HC, Tauxe RV. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blazer M, editors. Campylobacter, 2nd ed, Vol 1. Washington: ASM Press; 2000. pp. 121–138. [Google Scholar]
- 2.Kist M, Bereswill S. Campylobacter jejuni. Contrib Microbiol. 2001;8:150–165. doi: 10.1159/000060405. [DOI] [PubMed] [Google Scholar]
- 3.Bereswill S, Kist M. Recent developments in Campylobacter pathogenesis. Curr Opin Infect Dis. 2003;16:487–491. doi: 10.1097/00001432-200310000-00017. [DOI] [PubMed] [Google Scholar]
- 4.van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, et al. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut. 1985;26:945–951. doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker RI, Caldwell MB, Lee EC, Guerry P, Trust TJ, et al. Pathophysiology of Campylobacter enteritis. Microbiol Rev. 1986;50:81–94. doi: 10.1128/mr.50.1.81-94.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143(Pt 1):5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 7.Tsang RS. The relationship of Campylobacter jejuni infection and the development of Guillain-Barre syndrome. Curr Opin Infect Dis. 2002;15:221–228. doi: 10.1097/00001432-200206000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300:205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Dorrell N, Wren BW. The second century of Campylobacter research: recent advances, new opportunities and old problems. Curr Opin Infect Dis. 2007;20:514–518. doi: 10.1097/QCO.0b013e3282a56b15. [DOI] [PubMed] [Google Scholar]
- 10.Chang C, Miller JF. Campylobacter jejuni colonization of mice with limited enteric flora. Infect Immun. 2006;74:5261–5271. doi: 10.1128/IAI.01094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodgson AE, McBride BW, Hudson MJ, Hall G, Leach SA. Experimental campylobacter infection and diarrhoea in immunodeficient mice. J Med Microbiol. 1998;47:799–809. doi: 10.1099/00222615-47-9-799. [DOI] [PubMed] [Google Scholar]
- 12.Yrios JW, Balish E. Immune response of athymic and euthymic germfree mice to Campylobacter spp. Infect Immun. 1986;54:339–346. doi: 10.1128/iai.54.2.339-346.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yrios JW, Balish E. Colonization and infection of athymic and euthymic germfree mice by Campylobacter jejuni and Campylobacter fetus subsp. fetus. Infect Immun. 1986;53:378–383. doi: 10.1128/iai.53.2.378-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yrios JW, Balish E. Pathogenesis of Campylobacter spp. in athymic and euthymic germfree mice. Infect Immun. 1986;53:384–392. doi: 10.1128/iai.53.2.384-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jesudason MV, Hentges DJ, Pongpech P. Colonization of mice by Campylobacter jejuni. Infect Immun. 1989;57:2279–2282. doi: 10.1128/iai.57.8.2279-2282.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youssef M, Corthier G, Goossens H, Tancrede C, Henry-Amar M, et al. Comparative translocation of enteropathogenic Campylobacter spp. and Escherichia coli from the intestinal tract of gnotobiotic mice. Infect Immun. 1987;55:1019–1021. doi: 10.1128/iai.55.4.1019-1021.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savidge TC, Smith MW, James PS, Aldred P. Salmonella-induced M-cell formation in germ-free mouse Peyer's patch tissue. Am J Pathol. 1991;139:177–184. [PMC free article] [PubMed] [Google Scholar]
- 18.Shroff KE, Cebra JJ. Development of mucosal humoral immune responses in germ-free (GF) mice. Adv Exp Med Biol. 1995;371A:441–446. doi: 10.1007/978-1-4615-1941-6_92. [DOI] [PubMed] [Google Scholar]
- 19.Watson RO, Novik V, Hofreuter D, Lara-Tejero M, Galan JE. A MyD88-deficient mouse model reveals a role for Nramp1 in Campylobacter jejuni infection. Infect Immun. 2007;75:1994–2003. doi: 10.1128/IAI.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Zoete MR, Keestra AM, Roszczenko P, van Putten JP. Activation of human and chicken toll-like receptors by Campylobacter spp. Infect Immun. 2010;78:1229–1238. doi: 10.1128/IAI.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathinam VA, Appledorn DM, Hoag KA, Amalfitano A, Mansfield LS. Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through Toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect Immun. 2009;77:2499–2507. doi: 10.1128/IAI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson RO, Galan JE. Signal transduction in Campylobacter jejuni-induced cytokine production. Cell Microbiol. 2005;7:655–665. doi: 10.1111/j.1462-5822.2004.00498.x. [DOI] [PubMed] [Google Scholar]
- 24.Friis LM, Keelan M, Taylor DE. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect Immun. 2009;77:1553–1560. doi: 10.1128/IAI.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, et al. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J Biol Chem. 2010;285:15828–15836. doi: 10.1074/jbc.M110.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watters TM, Kenny EF, O'Neill LA. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol. 2007;85:411–419. doi: 10.1038/sj.icb.7100095. [DOI] [PubMed] [Google Scholar]
- 27.Bell JA, St Charles JL, Murphy AJ, Rathinam VA, Plovanich-Jones AE, et al. Multiple factors interact to produce responses resembling spectrum of human disease in Campylobacter jejuni infected C57BL/6 IL-10−/− mice. BMC Microbiol. 2009;9:57. doi: 10.1186/1471-2180-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansfield LS, Bell JA, Wilson DL, Murphy AJ, Elsheikha HM, et al. C57BL/6 and congenic interleukin-10-deficient mice can serve as models of Campylobacter jejuni colonization and enteritis. Infect Immun. 2007;75:1099–1115. doi: 10.1128/IAI.00833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansfield LS, Patterson JS, Fierro BR, Murphy AJ, Rathinam VA, et al. Genetic background of IL-10(−/−) mice alters host-pathogen interactions with Campylobacter jejuni and influences disease phenotype. Microb Pathog. 2008;45:241–257. doi: 10.1016/j.micpath.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177:8785–8795. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 31.Heimesaat MM, Fischer A, Jahn HK, Niebergall J, Freudenberg M, et al. Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut. 2007;56:941–948. doi: 10.1136/gut.2006.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz M, Heimesaat MM, Danker K, Struck D, Lohmann U, et al. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J Exp Med. 2009;206:3047–3059. doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penner JL, Hennessy JN, Congi RV. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur J Clin Microbiol. 1983;2:378–383. doi: 10.1007/BF02019474. [DOI] [PubMed] [Google Scholar]
- 34.Poly F, Threadgill D, Stintzi A. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J Bacteriol. 2004;186:4781–4795. doi: 10.1128/JB.186.14.4781-4795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poly F, Threadgill D, Stintzi A. Genomic diversity in Campylobacter jejuni: identification of C. jejuni 81–176-specific genes. J Clin Microbiol. 2005;43:2330–2338. doi: 10.1128/JCM.43.5.2330-2338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 37.Tareen AM, Dasti JI, Zautner AE, Gross U, Lugert R. Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology. 2010;156:3123–3135. doi: 10.1099/mic.0.039438-0. [DOI] [PubMed] [Google Scholar]
- 38.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M, O'Hagan DT. Recent advances in vetinary vaccine adjuvants. Int J Parasitol. 2003;33:469–478. doi: 10.1016/s0020-7519(03)00053-5. [DOI] [PubMed] [Google Scholar]
- 40.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, et al. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut. 2010;59:1079–1087. doi: 10.1136/gut.2009.197434. [DOI] [PubMed] [Google Scholar]
- 41.Heimesaat MM, Fischer A, Siegmund B, Kupz A, Niebergall J, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;2:e662. doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR-based detection of gut bacteria in feces from conventionally colonized, but not gnotobiotic mice. PCR analysis with eubacterial primer amplifying 16S V6–8 region of DNA from fecal samples obtained from nine conventionally raised mice (conventional) and gnotobiotic mice (gnotobiotic) were performed and run on 1% agarose gels stained with ethidiumbromide.
(TIF)
C. jejuni colonization in gnotobiotic and conventional mice. Conventional wildtype mice (SPF, open circles), and gnotobiotic mice (GNOTO, filled circles, generated by antibiotic gut decontamination) were orally infected with C. jejuni strains 81–176, ATCC 43431 or B2 (as indicated on the x-axis) as described (see methods). The colonization capacity was determined by quantification of live C. jejuni in feces samples at day 12 p.i. by cultural analysis (CFU, colony forming units). Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) and significance levels (P-values, as compared to SPF animals) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments.
(TIF)
Histopathology in colon sections following C. jejuni infection. Paraffin sections of colon samples were HE-stained as described (see methods). (A) In naïve, uninfected wildtype mice many goblet cells (dashed arrows), normal crypt architecture, and no immune cell infiltration were observed. (B) TLR4-deficient mice displayed mild immune cell infiltration (solid arrow), whereas in wildtype animals (C) loss of goblet cells (dashed arrows), crypt elongation, and moderate immune cell infiltration into the lamina propria (solid arrow) could be detected at day 12 following C. jejuni ATCC 43431-infection. Representative photomicrographs (magnification ×200) from three independent experiments are shown.
(TIF)
Quantitative molecular analysis of fecel samples obtained from mfa and hfa mice. (A) Quantitative Real-Time-PCR amplifying bacterial 16S rRNA variable regions. 16S rRNA gene numbers / ng DNA from luminal colon content of mfa (M, open squares) or hfa (H, filled squares) recolonized mice after C. jejuni-infection (day 42 p.i.) of the following bacterial groups were determined: Enterobacteriaceae (EB), enterococci (EC), lactic acid Bacteria (LB), Bacteroides/ Prevotella spp. (BP), Clostridium leptum group (CL), Clostridium coccoides group (CC), mouse intestinal bacteroidetes (MIB), Bifidobacteria (BB), and total eubacterial load (TL). Medians (black bars) and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Quantitative loads of the respective bacterial groups at day 0 (after stable re-colonization; circles) and at day 42 (d42, squares) after C. jejuni-infection obtained from MFA (B; open symbols) and HFA (C; filled symbols) mice were analyzed by qRT-PCR. Numbers of animals harboring the respective bacterial rRNA are given in parentheses. Medians are indicated as black bars. Data shown were pooled from three independent experiments.
(TIF)
Impact of C. jejuni formic acid metabolism and perception on colonization in “humanized” mice. Human flora associated mice (hfa) were generated as described (see methods) and orally infected with the C. jejuni wildtype (WT; A) strain B2, or with isogenic mutants deficient in the formate dehydrogenase subunit D (B2ΔfdhD; B) or the formic acid receptor (B2Δcj0952c; C). The kinetic analysis of colonization capacity was determined by quantification of live C. jejuni in luminal colon samples until day 12 post infection by culture (CFU, colony forming units). Medians (black bars) and days post infection (on x-axis) are indicated. Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Data shown were pooled from three independent experiments.
(TIF)