Abstract
Etoposide, a topoisomerase II inhibitor widely used in cancer therapy, is suspected of inducing secondary tumors and affecting the genetic constitution of germ cells. A better understanding of the potential heritable risk of etoposide is needed to provide sound genetic counseling to cancer patients treated with this drug in their reproductive years. We used a mouse model to investigate the effects of clinical doses of etoposide on the induction of chromosomal abnormalities in spermatocytes and their transmission to zygotes by using a combination of chromosome painting and 4′,6-diamidino-2-phenylindole staining. High frequencies of chromosomal aberrations were detected in spermatocytes within 64 h after treatment when over 30% of the metaphases analyzed had structural aberrations (P < 0.01). Significant increases in the percentages of zygotic metaphases with structural aberrations were found only for matings that sampled treated pachytene (28-fold, P < 0.0001) and preleptotene spermatocytes (13-fold, P < 0.001). Etoposide induced mostly acentric fragments and deletions, types of aberrations expected to result in embryonic lethality, because they represent loss of genetic material. Chromosomal exchanges were rare. Etoposide treatment of pachytene cells induced aneuploidy in both spermatocytes (18-fold, P < 0.01) and zygotes (8-fold, P < 0.05). We know of no other report of an agent for which paternal exposure leads to an increased incidence of aneuploidy in the offspring. Thus, we found that therapeutic doses of etoposide affect primarily meiotic germ cells, producing unstable structural aberrations and aneuploidy, effects that are transmitted to the progeny. This finding suggests that individuals who undergo chemotherapy with etoposide may be at a higher risk for abnormal reproductive outcomes especially within the 2 months after chemotherapy.
Etoposide (ET), also known as VP-16, is one of the most commonly used agents in cancer chemotherapy (1, 2) and has improved significantly the treatment of leukemia, lymphomas, and many solid tumors, including testicular and ovarian cancers (3). However, evidence is accumulating that ET is genotoxic (4–6), and that it can induce secondary tumors (7) and impair fertility (6). ET is a semisynthetic derivative of podophyllotoxin and acts as a topoisomerase II (topo II) inhibitor (1). Topo II enzymes function by transiently introducing DNA double-strand breaks, allowing the passage of one double helix through another, and resealing the double-strand break (8). Topo II activity is needed for removing regions of DNA catenation during DNA replication and before chromosome segregation (9–11) and for chromosome condensation (12). It is an abundant nuclear protein that is associated with the chromosome core or scaffold of metaphase chromosomes (13, 14) and with elements of the synaptonemal complex (15). ET inhibits topo II activity by forming a ternary complex, DNA–topo II–ET, that prevents the ligation of the double-strand breaks (2, 4–6). Stabilization of this complex results in the disruption of chromosomal integrity and formation of chromosomal aberrations (16–18).
ET is a somatic-cell mutagen capable of inducing both numerical and structural chromosome aberrations (5, 6, 19). It is also a germ-cell mutagen with unique properties in both maternal and paternal gametes of mice. ET induced both aneuploidy and chromosomal structural aberrations in female germ cells (20, 21) and, unlike any other chemical shown to induce aneuploidy in female germ cells (22), ET did so without causing a delay in meiotic progression (20). In male germ cells, ET induced fragmentation of centromeric DNA and micronuclei after exposure of primary spermatocytes (23, 24). Diplotene–diakinesis I cells were the most sensitive to the action of ET (25). ET induced specific locus mutations (26) and dominant lethality (27) only in early and late meiotic stages. This pattern of sensitivity has not been reported before (28). Surprisingly, ET is a weak inducer of heritable translocations (27), suggesting that it induces unstable and stable chromosomal aberrations in differing proportions in male germ cells. The mechanisms underlying this differential susceptibility are not understood.
Studies in humans have reported that certain chemotherapy regimens increase the frequencies of aneuploid sperm (29, 30), suggesting that such patients may be at higher risk for abnormal reproductive outcomes. However, direct epidemiological evidence for transmitted chromosomal damage after paternal chemotherapy is lacking still. We applied PAINT/DAPI analysis (31, 32) in the mouse to model the human response. The PAINT/DAPI assay uses chromosome painting (PAINT) and 4′,6-diamidino-2-phenylindole (DAPI) to characterize the time course of induction of chromosomal aberrations and aneuploidy in male germ cells and the transmission of cytogenetic damage to first-cleavage zygotes after paternal exposure to doses of ET that were equivalent to human therapeutic doses. Our design allows us to measure the differential sensitivity of postmeiotic, meiotic, and premeiotic cells, and to determine whether there is selection against chromosomally abnormal sperm at fertilization.
Materials and Methods
Animals and Chemical Treatment of Males.
B6C3F1 mice (Harlan–Sprague–Dawley), 6 to 8 weeks of age at the beginning of the experiments, were maintained under a 14-h light/10-h dark photoperiod at a room temperature of 21–23°C and a relative humidity of 50 ± 5%. Pelleted food and sterilized tap water were provided ad libitum.
Male mice received 80 mg/kg ET (CAS no. 33419-42-0, Sigma) dissolved in DMSO (Sigma). ET was administered i.p. at the final volume of 0.1 ml/30 g of body weight. Control mice received similar amounts of DMSO only. For human chemotherapy, ET is administered typically at doses of 50–150 mg/m2 per day for 3 to 5 days (33). However, daily doses up to 750 mg/m2 also are used (34). The mouse has a body weight/surface area ratio of ≈3 kg/m2 (35); therefore the dose used in our study corresponds to ≈240 mg/m2 and is within the dose range used for human chemotherapy.
Twenty of the 83 males (24%) that were treated with ET died. The average time of death was 24.7 days after treatment (range: 6 to 40 days). The cause of death was not ascertained; however, a previous study (36) reported macroscopic intestinal bleeding, necrosis of the intestinal mucosa, and pulmonary congestion in autopsies of animals exposed to similar doses of ET. This result raises the question of whether mice may be more sensitive than humans to the toxic effects of ET.
Analysis of Meiosis I and II Metaphases.
Males were euthanized by CO2 inhalation 6, 16, 40, and 64 h and 10 days after treatment (Table 1). Testis preparations were made according to a standard method (37). Slides were coded and stained with 0.25 μg/ml DAPI diluted in Vectashield mounting medium (Vector Laboratories). Each slide was examined by using a ×40 objective for localizing metaphase I (MI) and metaphase II (MII) spermatocytes, and then with a ×100 objective for identifying chromosome structural and numerical abnormalities. MI and MII metaphases were analyzed for chromosome structural aberrations, whereas only MII spermatocytes were analyzed for numerical abnormalities. For each time point, the data from four animals were combined, and the mean plus the standard error of the mean was calculated.
Table 1.
Germ-cell stage at the time of ET treatment for cells that were analyzed as spermatocytes or zygotes
| Time* | Spermatocyte analysis | Zygote analysis |
|---|---|---|
| 6 h | Diakinesis | |
| 16 h | Diplotene/diakinesis | |
| 40 h | Pachytene/diplotene | |
| 64 h | Pachytene | |
| 6.5 d | Testicular sperm | |
| 10 d | Leptotene/zygotene | |
| 24.5 d | Pachytene | |
| 34.5 d | Preleptotene | |
| 41.5 d | A spermatogonia |
For spermatocyte analysis: time between exposure and sacrifice; for zygote analysis: time between exposure and estimated time of fertilization.
A subgroup of these slides was hybridized with a probe mixture containing four biotin-labeled painting probes specific for chromosomes 1, 6, 16, and X, and a digoxigenin-labeled probe specific for chromosome Y (Applied Genetics Laboratories). This probe combination detects ≈32% of all possible chromosomal exchanges. Hybridization and washing conditions were as described (31). Fluorescent signals were amplified by using the Biotin-Digoxigenin Dual Color Detection kit (Oncor) and by following the manufacturer's specifications. DAPI (0.25 μg/ml) was used as counterstaining. These slides were analyzed for the presence of chromatid exchanges.
Analysis of First-Cleavage Zygote Metaphases.
Males were mated with untreated females at 6.5, 24.5, 34.5, and 41.5 days after administration of ET or DMSO only. These time points correspond to fertilization with sperm that were testicular spermatozoa, pachytene spermatocytes, preleptotene spermatocytes, or differentiating spermatogonia at the time of treatment (38), respectively (Table 1). For each time point, the data from at least three repetitions (each using a different group of males) were combined, and the mean plus the standard error of the mean were calculated. Female mice were superovulated by an i.p. injection of 7.5 units of pregnant mare's serum (Sigma) followed 48h later by an i.p. injection of 5.0 units of human chorionic gonadotrophin (hCG, Sigma). They were caged with males (1:1) immediately after the injection of hCG. The mating pairs were separated 8 h later and females were checked for the presence of vaginal plugs. Twenty-four hours after the hCG injection, mated females received an i.p. injection of 0.08 mg of colchicine (CAS no. 64-86-8, Sigma) in 0.2 ml of distilled water to prevent the union of the two parental pronuclei of the zygote and arrest development at the metaphase stage of the first-cleavage division. Females were euthanized by CO2 inhalation 6 h later. Zygotes collected from 10–15 females were pooled and processed by using the mass harvest procedure (39).
Prepared slides were examined with a ×10 objective to obtain the fertilization rate (number of eggs that were fertilized) and the rate of zygotic development (number of fertilized eggs that reached the first-cleavage metaphase stage). Slides then were hybridized with a probe mixture containing four biotin-labeled probes specific for chromosomes 1, 2, 3, or X, plus a digoxigenin-labeled probe specific for chromosome Y (Applied Genetics Laboratories). This probe combination detects ≈37% of all possible exchanges (31). Hybridization, washing, and amplification of the signals were performed as described for meiotic preparations. Scoring of the chromosomal aberrations was done as described (31).
Statistical Analysis.
A χ2 test with adjustment for overdispersion (40) was used for the analysis of the results in spermatocytes, because the observations within the 6- and 64-h groups did not follow a Poisson distribution. The χ2 test also was used for evaluating the effects on fertilization rate and zygotic development, whereas Fisher's exact test (FET) was used for evaluating the induction of chromosomal abnormalities and aneuploidy in zygotes.
Results
Analyses of Meiotic Metaphases.
Chromosomal structural aberrations in MI and MII.
Significant increases (P < 0.01) in the percentages of meiotic metaphases with chromosomal aberrations relative to control values were found at all time points tested except in MII spermatocytes collected 10 days after treatment (Table 2). The highest frequencies of chromosomal aberrations were found 40 h after treatment, when 53.9% of MI spermatocytes and 56.3% of MII spermatocytes analyzed had chromosomal aberrations. The proportions of MI and MII spermatocytes with chromosomal aberrations found at each time point were not different from each other. Common features between MI and MII spermatocytes (Table 3) were as follows: (i) the presence of a substantial fraction of metaphases that were heavily damaged (registered as multiple aberrations, Fig. 1B), and (ii) the presence of chromosomal damage localized at the centromeric region (breaks at the centromere). However, other types of aberrations were represented differentially in the two metaphases. Chromosome fragments were seen mostly in MI spermatocytes, whereas chromatid fragments were seen mostly in MII spermatocytes (Fig. 1C). In addition, an unusual type of damage was seen in MI spermatocytes collected 6 h after ET treatment. Over 18% of the MI spermatocytes analyzed at this time point had extensive fragmentation localized in the bright DAPI heterochromatic region (centromere fragmentation). No such type of damage was seen in MII spermatocytes. A few quadrivalent configurations, an indication of chromatid exchange, were seen in MI spermatocytes in all treated groups. However, PAINT analysis of 193 MII spermatocytes (equivalent to the analysis of 60 metaphases, considering the fraction of the genome painted) failed to identify a single metaphase with a chromatid exchange.
Table 2.
Structural chromosome aberrations detected by DAPI analysis in MI and MII spermatocytes at various times after exposure of male mice to ET
| Dose, mg/kg | Harvest time | Metaphase | No. cells analyzed | Aberrations
|
|
|---|---|---|---|---|---|
| No. | % | ||||
| 0 | MI | 315 | 2 | 0.6 ± 0.4 | |
| MII | 268 | 5 | 1.9 ± 0.8 | ||
| 80 | 6 h | MI | 261 | 95 | 36.4 ± 14.6* |
| MII | 242 | 102 | 42.1 ± 12.3* | ||
| 80 | 16 h | MI | 320 | 141 | 44.1 ± 4.6* |
| MII | 229 | 99 | 43.2 ± 7.9* | ||
| 80 | 40 h | MI | 358 | 193 | 53.9 ± 8.5* |
| MII | 158 | 89 | 56.3 ± 6.3* | ||
| 80 | 64 h | MI | 303 | 83 | 27.4 ± 6.5* |
| MII | 210 | 66 | 31.4 ± 4.8* | ||
| 80 | 10 d | MI | 200 | 19 | 9.5 ± 2.6* |
| MII | 219 | 12 | 5.5 ± 2.7 | ||
Results are presented ±SE. *, P < 0.01 vs. controls (χ2).
Table 3.
Specific types of structural aberrations detected by DAPI analysis of MI and MII spermatocytes after exposure of male mice to ET
| Dose, mg/kg | Harvest time | Metaphase | No. cells analyzed | Types of chromosomal aberrations
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Multiple aberrations
|
Centromere
fragmentation
|
Breaks at
centromere
|
Chromatid fragments
|
Chromatid
exchanges
|
Chromosome fragments
|
||||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | ||||
| 0 | MI | 315 | 0 | 0 | 0 | 0 | 1 | 0.3 | 0 | 0 | 0 | 0 | 1 | 0.3 | |
| MII | 268 | 0 | 0 | 0 | 0 | 1 | 0.4 | 4 | 1.5 | 0 | 0 | 0 | 0 | ||
| 80 | 6 h | MI | 261 | 23 | 8.8 | 48 | 18.4 | 1 | 0.4 | 7 | 2.7 | 5 | 1.9 | 11 | 4.2 |
| MII | 242 | 26 | 10.7 | 0 | 0 | 6 | 2.5 | 68 | 28.1 | 0 | 0 | 2 | 0.8 | ||
| 80 | 16 h | MI | 320 | 71 | 22.1 | 10 | 3.1 | 15 | 4.7 | 6 | 1.9 | 23 | 7.2 | 16 | 5.0 |
| MII | 229 | 28 | 12.2 | 0 | 0 | 18 | 4.7 | 53 | 23.1 | 0 | 0 | 0 | 0 | ||
| 80 | 40 h | MI | 358 | 112 | 31.3 | 4 | 1.1 | 11 | 3.1 | 3 | 0.8 | 32 | 8.9 | 29 | 8.1 |
| MII | 158 | 21 | 13.3 | 0 | 0 | 7 | 4.4 | 61 | 38.6 | 0 | 0 | 0 | 0 | ||
| 80 | 64 h | MI | 303 | 32 | 10.6 | 0 | 0 | 9 | 3.0 | 7 | 2.3 | 18 | 5.9 | 15 | 4.9 |
| MII | 210 | 7 | 3.3 | 0 | 0 | 10 | 4.8 | 48 | 22.8 | 0 | 0 | 1 | 0.5 | ||
| 80 | 10 d | MI | 200 | 6 | 3.0 | 2 | 1.0 | 2 | 1.0 | 0 | 0 | 5 | 2.5 | 4 | 2.0 |
| MII | 219 | 0 | 0 | 0 | 0 | 5 | 2.3 | 7 | 2.3 | 0 | 0 | 0 | 0 | ||
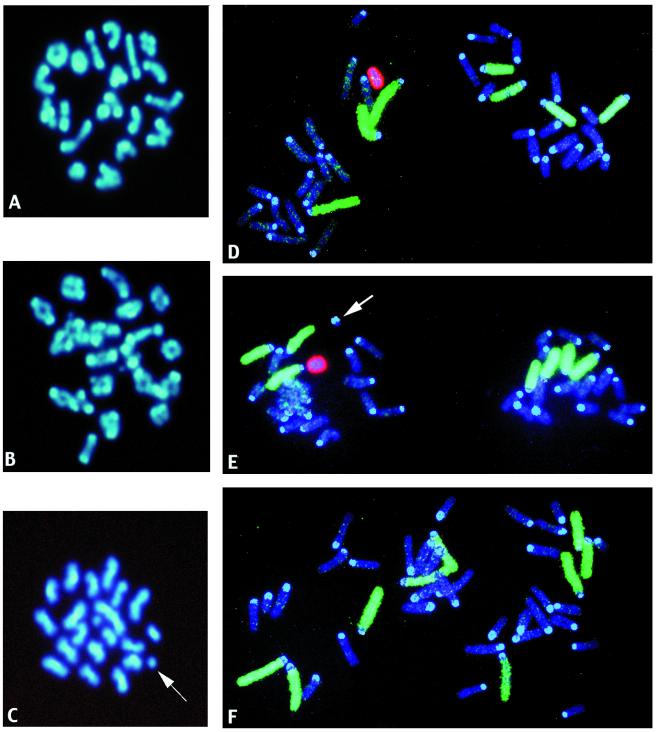
Figure 1.
Photographs of mouse MI and II spermatocytes after DAPI staining (A–C) and first-cleavage (1-Cl) zygotes after hybridization with chromosome-specific painting probes for chromosomes 1–3 and X (labeled with biotin and signaled with FITC) and chromosome Y [labeled with digoxigenin and signaled with rhodamine (D–F)]. Images were taken by using a Vysis (Downers Grove, IL) QUIPS Imaging Analysis System, and the final composite figure was made in Adobe PHOTOSHOP. (A) Normal MI spermatocyte. (B) MI spermatocyte with multiple chromosome structural aberrations. (C) MII spermatocyte with chromatid acentric fragments (one is indicated by the arrow, the second is in the center of the metaphase). (D) Normal 1-Cl zygote metaphase with the Y-bearing sperm-derived chromosomes on the left. (E) 1-Cl zygote with a centric fragment in the paternal chromosomes (arrow). (F) Hyperploid 1-Cl zygote. Note the presence ofa an extra chromosome (green) in the X-bearing sperm-derived chromosomes.
Numerical abnormalities in MII.
Significant increases (15- to 18-fold) in the number of hyperhaploid spermatocytes were found at 16 and 40 h (Table 4). This is the highest level of chemical-induced hyperhaploidy ever reported in MII spermatocytes (41, 42). Diploidy was increased also with respect to control values at 16 , 40, and 64 h (P < 0.01). The highest frequency of diploid MII spermatocytes was found 40 h after ET administration, when 19% of the spermatocytes analyzed had 40 chromosomes. A 2-fold increase in the frequencies of diploid spermatocytes was reported previously after administration of 20 mg/kg ET (43).
Table 4.
Numerical abnormalities in MII spermatocytes after exposure of male mice to ET
| Dose, mg/kg | Harvest time | Aneuploidy
|
Diploidy (N =
40)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. spermatocytes | Hypohaploidy
(N = 16–19)
|
Haploidy
(N = 20)
|
Hyperhaploidy
(N = 21)
|
Total no. spermatocytes | No. | % | |||||
| No. | % | No. | % | No. | % | ||||||
| 0 | 257 | 28 | 10.9 ± 2.9 | 228 | 88.7 | 1 | 0.4 ± 0.4 | 268 | 11 | 4.1 ± 1.4 | |
| 80 | 6 h | 184 | 15 | 8.2 ± 2.3 | 169 | 91.8 | 0 | 0 | 189 | 5 | 2.6 ± 0.8 |
| 80 | 16 h | 164 | 27 | 16.5 ± 4.8 | 127 | 77.4 | 10 | 6.1 ± 2.3* | 187 | 23 | 12.3 ± 3.5* |
| 80 | 40 h | 111 | 16 | 14.4 ± 3.1 | 87 | 78.4 | 8 | 7.2 ± 1.9* | 137 | 26 | 19.0 ± 4.5* |
| 80 | 64 h | 177 | 21 | 11.9 ± 3.4 | 153 | 86.4 | 3 | 1.7 ± 1.0 | 195 | 18 | 9.2 ± 0.9* |
| 80 | 10 d | 211 | 43 | 20.4 ± 3.1 | 167 | 79.1 | 1 | 0.5 ± 0.5 | 217 | 6 | 2.8 ± 1.2 |
Results are presented ±SE. *, P < 0.01 vs. controls (χ2).
Analyses of First-Cleavage Metaphases.
Transmitted chromosomal structural aberrations.
Overall, the fertilization rate and zygotic development were not affected by ET treatment of male germ cells (Table 5). There was a small reduction in the fertilization rate with respect to the control value (P < 0.01, χ2) only at day 24.5. Interestingly, this was the time point with the highest percentage of zygotes with chromosomal aberrations. Specifically, ET treatment of pachytene spermatocytes resulted in structural aberrations in 16.5% of the zygotes analyzed (P < 0.001; FET; Table 6). An elevated frequency of chromosomally abnormal zygotes was found also after treatment of preleptotene spermatocytes (34.5 days, 7.5%, P < 0.001, FET). Conversely, chromosome aberrations were not increased significantly (P > 0.05, FET) with respect to the control value after treatment of either early spermatozoa (6.5 days) or differentiating spermatogonia (41.5 days).
Table 5.
Effect of ET treatment of male germ cells on fertilization rate and zygotic development
| Dose, mg/kg | Days post ET | No. eggs | Fertilized
eggs*
|
First-cleavage
metaphases†
|
||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| 0 | 698 | 568 | 81.4 ± 2.1 | 459 | 80.8 ± 4.9 | |
| 80 | 6.5 | 627 | 522 | 83.3 ± 3.9 | 451 | 86.4 ± 9.7 |
| 80 | 24.5 | 835 | 630 | 75.4 ± 2.0‡ | 486 | 77.1 ± 8.2 |
| 80 | 34.5 | 318 | 242 | 76.1 ± 3.2 | 201 | 83.1 ± 8.0 |
| 80 | 41.5 | 321 | 233 | 72.6 ± 12.8 | 188 | 80.7 ± 9.4 |
Results are presented ± SE.
Fertilization rate, indicator of prefertilization toxicity.
Zygotic development (first-cleavage metaphases/fertilized eggs), indicator of postfertilization toxicity.
P < 0.01 vs. controls (χ2).
Table 6.
Structural and numerical chromosome abnormalities detected by PAINT/DAPI analysis in mouse first-cleavage zygotes after exposure of male mice to ET
| Dose, mg/kg | Days post ET | Structural aberrations
|
Numerical
abnormalities
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. zygotes | No. with aberrations | % ± SEM | Total no. zygotes | Hypoploidy
(N = 36–39)
|
Diploidy
(N = 40)
|
Hyperploidy
(N = 41)
|
|||||
| No. | % | No. | % | No. | % | ||||||
| 0 | 318 | 2 | 0.6 ± 0.3 | 303 | 29 | 9.6 ± 2.3 | 268 | 88.4 | 1 | 0.3 ± 0.5 | |
| 80 | 6.5 | 180 | 2 | 1.1 ± 1.0 | 170 | 17 | 10.0 ± 6.1 | 153 | 90.0 | 0 | 0 |
| 80 | 24.5 | 254 | 42 | 16.5 ± 0.8* | 249 | 29 | 11.6 ± 1.8 | 213 | 85.5 | 6 | 2.4 ± 1.0† |
| 80 | 34.5 | 146 | 11 | 7.5 ± 1.9* | 140 | 6 | 4.3 ± 1.9 | 129 | 92.1 | 4 | 2.9 ± 3.8† |
| 80 | 41.5 | 152 | 4 | 2.6 ± 1.0 | 145 | 11 | 7.6 ± 2.7 | 131 | 90.3 | 2 | 1.4 ± 0.9 |
Results are presented ±SE.
P < 0.001 vs. controls (FET).
P < 0.05 vs. controls (FET).
The specific types of chromosomal aberrations observed by PAINT/DAPI analysis in zygotes are listed in Table 7. The majority of the aberrations detected consisted of chromosome acentric fragments. Although a few chromosomal exchanges were observed, no reciprocal translocations were found. Among the chromosomal exchanges, several originated from breaks occurring within the heterochromatin region near the centromere (Robertsonian fusion-like chromosomes). At 24.5 days, a considerable fraction of the abnormal metaphases (≈20%) had centric fragments, that is, chromosomes in which the bright DAPI heterochromatin region was as long as the rest of the chromosome, suggesting the presence of an extensive chromosomal deletion (Fig. 1E).
Table 7.
Numbers and types of structural aberrations detected by PAINT/DAPI analysis in mouse zygotes after exposure of male mice to ET
| Dose, mg/kg | Days post ET | DAPI
analysis
|
PAINT analysis*
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total cells | Dicentrics | Fragments
|
RFL† | Other | Cell eq‡ | Translocations
|
dic(AB) | Acentric
fragments
|
|||||
| Acentric | Centric | t(Ab) | t(Ba) | ace(ab) | ace(b) | ||||||||
| 0 | 318 | 0 | 2 | 0 | 0 | 0 | 114 | 0 | 0 | 0 | 0 | 0 | |
| 80 | 6.5 | 180 | 0 | 1 | 0 | 0 | 1 | 63 | 0 | 0 | 0 | 0 | 0 |
| 80 | 24.5 | 254 | 9 | 33 | 8 | 12 | 3 | 94 | 4 | 1 | 1 | 5 | 9 |
| 80 | 34.5 | 146 | 3 | 11 | 0 | 0 | 1 | 54 | 0 | 1 | 3 | 0 | 0 |
| 80 | 41.5 | 152 | 1 | 2 | 1 | 0 | 1 | 57 | 0 | 1 | 0 | 0 | 1 |
A detailed description of these aberrations can be found in Marchetti et al. (31).
Robertsonian fusion-like chromosomes.
Cell-equivalent (number of cells analyzed/percent of exchanges that can be detected with our probe combination).
Transmitted numerical abnormalities.
ET treatment of male germ cells resulted in an increase in zygotes with numerical abnormalities (Table 6). The number of hyperploid zygotes (Fig. 1F) was increased (8- to 10-fold) with respect to the control value at both 24.5- and 34.5-day mating times (P < 0.05, FET). However, although all six cases observed at 24.5 days were of paternal origin, only two of four cases found at 34.5 days were unequivocally paternal. In the other two cases, the parental origin was not determined, because either the two sets of chromosomes had already joined or it was not possible to distinguish objectively the paternal from the maternal chromosomes on the basis of the degree of chromosome condensation. One hyperploid zygote in the control group and one at 41.5 days were of maternal origin. These results, together with those in MII spermatocytes, show that ET induced aneuploidy in treated pachytene spermatocytes.
Discussion
We applied PAINT/DAPI analyses to meiotic cells as well as first-cleavage zygotes to investigate the induction of chromosomal abnormalities in male germ cells and their subsequent transmission to zygotes after paternal treatment with ET. Our results showed that ET induced (i) its clastogenic effects only in meiotic cells with relatively little to no detectable effects on spermatogonia or postmeiotic cells; (ii) mostly chromosomal fragmentation and very few chromosomal exchanges; and (iii) aneuploidy after treatment of pachytene spermatocytes, which was detected in both spermatocytes and zygotes.
Chromosomal Structural Aberrations.
The time course study of ET-induced effects in male germ cells shows that only meiotic cells are sensitive to ET and is in agreement with previous findings (26, 27). Two unique features of this pattern are the peak response in pachytene spermatocytes and the lack of an effect in postmeiotic stages (28). The highest induction of chromosomal aberrations in our study was during the stage of meiosis with the highest activity of topo II (15, 44). Chromosomal aberrations in pachytene spermatocytes may arise, because inhibition of topo II activity can affect both chromatin condensation (12) and resolution of chiasmata at the transition from meiotic prophase I to MI (44).
The majority of chromosomal aberrations were breaks, whereas very few exchanges were detected. This finding suggests that unstable aberrations were produced at higher rates. It is possible that ET inhibited the rejoining of the double-strand break formed by topo II and that the presence of the ET–topo II–DNA complex at the site of the break prevented accessibility of the DNA repair proteins to the strand break, creating the chromosome fragmentation and at the same time preventing the formation of chromosomal exchanges.
It has been reported that exposure of germ cells to ET results in a preferential localization of the aberrations near the centromere (20, 23, 24). We have observed a similar phenomenon in both MI and MII spermatocytes. In addition, 6 h after exposure (that is, after treatment of diakinesis spermatocytes), about 20% of MI spermatocytes analyzed had extensive fragmentation localized in the pericentromeric region. As already mentioned, there is a functional requirement for topo II activity in the transition from meiotic prophase to meiotic metaphase (44), and analysis of mitotic chromosomes has shown that topo II is distributed throughout the chromosome in prophase but it accumulates in the centromeric regions as it nears metaphase (14). This localized accumulation of topo II may make the centromeric region a preferential target for the action of ET. Supporting this notion, studies in somatic cells have shown a direct correlation between the nuclear levels of topo II and the amount of chromosomal aberrations induced after exposure to ET (45).
ET-Induced Numerical Abnormalities.
The analysis of MII spermatocytes and zygotes showed that ET is also an aneugen in male germ cells. Unlike spindle inhibitors, the most common class of aneugens, ET induces numerical abnormalities without interacting directly with cellular organelles responsible for chromosome movement (i.e., kinetochores, centromeres, spindle fibers, etc.). ET may prevent the segregation of the homologous chromosomes because topo II is required for the resolution of recombined chromosomes and the proper segregation of sister chromatids (10). Recently, it was reported that ET treatment decreased meiotic recombination frequencies in male germ cells (46). The frequency of hyperhaploidy found in MII spermatocytes after treatment of pachytene spermatocytes is the highest ever reported for an aneugen in male germ cells (41, 42). Also, unlike model aneugens, such as colchicine and vinblastine, for which male germ cells seem to be less sensitive than female germ cells (47), the frequency of hyperhaploidy found in MII spermatocytes in this study was similar to that found in MII oocytes (20).
An important finding of this study was the significant increase in the frequencies of hyperploid zygotes of paternal origin in matings occurring 24.5 days after ET treatment (Table 6). We know of no other report of a male germ-cell exposure that produced a significant increase in the incidence of aneuploidy in the offspring.
Selection Against Chromosomally Defective Spermatocytes.
Comparisons between the data in spermatocytes and zygotes for cells that were at the same stage of meiosis at the time of treatment (Table 1) suggest that there is selection against damaged spermatocytes. In fact, both the frequencies of cells with chromosomal structural aberrations (P < 0.01, 64 h vs. 24.5 days, FET) and aneuploidy (P < 0.05, 16 and 40 h vs. 24.5 days, FET) were higher in spermatocytes than in zygotes. Damaged spermatocytes may have been eliminated by apoptosis during spermiogenesis or, alternatively, ET-induced chromosomal damage may have interfered with the sperm fertilizing ability. In support of the first possibility, studies in rats have shown that ET is a potent inducer of apoptosis and that the most sensitive cells included pachytene and dividing spermatocytes (48). Also, we have shown that aneuploidy per se is not sufficient to affect sperm development and fertilization functions (49).
Comparisons with Dominant-Lethal and Heritable-Translocation Data.
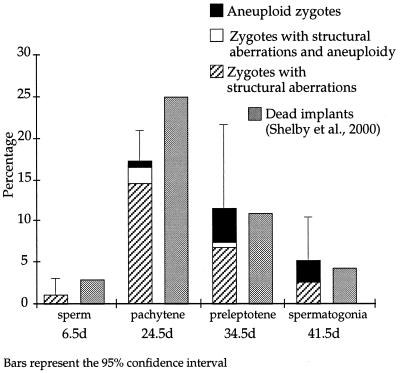
In a previous study with the model germ-cell clastogen acrylamide, we reported a strong correlation between the frequencies of unstable and stable aberrations in zygotes, and the frequencies of dead implants and of offspring with reciprocal translocations, respectively (32). Therefore, we determined whether a similar correlation existed after paternal exposure to ET. A study of dominant lethality after paternal exposure to 80 mg/kg ET (27) reported that the frequencies of dead implants were increased with respect to controls only at mating intervals 20.5–27.5 and 32.5–35.5 days. As shown in Fig. 2, the proportion of zygotes with chromosomal abnormalities that are expected to result in embryonic lethality, i.e., unstable aberrations and aneuploidy, paralleled the findings of the dominant-lethal study in both time course and magnitude. Additionally, at 24.5 days we analyzed 254 zygotes by PAINT (Table 7). Taking into account the percentage of the genome painted with our probe combination and the number of zygotes with unstable aberrations that would result in dominant lethality, this was equivalent to analyzing the complete genomes of 75 offspring. PAINT analysis of this sample failed to reveal a single reciprocal translocation. This result is consistent with the analysis of offspring from matings occurring between 22.5 and 26.5 days after paternal exposure to ET, which indicated that only 10 of 693 F1 males (1.4%) carried reciprocal translocations (27).
Figure 2.
Comparison of the proportions of zygotes with unstable chromosomal aberrations and/or aneuploidy vs. proportions of dead implants. d, days.
Conclusions
We found that ET is a unique germ-cell mutagen affecting only meiotic germ cells, in which it produces mostly unstable structural aberrations and aneuploidy, effects that are transmitted to the progeny. The ability of ET to induce heritable chromosomal abnormalities in male germ cells at clinically relevant doses suggest that individuals who undergo chemotherapy with this drug may be at higher risk for abnormal reproductive outcomes, especially in the second month after chemotherapy, when the products of treated meiotic cells appear in the ejaculate.
Acknowledgments
This work was funded by the National Institute of Environmental Health Sciences through an National Institute of Environmental Health Sciences/Department of Energy Interagency Agreement (Y01-ES-10203-00), a National Institute of Environmental Health Sciences Small Business Innovation Research Grant N44-ES72003, and was performed under the auspices of the U.S. Department of Energy by the Lawrence Livermore National Laboratory through Contract W-7405-END-48.
Abbreviations
- ET
etoposide
- topo II
topoisomerase II
- PAINT
chromosome painting
- DAPI
4′,6-diamidino-2-phenylindole
- MI
metaphase I
- FET
Fisher's exact test
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Liu L F. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 2.Smith P J. BioEssays. 1990;12:167–172. doi: 10.1002/bies.950120405. [DOI] [PubMed] [Google Scholar]
- 3.Henwood J M, Brogden R N. Drugs. 1990;39:438–490. doi: 10.2165/00003495-199039030-00008. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson L R, Baguley B C. Environ Mol Mutagen. 1994;24:245–261. doi: 10.1002/em.2850240402. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson L R, Baguley B C. Mutat Res. 1996;355:91–101. doi: 10.1016/0027-5107(96)00024-3. [DOI] [PubMed] [Google Scholar]
- 6.Anderson R D, Berger N A. Mutat Res. 1994;309:109–142. doi: 10.1016/0027-5107(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 7.Murphy S B. J Clin Oncol. 1993;11:199–201. doi: 10.1200/JCO.1993.11.2.199. [DOI] [PubMed] [Google Scholar]
- 8.Wang J C, Caron P R, Kim R A. Cell. 1990;62:403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- 9.DiNardo S, Voelkel K, Sternglanz R. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose D, Thomas W, Holm C. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- 11.Downes C S, Mullinger A M, Johnson R T. Proc Natl Acad Sci USA. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi Y, Luke M, Laemmli U K. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 13.Poljak L, Kas E. Trends Cell Biol. 1995;5:348–354. doi: 10.1016/s0962-8924(00)89068-6. [DOI] [PubMed] [Google Scholar]
- 14.Sumner A T. Chromosome Res. 1996;4:5–14. doi: 10.1007/BF02254938. [DOI] [PubMed] [Google Scholar]
- 15.Moens P B. J Cell Sci. 1990;97:1–3. doi: 10.1242/jcs.97.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki H, Ikeda T, Yamgishi T, Nakaike S, Nakane S, Ohsawa M. Mutat Res. 1995;328:151–161. doi: 10.1016/0027-5107(95)00005-4. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H, Tarumoto Y, Ohsawa M. Mutagenesis. 1997;12:29–33. doi: 10.1093/mutage/12.1.29. [DOI] [PubMed] [Google Scholar]
- 18.Pommier Y, Kohn K W. In: Developments in Cancer Chemotherapy. Glazer R I, editor. Boca Raton, FL: CRC Press; 1989. pp. 175–196. [Google Scholar]
- 19.Cimini D, Antoccia A, Tanzarella C, Degrassi F. Cytogenet Cell Genet. 1997;76:61–67. doi: 10.1159/000134517. [DOI] [PubMed] [Google Scholar]
- 20.Mailhes J B, Marchetti F, Phillips G L J, Barnhill D R. Teratog Carcinog Mutagen. 1994;14:39–51. doi: 10.1002/tcm.1770140106. [DOI] [PubMed] [Google Scholar]
- 21.Mailhes J B, Marchetti F, Young D, London S N. Mutagenesis. 1996;11:357–361. doi: 10.1093/mutage/11.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Mailhes J B, Marchetti F. Mutat Res. 1994;320:87–111. doi: 10.1016/0165-1218(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 23.Kallio M, Lähdetie J. Mutagenesis. 1993;8:561–567. doi: 10.1093/mutage/8.6.561. [DOI] [PubMed] [Google Scholar]
- 24.Kallio M, Lähdetie J. Mutagenesis. 1996;11:435–443. doi: 10.1093/mutage/11.5.435. [DOI] [PubMed] [Google Scholar]
- 25.Lähdetie J, Keiski A, Suutai A, Toppari J. Environ Mol Mutagen. 1994;24:192–202. doi: 10.1002/em.2850240308. [DOI] [PubMed] [Google Scholar]
- 26.Russell L B, Hunsicker P R, Johnson D K, Shelby M D. Mutat Res. 1998;400:279–286. doi: 10.1016/s0027-5107(98)00036-0. [DOI] [PubMed] [Google Scholar]
- 27.Shelby, M. D., Bishop, J. B., Hughes, L., Morris, R. W. & Generoso, W. M. (2001), in press.
- 28.Shelby M D. Mutat Res. 1996;325:159–167. doi: 10.1016/0027-5107(95)00222-7. [DOI] [PubMed] [Google Scholar]
- 29.Robbins W A, Meistrich M L, Moore D, Hagemeister F B, Weier H U, Cassel M J, Wilson G, Eskenazi B, Wyrobek A J. Nat Genet. 1997;16:74–78. doi: 10.1038/ng0597-74. [DOI] [PubMed] [Google Scholar]
- 30.Martin R, Ernst S, Rademaker A, Barclay L, Ko E, Summers N. Cancer Genet Cytogenet. 1999;108:133–136. doi: 10.1016/s0165-4608(98)00125-3. [DOI] [PubMed] [Google Scholar]
- 31.Marchetti F, Lowe X, Moore D I, Bishop J, Wyrobek A J. Chromosome Res. 1996;4:604–613. doi: 10.1007/BF02261723. [DOI] [PubMed] [Google Scholar]
- 32.Marchetti F, Lowe X, Bishop J, Wyrobek A J. Environ Mol Mutagen. 1997;30:410–417. doi: 10.1002/(sici)1098-2280(1997)30:4<410::aid-em6>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Einhorn L H. Clin Cancer Res. 1997;3:2630–2632. [PubMed] [Google Scholar]
- 34.Broun E R, Nichols C R, Gize G, Cornetta K, Hromas R A, Schacht B, Einhorn L H. Cancer. 1997;79:1605–1610. doi: 10.1002/(sici)1097-0142(19970415)79:8<1605::aid-cncr25>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 35.Freireich E J, Gehan E A, Rall D P, Schimdt L H, Skiupper H E. Cancer Chemother Rep. 1966;50:219–244. [PubMed] [Google Scholar]
- 36.Lee J S, Takahashi T, Hagiwara A, Yoneyama C, Itoh M, Sasabe T, Muranishi S, Tashima S. Cancer Chemother Pharmacol. 1995;36:211–216. doi: 10.1007/BF00685848. [DOI] [PubMed] [Google Scholar]
- 37.Evans E P, Breckon G, Ford C E. Cytogenetics. 1964;3:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
- 38.Oakberg E F. Am J Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- 39.Mailhes J B, Yuan Z P. Gamete Res. 1987;18:77–83. doi: 10.1002/mrd.1120180109. [DOI] [PubMed] [Google Scholar]
- 40.Collett D. Modelling Binary Data. London: Chapman & Hall; 1991. [Google Scholar]
- 41.Allen J W, Liang J C, Carrano A V, Preston R J. Mutat Res. 1986;167:123–137. doi: 10.1016/0165-1110(86)90013-8. [DOI] [PubMed] [Google Scholar]
- 42.Miller B M, Adler I D. Mutagenesis. 1992;7:69–76. doi: 10.1093/mutage/7.1.69. [DOI] [PubMed] [Google Scholar]
- 43.Kallio M, Lähdetie J. Environ Mol Mutagen. 1997;29:16–27. doi: 10.1002/(sici)1098-2280(1997)29:1<16::aid-em3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 44.Cobb J, Reddy R K, Park C, Handel M A. Mol Reprod Dev. 1997;46:489–498. doi: 10.1002/(SICI)1098-2795(199704)46:4<489::AID-MRD7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Davies S M, Robson C N, Davies S L, Hickson I D. J Biol Chem. 1988;263:17724–17729. [PubMed] [Google Scholar]
- 46.Russell L B, Hunsicker P R, Hack A M, Ashley T. Mutat Res. 2000;464:201–212. doi: 10.1016/s1383-5718(99)00185-0. [DOI] [PubMed] [Google Scholar]
- 47.Mailhes J B. Mutat Res. 1995;339:155–176. doi: 10.1016/0165-1110(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 48.Sjöblom T, West A, Lähdetie J. Environ Mol Mutagen. 1998;31:133–148. doi: 10.1002/(sici)1098-2280(1998)31:2<133::aid-em5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 49.Marchetti F, Lowe X, Bishop J, Wyrobek J. Biol Reprod. 1999;61:948–954. doi: 10.1095/biolreprod61.4.948. [DOI] [PubMed] [Google Scholar]