Abstract
Homocitrate synthase (HCS) catalyzes the first step of L-lysine biosynthesis in fungi by condensing acetyl-Coenzyme A and 2-oxoglutarate to form 3R-homocitrate and Coenzyme A. Due to its conservation in pathogenic fungi, HCS has been proposed as a candidate for antifungal drug design. Here we report the development and validation of a robust, fluorescent assay for HCS that is amenable to high-throughput screening for inhibitors in vitro. Using this assay, Schizosaccharomyces pombe HCS was screened against a diverse library of ~41,000 small molecules. Following confirmation, counter screens, and dose-response analysis, we prioritized over 100 compounds for further in vitro and in vivo analysis. This assay can be readily adapted to screen for small molecule modulators of other acyl-CoA-dependent acyltransferases or enzymes that generate a product with a free sulfhydryl group, including histone acetyltransferases, aminoglycoside N-acetyltransferases, thioesterases and enzymes involved in lipid metabolism.
Keywords: homocitrate synthase, antifungal, high-throughput screen, fluorescent assay, MMBC, ThioGlo 1, acetyltransferase
Introduction
Invasive fungal infections represent an escalating threat to human health. Although these types of infections generally pose a low risk to healthy individuals, they occur frequently and have a high rate of morbidity and mortality in immunocompromised individuals, such as burn patients, transplant recipients, cancer patients undergoing chemotherapy and individuals suffering from HIV/AIDS or other immunodeficiency syndromes [1–6]. While progress has been made in devising new therapies to treat fungal infections, there remains a pressing need to develop novel antifungal agents. Several classes of antifungal drugs are used clinically to treat invasive fungal infections, including polyenes, azoles and echinocandins, which target components of the cell membrane or cell wall [7–10]. However, many of these drugs are associated with severe cytotoxic side effects or can interfere with other medications. Furthermore, there has been an alarming rise in drug resistant fungal strains that do not respond to conventional therapies. [11]. For example, treatment of invasive aspergillosis, most commonly caused by the pathogen Aspergillus fumigatus, with a single drug has a cure rate of only 50% [10], underscoring the need to develop new drugs and combinatorial therapies to combat fungal pathogens.
Several metabolic pathways have been proposed as targets for novel antifungal drug design. The α-aminoadipate (AAA) pathway represents one such target. This pathway is responsible for L-lysine biosynthesis in certain archaebacteria and fungi, including pathogens from the genera Candida, Cryptococcus and Aspergillus [12]. In fungi, the AAA pathway consists of eight steps catalyzed by seven enzymes. The divalent metal-dependent enzyme homocitrate synthase (HCS) catalyzes the first reaction in this pathway by condensing 2-oxoglutarate (2-OG) and acetyl-Coenzyme A (AcCoA) to yield 3R-homocitrate and Coenzyme A (CoA) [13] (Supplemental Fig. 1). In the second reaction, homoaconitase has been proposed to catalyze a two-step isomerization of 3R-homocitrate to form 2R,3S-homoisocitrate, which is subsequently oxidized by homoisocitrate dehydrogenase to produce 2-oxoadipate. The enzymes 2-aminoadipate aminotransferase, 2-aminoadipate reductase, saccharopine reductase and saccharopine dehydrogenase catalyze the remaining four reactions in which 2-oxoadipate is converted to L-lysine (Supplemental Fig. 1). Homologs of these latter four enzymes catalyze lysine catabolism in mammals and are thus unfavorable candidates for drug design [14].
Of the enzymes in the AAA pathway, HCS represents a prime target for inhibition for several reasons. Notably, HCS catalyzes the first and committed reaction in the AAA pathway and thus has a key role in regulating lysine homeostasis in fungi. HCS homologs from several fungi have been shown to be feedback inhibited by L-lysine, which has an essential role in modulating the metabolic flux through the AAA pathway [15–18]. Recent structural studies have illustrated that L-lysine is a competitive inhibitor of the substrate 2-OG, highlighting the potential for small molecules to regulate HCS activity [19; 20]. In addition to feedback inhibition, genetic studies of a Schizosaccharomyces pombe mutant lacking Cu/Zn superoxide dismutase have revealed the activity and protein level of HCS within cells is diminished under conditions of oxidative stress, suggesting that the enzyme is also subject to redox regulation [21]. At the transcriptional level, HCS expression in Saccharomyces cerevisiae is regulated through general control of amino acid biosynthesis [22; 23] as well as by the transcription factor Lys14, which is activated upon binding the AAA pathway intermediate 2-aminoadipate semialdehyde [24–26]. Finally, a very recent study by Schobel et al. reported that the deletion of the HCS gene in the pathogen A. fumigatus virtually abolished virulence in a mouse model for bronchopulmonary aspergillosis, whereas the virulence of the knockout strain was unaffected in a disseminated model for invasive aspergillosis [27]. Because the major route of A. fumigatus infections is through inhalation into the lungs, these findings imply that HCS inhibitors may find clinical applications in treating allergic bronchopulmonary aspergillosis, aspergilloma, and chromic pulmonary aspergillosis.
In an effort to discover small molecule inhibitors of HCS that may prove useful in characterizing its functions in vitro and in vivo, we have developed and validated an in vitro assay for HCS that is amenable to high-throughput screening (HTS). This method was adapted from a fluorescent assay for histone acetyltransferases (HATs) [28] and detects the formation of CoA produced through reaction of its free sulfhydryl group with the sulfhydryl-sensitive fluorophore MMBC in a 384-well plate format. The utility of this assay was demonstrated by screening a diverse chemical library composed of ~41,000 compounds to identify inhibitors of S. pombe HCS (SpHCS), with dose response studies identifying several potent inhibitors. This HTS assay will not only aid in discovering novel inhibitors of HCS but is also broadly applicable to other acyl-CoA-dependent acyltransferases that are potential drug targets.
Materials and Methods
Reagents and protein purification
All reagents used were of the highest grade commercially available. The disodium salt of 2-OG, trilithium salts of AcCoA and CoA and HEPES were purchased from Sigma. AcCoA was treated with acetic anhydride (Fluka) to acetylate trace amounts of free CoA as previously described [28] and was quenched with 100 mM HEPES (pH 7.5). AcCoA was diluted 1:2 in 1 M HEPES (pH 7.5) to bring the pH to 5 prior to using in assays. The fluorophore MMBC [10-(2,5-dihydro-2, 5-di-oxo-1H-pyrrol-1-yl)-9-methoxy-3-oxo-, methyl ester 3H-naphthol(2,1-b) pyran-S-carboxylic acid, also known as ThioGlo 1] was purchased from Berry & Associates (Ann Arbor, MI) and was prepared by dissolution in anhydrous dimethyl sulfoxide (DMSO). The dye concentration was determined by measuring absorbance at 381 nm (ε=15,100 M−1 cm−1, Covalent Associates). MnCl2-tetrahydrate was purchased from Acros Organics. Costar black 384-well polypropylene plates (Corning Life Sciences) and anhydrous DMSO (Fluka or Acros) were used in all assays. Full length SpHCS was recombinantly expressed in Escherichia coli Rosetta 2 DE3 cells (EMD Biosciences) and purified using a Zn(II)-charged immobilized metal affinity sepharose column (GE Healthcare) followed by gel filtration chromatography as previously described [29].
Small molecule libraries
In the primary screen, approximately 41,000 compounds were tested at the Center for Chemical Genomics (CCG) in the Life Sciences Institute at the University of Michigan. This library comprises several commercially available compound collections, including the Maybridge Hit-finder Chemical Collection, a diversity collection from Chembridge, the MicroSource Spectrum 2000 Library, the NIH Clinical Compound set and a diversity set from ChemDiv.
Homocitrate synthase HTS assay
Primary screening was performed at room temperature by adding 100 mM HEPES (pH 7.5) with 160 μM 2-OG (20 μl) to the 384-well microplates using a Multidrop 384 (Thermo Scientific). Inhibitor compounds (0.2 μl of 1.2–2 mM stocks, n=1) or DMSO (0.2 μl for negative and positive controls for inhibition n=32 per plate) were added using the pin-tool application on a Biomek FX liquid handling robot (Beckman). A mixture of 100 mM HEPES (pH 7.5) and 10.7 μM AcCoA (20 μl) was added to the positive controls for inhibition (n=16 per plate). A solution of 100 mM HEPES (pH 7.5) 10 nM SpHCS and 10.7 μM AcCoA (20 μl) was added to the remaining wells with the Multidrop 384 to initiate the assay, yielding final concentrations of 100 mM HEPES (pH 7.5), 80 μM 2-OG, 5.35 μM AcCoA and 5 nM SpHCS. Plates were incubated at room temperature for 20 min and the reactions were terminated with the addition of the detection reagent (40 μl of 25 μM MMBC in DMSO). The plates were covered and after a 10 min incubation the fluorescence of the MMBC-CoA adduct was measured at 470 nm using an excitation wavelength of 380 nm using a PHERAstar plate reader (BMG Labs).
Data analysis
To validate the HCS assay, the Z′-factor, (Z′, Eq. 1) [30] coefficient of variation (CV, Eq. 2) and signal to noise (S/N) ratio were calculated from a single 384-well plate containing the negative controls for inhibition (assay solution in the absence of inhibitors; n=192) and the positive controls for inhibition (assay solution without SpHCS; n=191 with one outlier removed).
| Eq. 1 |
| Eq. 2 |
Compounds were considered initial hits if they: 1) exhibited ≥ 30.0% inhibition by plate, where 0% inhibition is defined as the average of the negative controls for inhibition (inhibitor omitted) and 100% is the average of the positive controls for inhibition (SpHCS omitted) or 2) had an inhibition value that exceeded an SD of 3.0 by plate (calculated from the negative controls and samples). Additional triage removed compounds with unfavorable properties, including compounds flagged for cytotoxicity, promiscuous compounds (> 3 SD in 10 or more previous screens) and molecules with maleimide groups that could react with the free sulfhydryl group of CoA. Potential hits meeting these initial criteria (1399 molecules) were confirmed by re-screening in triplicate at the same concentration used in primary screening using a Mosquito X1 liquid handling robot (TTP Labtech). Compounds with ≥ 30.0% inhibition by plate were considered confirmed hits. Additional triage criteria removed generally promiscuous compounds (≥ 30.0% inhibition in 10 or more previous screens) and large compounds with topological polar surface area ≥ 140 Å2.
MMBC-CoA quenching and sulfhydryl-reacting counter screen
Compounds with confirmed activity were assayed to ensure that they were not false positives due to intrinsic reactivity with the sulfhydryl group of CoA or due to fluorescence quenching of the MMBC-CoA conjugate. This screen was conducted in triplicate essentially as described above except that 1.0 μM CoA was substituted for SpHCS. The reporter reagent was added, and the plates were covered and allowed to incubate at room temperature for 10 min before the fluorescence of the MMBC-CoA adduct was measured. Compounds that had median percent inhibition value of > 30.0 % were rejected as probable false positives.
Metal chelation counter screen
To eliminate small molecule that indirectly inhibit HCS through chelation of its active site divalent metal ion, a second counter screen was performed in triplicate as described for the primary screen with the exception that a mixture of 100 mM HEPES (pH 7.5), 160 μM 2-OG and 20 μM MnCl2 (20 μl) was added to the assay. Compounds that displayed median percent inhibition values < 30.0% were removed as probable metal chelators.
Dose response analysis
Compounds classified as active hits after counter-screening triages were analyzed in dose response studies (n=2) following the primary screening protocol. Compounds were serially diluted two-fold using the Mosquito X1 (TTP Labtech) and Biomek FX (Beckman) robots, yielding concentrations ranging from 0.586–75 μM in the assays. The fluorescence was plotted versus the concentration of the inhibitor compounds (I) and a nonlinear regression (Eq. 3) was fit to the data for compounds that exhibited a sigmoidal curve.
| Eq. 3 |
In this equation, “min” is the lower limit of the assay, “max” is the upper limit, [I] is the log[compound], M1 is the log(IC50) and M2 is the Hill slope. For compounds in which the concentration did not properly define either the upper or lower plateaus of the curve, the value was entered manually using the negative or positive controls of the plate, respectively. Subsequent triage eliminated compounds that did not exhibit sigmoidal dose response curves or that had a pAC50 values < 4.5 or Hill slopes < −2.
Results
Homocitrate synthase fluorescence assay
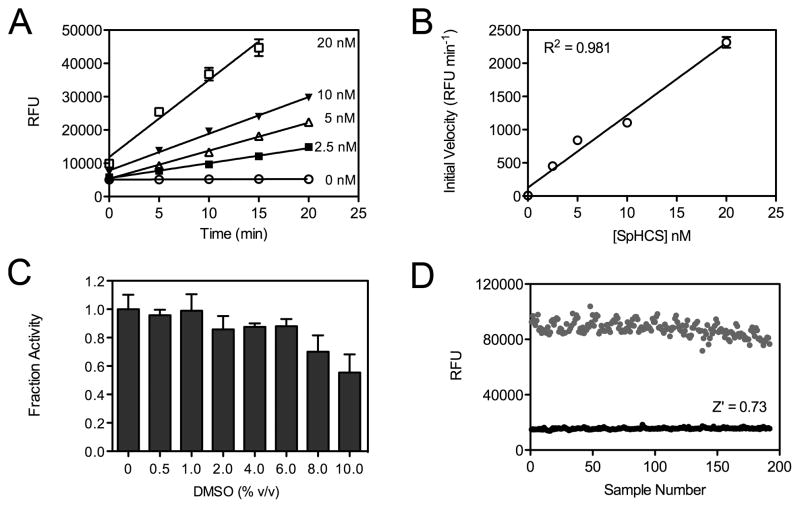
In prior structural and functional studies, we characterized the kinetic parameters of SpHCS using a fluorescent assay that was adapted from a previously reported HAT assay [19; 28; 29]. This assay monitors CoA production by reacting its sulfhydryl group with the thiol-sensitive dye MMBC to form a highly fluorescent MMBC-CoA adduct (Fig. 1) [31]. Using this method, we measured the steady state kinetic parameters for the Zn(II)-bound form of the enzyme (kcat = 303 ± 4 min−1, Km(AcCoA) = 10.7 ± 0.6 μM and Km(2-OG) = 159 ± 15 μM) [29]. Based on this initial work, we optimized the assay for HTS using SpHCS as a model due to its relative stability compared to other HCS homologs that can rapidly lose activity or require a complex stabilization buffer to preserve enzymatic activity [27; 32; 33]. To adapt the assay for screening in a 384-well plate format, a series of controls were performed to optimize the assay conditions. Fluorescent assays were performed using substrate concentrations at half of their respective Km values (5.35 μM AcCoA and 80 μM 2-OG) to facilitate the identification of compounds that competitively inhibit HCS. Under these conditions, we determined the reaction linearity with respect to time and enzyme concentration and observed that up to 10 nM SpHCS produced a linear velocity for up to 20 min (Fig. 2A). The initial velocities were also linear with respect to enzyme concentration between 0–20 nM (Fig. 2B). In the HTS assays, 5 nM SpHCS was used because this concentration yielded a reasonable S/N ratio at 20 min while limiting AcCoA consumption.
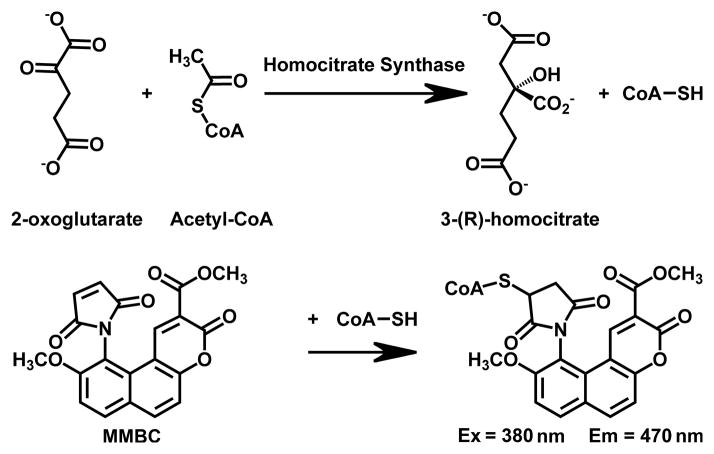
Fig. 1.
Schematic of the fluorescent HCS assay. The CoA produced in the HCS reaction is quantified through its reaction with the sulfhydryl-sensitive fluorophore MMBC (also known as ThioGlo 1). The reaction yields an MMBC-CoA adduct that fluoresces strongly at 470 nm when excited at 380 nm.
Fig. 2.
Optimization of the fluorescent HCS assay for HTS. (A) Linearity of SpHCS activity versus time with substrate concentrations equal to half Km values. Relative fluorescence units (RFU) were plotted versus time, and linear regressions were fit to the data points (n=8) with error bars indicating one SD. (B) Plot of initial velocity versus enzyme concentration. A linear regression was fit to the data with error bars representing the curve fitting error of the slope of the linear plots in panel A. (C) Activity of SpHCS in the presence of increasing concentrations of DMSO. The fraction activity was calculated relative to the activity of the enzyme in the absence of DMSO. Data are the average of three replicates with the error bars indicating one SD. (D) Scatter plot of the RFUs for an equivalent number of positive (black circles, n=191) and negative controls (gray circles, n=192) for inhibition from a single 384 well plate. These data were used to calculate the Z′ value of the optimized HTS assay.
Additional controls were conducted to optimize the fluorescent assay for HTS. The compounds in the small molecule library are dissolved in DMSO and were assayed at final DMSO concentrations of 0.5% (v/v) in the primary assay and up to 3% (v/v) in dose response analysis. Thus, it was necessary to determine to what extent DMSO affected SPHCS activity. Control assays with increasing concentrations of DMSO demonstrated that SpHCS retained 100% activity in 1% DMSO and ~90% activity up to 6% DMSO, indicating that the solvent does not appreciably inhibit the enzyme under the concentrations used in HTS (Fig. 2C). We also verified that SpHCS retains full activity for at least 1 hour at 24 °C using standard assay conditions and saturating substrate concentrations (200 μM AcCoA and 5 mM 2-OG), enabling us to conduct the HTS at room temperature without concern for a loss in enzymatic activity (data not shown).
After completing the initial enzymatic controls, we statistically validated the HCS assay for HTS by performing an equal number of negative controls (assay solution without compounds representing 0% inhibition) and positive controls (assays with SpHCS omitted representing 100% inhibition) in a single 384-well microplate (Fig. 2D). Analysis of the control data yielded a Z′ value of 0.73, indicating an excellent assay in which there is a large separation between the variations in the sample and control signals [30]. The CV value and an S/N ratio from the control plate were 6.7% and 5.6 respectively, which are acceptable statistical values for HTS [34].
HTS for inhibitors of HCS
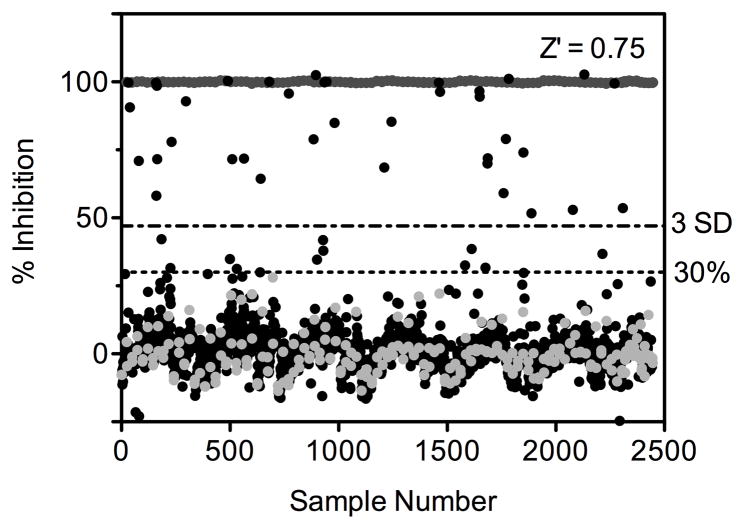
A diverse library of 40,879 compounds was screened for small molecule inhibitors of SpHCS. Each compound was added at a single concentration (6–10 μM) to black 384-well plates containing the 2-OG substrate. The assay was initiated with the addition of a mixture of AcCoA and SpHCS, with negative and positive controls included in each plate to simplify the analysis of the inhibition data and to provide plate-by-plate statistics to detect any irregularities in the assay. The reporter reagent MMBC was added after a 20 min incubation and fluorescence intensity was read in a PHERAstar plate reader (Table 1). A scatter plot of the MicroSource Spectrum 2000 library shown in Fig. 3 is representative of the data for the complete primary screen. The Z′ value was calculated for each plate and averaged 0.75 for this particular assay with all primary assays averaging Z′ values between 0.73–0.82. Additionally, the S/N ratio and CV value averaged between 7.9–14.9 and 4.9%–6.2%, consistent with the values determined from the initial control plate. Following primary screening, a total of 1665 compounds were categorized as actives and additional triage excluded undesirable compounds as outlined in the methods (Table 2). After triage, a total of 1399 compounds were selected for confirmation assays that were performed in triplicate for each compound. Re-screening confirmed 595 active compounds and triage eliminated additional compounds (Table 2), yielding a final total of 496 (~1 %) positive hits.
Table 1.
Homocitrate synthase HTS protocol.
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | 2-OG substrate | 20 μLa | 100 mM HEPES (pH 7.5), 160 μM 2-OG |
| 2 | Library compounds | 0.2 μL | 1.2–2 mM in DMSO |
| 3 | Positive inhibition control | 20 μL | 100 mM HEPES (pH 7.5), 10.7 μM AcCoA |
| 4 | Assay initiation | 20 μL | 100 mM HEPES (pH 7.5), 10.7 μM AcCoA, 10 nM SpHCS |
| 5 | Incubation time | 20 min | room temperature |
| 6 | Detection reagent | 40 μL | 25μM MMBC in DMSO |
| 7 | Incubation time | 10 min | room temperature |
| 8 | Assay readout | 380 nm ex/ 470 nm em | PHERAstar (BMG Labtech) plate reader, fluorescent module |
| Step | Notes | ||
|---|---|---|---|
| 1 | 384-well black plates (Corning), 8-tip Multidrop dispenser (Thermo Scientific), added to all columns | ||
| 2 | Pintool transfer with Biomek FX (Beckman), DMSO added to control columns 1–2 and 23–24 | ||
| 3 | 8-tip Multidrop dispenser (Thermo Scientific), added to columns 23–24 | ||
| 4 | 8-tip Multidrop dispenser (ThermoScientific), added to columns 1–22 | ||
| 5 | |||
| 6 | 8-tip Multidrop dispenser (Thermo Scientific), added to all columns | ||
| 7 | Plates covered with opaque (tinfoil) cover | ||
| 8 | |||
Amount added per well. The notes listed in the bottom of the table refer to the steps in the upper part of the table.
Fig. 3.
HTS results for the MicroSource Spectrum 2000 compound library. The x-axis represents the compounds screened (black circles), the positive controls (dark gray circles), and the negative controls for inhibition (light gray circles). The y-axis corresponds to the RFU values for the compounds and controls. Dashed lines represent the two criteria used to identify potential inhibitors and represent: 1) 3.0 SD as calculated from the negative controls and samples and 2) a 30% inhibition threshold.
Table 2.
Screening results identifying inhibitors of SpHCS.
| Compound Category | # of Compounds |
|---|---|
| Total compounds in primary screen | 40,879 |
|
| |
| Total hits (SD >3 or % inhibition >30%) | 1665 |
| Flagged cytoxic compounds | −27a |
| Promiscuous compounds (SD > 3 in ≥ 10 previous screens) | −216 |
| Maleimide containing compounds | −23 |
|
| |
| Total compounds in confirmation screen | 1399 (3.4 %) |
|
| |
| Total hits (median % inhibition > 30%) | 595 |
| Generally promiscuous compounds (> 30% inhibition in ≥ 10 previous screens) | −67 |
| Large compounds (topological polar surface area ≥ 140 Å2) | −14 |
| Compounds with limited availability | −18 |
|
| |
| Total compounds for counter screens | 496 (1.2 %) |
|
| |
| Sulfhydryl-reacting or MMBC-CoA quenching compounds (% inhibition ≥ 30%) | −54 |
| Metal chelators (% inhibition < 30%) | −81 |
| 8-hydroxyquinoline containing compounds | −20 |
| Compounds with limited availability | −3 |
|
| |
| Total compounds analyzed for dose response | 338 (0.8 %) |
|
| |
| Total hits (compounds with sigmoidal dose response curves) | 266 |
| Compounds with pAC50 values <4.5 (IC50 >31.6 μM) | −86 |
| Compounds with Hill slope values <−2.0 | −59 |
|
| |
| Compounds for follow-up studies | 122 (0.3 %) |
A minus (−) sign before the number indicates that the number of compounds eliminated from further consideration at this step.
Counter screen to detect MMBC-CoA quenching and sulfhydryl-reactive compounds
Inspection of the chemical structures of the 496 positive hits revealed that several of the compounds had large aromatic systems with extended conjugation that may quench the fluorescence of the MMBC-CoA adduct. In addition, certain compounds possessed moieties that could react with the free sulfhydryl group of CoA. To eliminate these potential false positives, an MMBC-CoA counter screen was conducted by substituting SpHCS in the initiation step with 1.0 μM CoA (Table 1). This concentration of CoA was used to mimic the approximate amount of CoA generated in the negative control reactions. The CoA counter screen data identified 54 compounds as either suspected fluorescence quenchers or as being sulfhydryl-reactive (Table 2). Representative compounds eliminated in this counter screen are shown in Supplemental Table S1.
Counter screen to detect potential metal chelators
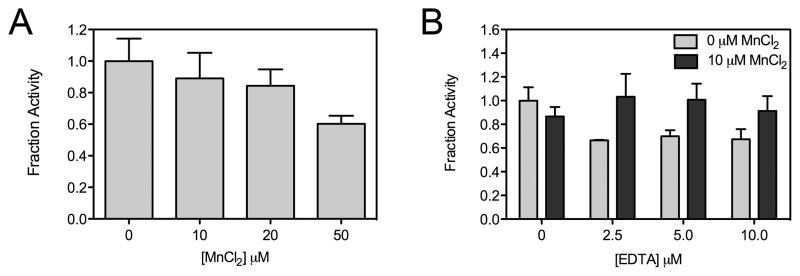
A second potential source of false positive hits in the HTS involve compounds that indirectly inhibit HCS through chelation of its active site divalent metal ion that is required for catalysis [35]. To isolate potential metal chelators, a counter screen was devised in which excess Mn(II) was added to the HTS assay. This transition metal was chosen for this counter screen because: 1) we were unable to reconstitute the activity of the SpHCS apoenzyme with Mn(II) in vitro and 2) inductively coupled plasma mass spectrometry analysis of the recombinant enzyme purified under divalent metal free conditions indicated that it does not bind Mn(II) in vivo (unpublished data). Control assays demonstrated that the addition of 10 μM MnCl2 to 5 nM SpHCS under the HTS conditions caused only a modest (~10%) decrease in activity compared to enzyme assayed in the absence of the metal ion (Fig. 4A). To evaluate whether the addition of 10 μM MnCl2 could preclude inactivation by metal chelators (tested at 6–10 μM), SpHCS was assayed in the presence of increasing concentrations of the metal chelator EDTA in the absence or presence of 10 μM MnCl2. The results demonstrated that the addition of MnCl2 effectively blocked inactivation of SpHCS by concentrations of EDTA up to 10 μM (Fig. 4B). This counter screen was then applied to the 496 confirmed hits using the HTS assay protocol (Table 1) by including 10 μM MnCl2 in the enzyme and 2-OG solution used to initiate the assay. Analysis of the data identified 81 compounds as probable metal chelators (Table 2), with representative compounds eliminated by this counter screen illustrated in Supplemental Table S2. Furthermore, an inspection of the remaining hits revealed 20 compounds that contained a 8-hydroxyquinoline functional group, a reported metal chelator that can bind a variety of biologically relevant divalent metal ions [36]. These compounds were also excluded from further analysis (Table 2).
Fig. 4.
Control assays for the metal-chelation counter screen. (A) SpHCS activity in the presence of increasing concentration of MnCl2. The fraction activity was calculated relative to the activity of the enzyme in the absence of MnCl2. The averages of three measurements with an error of one SD are illustrated. (B) Fractional activity of SpHCS incubated with 0 or 10 μM MnCl2 in the presence of increasing concentrations of EDTA. Data represent the average of triplicate measurements normalized to SpHCS activity in the absence of MnCl2 and EDTA with error bars indicating one SD.
Identifying potent inhibitor compounds of HCS
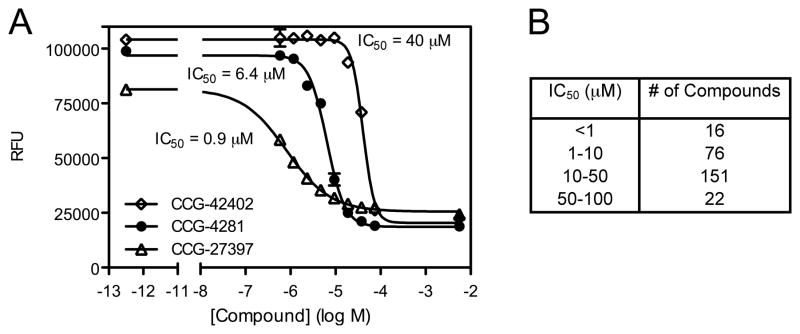
In order to quantify the inhibition of SpHCS, dose response curves were measured for the 338 compounds remaining after the CoA and Mn(II) counter screen triages. Each compound was assayed in duplicate in serial two-fold dilutions ranging from 0.586 – 75.0 μM following the primary assay protocol (Table 1). Of the compounds analyzed by dose response, 266 exhibited sigmoidal behavior for which IC50 values were calculated. Representative data for the dose responses plots for three compounds with differing IC50 values are illustrated in Fig. 5A. The 266 compounds had IC50 values ranging from <1.0 – 135 μM with 16 compounds being particularly potent with sub-micromolar values (Fig. 5B). Compounds that exhibited reasonable potency (pAC50 > 4.5) and a Hill slope between 0 and −2 were prioritized for future analysis.
Fig. 5.
Dose response analysis of small molecule inhibitors of SpHCS. (A) Dose response curves for three representative compounds with differing IC50 values. A sigmoidal nonlinear regression was fit to the data points (n=2) with error bars indication one SD. (B) Range of the IC50 values for compounds that exhibited sigmoidal dose-response curves.
Discussion
The enzymes in the first half of the AAA pathway for L-lysine biosynthesis are highly conserved in fungal pathogens and represent potential targets for antifungal drug design. Initial efforts to inhibit fungal lysine biosynthesis have focused on the rational design of inhibitors of homoaconitase and homoisocitrate dehydrogenase, the second and third enzymes in this pathway, respectively (Supplemental Fig. 1) [37; 38]. The results presented here represent the first effort to employ HTS to identify small molecule inhibitors of HCS, which is a nexus for the biochemical and transcriptional regulation of lysine biosynthesis in fungi. The fluorescent HCS assay used for HTS is easy, reproducible and amenable to a 384-well plate format and also assay displays excellent statistical parameters for HTS. Two counter screens were devised to eliminate false positive hits due to quenching of the fluorescence of the MMBC-CoA adduct or indirect inhibition of SpHCS activity by metal ion chelators. The latter counter screen failed to eliminate many compounds containing an 8-hydroxyquinoline moiety, which has previously been reported to exhibit metal chelation properties [36]. This group was shown to bind Mn(II) at pH 7.0 at a 10:1 ratio of compound suggesting that its binding affinity may be too weak to effectively chelate Mn(II) under the HTS conditions. Due to the uncertainty as to whether 8-hydroxyquinoline-bearing compounds inhibited SpHCS through metal chelation, they were categorized as false positives and excluded from subsequent dose response analysis. Of the compounds that were analyzed by dose response, 78% exhibited sigmoidal plots for inhibition, with 122 compounds (0.3% of compounds screened) displaying desirable potency and reasonable Hill slopes. These compounds have been prioritized for future in vitro and in vivo studies designed to characterize their kinetic mechanisms of inhibition and to assess their efficacy in inhibiting HCS in cell-based assays. Moreover, these compounds may prove useful in elucidating the nuclear functions of HCS, which has recently been implicated in DNA damage response in S. cerevisiae [39].
In addition to its applicability to HCS, the HTS assay described here can also be readily adapted to identify chemical modulators of other acyl-CoA-dependent acyltransferases or enzymes that generate a product bearing a free sulfhydryl group. For example, this assay could be used to screen for inhibitors of 2-isopropylmalate synthase, an enzyme that is structurally and mechanistically related to HCS. 2-isopropylmalate synthase catalyzes the first step in leucine biosynthesis by reacting 2-oxoisovaletate and AcCoA to generate 2-isopropylmalate and CoA. The leucine biosynthetic pathway is essential in Mycobacterium tuberculosis, rendering this enzyme a potential target for anti-tubercular drug design [40]. Aminoglycoside N-acetyltransferases are another category of AcCoA-utilizing enzymes that are pharmacologically relevant drug targets. These enzymes are members of the GCN5-related N-acetyltransferase superfamily and detoxify antibiotics by catalyzing the AcCoA-dependent acetylation of the pendant amine groups of aminoglycosides, such as kanamycin and gentamicin [41]. HATs represent an additional class of biomedically important AcCoA-dependent N-acetyltransferases. These enzymes belong to various classes, including the GCN5, p300/CBP and MYST families, and acetylate the terminal ε-amine groups of specific lysines in histones and non-histone proteins [42]. HATs have been causally linked to cell proliferation and tumor suppression, and small molecules that modulate their activity may offer novel avenues for treating various forms of cancer. In addition, the yeast HAT Rtt109 has recently been reported to be essential to the virulence of the fungal pathogen C. albicans, suggesting that Rtt109-specific inhibitors may hold clinical value in combating candidiasis [43]. Finally, enzymes involved in lipid metabolism that produce CoA could be screened using this assay. One such class of enzymes is the acyl-CoA thioesterases, which catalyze the hydrolysis of fatty acyl-CoA esters to yield a free fatty acid and CoA. These enzymes represent important regulators of lipid metabolism and other cellular processes and include acyl-CoA thioesterase 7, which has been recently identified for its roles in inflammation and neurodegenerative diseases [44]. In summary, the fluorescent HCS assay offers a rapid and facile method for screening the activity of acyl-CoA-dependent acyltransferases implicated in disease and is readily scalable for HTS applications.
Supplementary Material
Acknowledgments
We kindly thank Dr. Paul Kirchhoff (Center for Chemical Genomics and Vahlteich Medicinal Chemistry Core, University of Michigan) and Dr. Scott Larsen (Department of Medicinal Chemistry and Vahlteich Medicinal Chemistry Core, University of Michigan) for their assistance with the HTS triages. We also acknowledge Dr. Lorraine Pillus, Dr. Cory Hogaboam and Rei Otsuka for reading the manuscript and providing useful comments. S.L.B. received support from an NIH CBTP Training grant (5T32GM008353-18) and from the Rackham Graduate School at the University of Michigan through a Predoctoral Fellowship and a Graduate Student Research Grant. Research for this project was funded in part through a University of Michigan Biomedical Research Council Pilot Grant and a Margolies Grant to R.C.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Branski LK, Al-Mousawi A, Rivero H, Jeschke MG, Sanford AP, Herndon DN. Emerging infections in burns. Surg Infect (Larchmt) 2009;10:389–97. doi: 10.1089/sur.2009.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan JE, Benson C, Holmes KH, Brooks JT, Pau A, Masur H. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm Rep. 2009;58:1–207. quiz CE1–4. [PubMed] [Google Scholar]
- 3.Mofenson LM, Brady MT, Danner SP, Dominguez KL, Hazra R, Handelsman E, Havens P, Nesheim S, Read JS, Serchuck L, Van Dyke R. Guidelines for the Prevention and Treatment of Opportunistic Infections among HIV-exposed and HIV-infected children: recommendations from CDC, the National Institutes of Health, the HIV Medicine Association of the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the American Academy of Pediatrics. MMWR Recomm Rep. 2009;58:1–166. [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TJ, Groll AH. Emerging fungal pathogens: evolving challenges to immunocompromised patients for the twenty-first century. Transpl Infect Dis. 1999;1:247–61. doi: 10.1034/j.1399-3062.1999.010404.x. [DOI] [PubMed] [Google Scholar]
- 5.Mahfouz T, Anaissie E. Prevention of fungal infections in the immunocompromised host. Curr Opin Investig Drugs. 2003;4:974–90. [PubMed] [Google Scholar]
- 6.Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET) Clin Infect Dis. 2010;50:1101–11. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 7.Caston-Osorio JJ, Rivero A, Torre-Cisneros J. Epidemiology of invasive fungal infection. Int J Antimicrob Agents. 2008;32(Suppl 2):S103–9. doi: 10.1016/S0924-8579(08)70009-8. [DOI] [PubMed] [Google Scholar]
- 8.Enoch DA, Ludlam HA, Brown NM. Invasive fungal infections: a review of epidemiology and management options. J Med Microbiol. 2006;55:809–18. doi: 10.1099/jmm.0.46548-0. [DOI] [PubMed] [Google Scholar]
- 9.Mathew BP, Nath M. Recent approaches to antifungal therapy for invasive mycoses. ChemMedChem. 2009;4:310–23. doi: 10.1002/cmdc.200800353. [DOI] [PubMed] [Google Scholar]
- 10.Rogers TR, Frost S. Newer antifungal agents for invasive fungal infections in patients with haematological malignancy. Br J Haematol. 2009;144:629–41. doi: 10.1111/j.1365-2141.2008.07412.x. [DOI] [PubMed] [Google Scholar]
- 11.Peman J, Canton E, Espinel-Ingroff A. Antifungal drug resistance mechanisms. Expert Rev Anti Infect Ther. 2009;7:453–60. doi: 10.1586/eri.09.18. [DOI] [PubMed] [Google Scholar]
- 12.Garrad RC, Bhattacharjee JK. Lysine biosynthesis in selected pathogenic fungi: characterization of lysine auxotrophs and the cloned LYS1 gene of Candida albicans. J Bacteriol. 1992;174:7379–84. doi: 10.1128/jb.174.22.7379-7384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Andi B, Qian J, West AH, Cook PF. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. doi: 10.1385/CBB:46:1:43. [DOI] [PubMed] [Google Scholar]
- 14.Zabriskie TM, Jackson MD. Lysine biosynthesis and metabolism in fungi. Nat Prod Rep. 2000;17:85–97. doi: 10.1039/a801345d. [DOI] [PubMed] [Google Scholar]
- 15.Demain AL, Masurekar PS. Lysine inhibition of in vivo homocitrate synthesis in Penicillium chrysogenum. J Gen Microbiol. 1974;82:143–51. doi: 10.1099/00221287-82-1-143. [DOI] [PubMed] [Google Scholar]
- 16.Gaillardin CM, Poirier L, Heslot H. A kinetic study of homocitrate synthetase activity in the yeast Saccharomycopsis lipolytica. Biochim Biophys Acta. 1976;422:390–406. doi: 10.1016/0005-2744(76)90150-9. [DOI] [PubMed] [Google Scholar]
- 17.Maragoudakis ME, Holmes H, Strassman M. Control of lysine biosynthesis in yeast by a feedback mechanism. J Bacteriol. 1967;93:1677–80. doi: 10.1128/jb.93.5.1677-1680.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wulandari AP, Miyazaki J, Kobashi N, Nishiyama M, Hoshino T, Yamane H. Characterization of bacterial homocitrate synthase involved in lysine biosynthesis. FEBS Lett. 2002;522:35–40. doi: 10.1016/s0014-5793(02)02877-6. [DOI] [PubMed] [Google Scholar]
- 19.Bulfer SL, Scott EM, Pillus L, Trievel RC. Structural basis for L-lysine feedback inhibition of homocitrate synthase. J Biol Chem. 2010;285:10446–53. doi: 10.1074/jbc.M109.094383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada T, Tomita T, Wulandari AP, Kuzuyama T, Nishiyama M. Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus. J Biol Chem. 2010;285:4195–205. doi: 10.1074/jbc.M109.086330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon ES, Jeong JH, Roe JH. Inactivation of homocitrate synthase causes lysine auxotrophy in copper/zinc-containing superoxide dismutase-deficient yeast Schizosaccharomyces pombe. J Biol Chem. 2006;281:1345–51. doi: 10.1074/jbc.M506611200. [DOI] [PubMed] [Google Scholar]
- 22.Wolfner M, Yep D, Messenguy F, Fink GR. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975;96:273–90. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- 23.Ramos F, Waime JM. Mutation affecting the specific reulatory control of lysine biosynthetic enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1985;200:291–294. doi: 10.1007/BF00425438. [DOI] [PubMed] [Google Scholar]
- 24.Becker B, Feller A, el Alami M, Dubois E, Pierard A. A nonameric core sequence is required upstream of the LYS genes of Saccharomyces cerevisiae for Lys14p-mediated activation and apparent repression by lysine. Mol Microbiol. 1998;29:151–63. doi: 10.1046/j.1365-2958.1998.00916.x. [DOI] [PubMed] [Google Scholar]
- 25.Feller A, Dubois E, Ramos F, Pierard A. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol Cell Biol. 1994;14:6411–8. doi: 10.1128/mcb.14.10.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos F, Dubois E, Pierard A. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur J Biochem. 1988;171:171–6. doi: 10.1111/j.1432-1033.1988.tb13773.x. [DOI] [PubMed] [Google Scholar]
- 27.Schobel F, Jacobsen ID, Brock M. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot Cell. 2010;9:878–93. doi: 10.1128/EC.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trievel RC, Li FY, Marmorstein R. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal Biochem. 2000;287:319–28. doi: 10.1006/abio.2000.4855. [DOI] [PubMed] [Google Scholar]
- 29.Bulfer SL, Scott EM, Couture JF, Pillus L, Trievel RC. Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis. J Biol Chem. 2009;284:35769–80. doi: 10.1074/jbc.M109.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Jaklitsch WM, Kubicek CP. Homocitrate synthase from Penicillium chrysogenum. Localization, purification of the cytosolic isoenzyme, and sensitivity to lysine. Biochem J. 1990;269:247–53. doi: 10.1042/bj2690247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andi B, West AH, Cook PF. Stabilization and characterization of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae. Arch Biochem Biophys. 2004;421:243–54. doi: 10.1016/j.abb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Achyuthan KE, Whitten DG. Design considerations for high-throughput screening and in vitro diagnostic assays. Comb Chem High Throughput Screen. 2007;10:399–412. doi: 10.2174/138620707781996475. [DOI] [PubMed] [Google Scholar]
- 35.Qian J, West AH, Cook PF. Acid-base chemical mechanism of homocitrate synthase from Saccharomyces cerevisiae. Biochemistry. 2006;45:12136–43. doi: 10.1021/bi060889h. [DOI] [PubMed] [Google Scholar]
- 36.Albert A, Gledhill WS. The choice of a chelating agent for inactivating trace metals: I. A survey of commercially available chelating agents. Biochem J. 1947;41:529–33. [PubMed] [Google Scholar]
- 37.Palmer DR, Balogh H, Ma G, Zhou X, Marko M, Kaminskyj SG. Synthesis and antifungal properties of compounds which target the alpha-aminoadipate pathway. Pharmazie. 2004;59:93–8. [PubMed] [Google Scholar]
- 38.Yamamoto T, Eguchi T. Thiahomoisocitrate: a highly potent inhibitor of homoisocitrate dehydrogenase involved in the alpha-aminoadipate pathway. Bioorg Med Chem. 2008;16:3372–6. doi: 10.1016/j.bmc.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Scott EM, Pillus L. Homocitrate synthase connects amino acid metabolism to chromatin functions through Esa1 and DNA damage. Genes Dev. 2010;24:1903–13. doi: 10.1101/gad.1935910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hondalus MK, Bardarov S, Russell R, Chan J, Jacobs WR, Jr, Bloom BR. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect Immun. 2000;68:2888–98. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vetting MW, SdCLP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–26. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–8. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci USA. 107:1594–9. doi: 10.1073/pnas.0912427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forwood JK, Thakur AS, Guncar G, Marfori M, Mouradov D, Meng W, Robinson J, Huber T, Kellie S, Martin JL, Hume DA, Kobe B. Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc Natl Acad Sci USA. 2007;104:10382–7. doi: 10.1073/pnas.0700974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.