Abstract
Suppressors and enhancers of position effect variegation (PEV) have been linked to the establishment and maintenance of heterochromatin. The presence of centromeres and other inheritance elements in hetero-chromatic regions suggests that suppressors and enhancers of PEV, Su(var) s and E(var)s [collectively termed Mod(var)s], may be required for chromosome inheritance. In order to test this hypothesis, we screened 59 ethyl methanesulfonate-generated Drosophila Mod (var)s for dominant effects on the partially compromised inheritance of a minichromosome (J21A) missing a portion of the genetically defined centromere. Nearly half of these Mod(var)s significantly increased or decreased the transmission of J21A. Analyses of homozygous mutant larval neuroblasts suggest that these mutations affect cell cycle progression and native chromosome morphology. Five out of six complementation groups tested displayed mitotic abnormalities, including phenotypes such as telomere fusions, overcondensed chromosomes, and low mitotic index. We conclude that Mod (var)s as a group are highly enriched for genes that encode essential inheritance functions. We propose that a primary function of Mod(var)s is to promote chromosome inheritance, and that the gene silencing phenotype associated with PEV may be a secondary consequence of the heterochromatic structures required to carry out these functions.
Introduction
The ability to distribute chromosomes properly during cell division is essential for the viability of all organisms. At the heart of the segregation process is the centromere, which serves as the nucleation site for the kinetochore (for review see Sullivan et al. 2001), where microtubules attach and help move sister chromatids or homologs to opposite poles during mitosis and meiosis. The centromere is typically embedded in heterochromatin, the portion of the genome rich in repeated sequences and poor in genes (for review see Hennig 1999). Heterochromatin is involved in other critical inheritance functions, including meiotic pairing (Dernburg et al. 1996; Karpen et al. 1996), sister chromatid cohesion (Bickel and Orr-Weaver 1996; Dej and Orr-Weaver 2000; Bernard et al. 2001; Hall et al. 2003) and interactions with the NOD kinesin-like protein (Afshar et al. 1995; Murphy and Karpen 1995a). Dissecting the roles of heterochromatin in centromere function and other inheritance processes will greatly increase our understanding of the cis and trans components of chromosome inheritance.
Significant information about heterochromatin has come from studying the phenomenon of position effect variegation (PEV; for review see Wallrath 1998). Position effect variegation refers to the clonal inactivation of gene expression that occurs when a gene is removed from its normal chromatin context (e.g. euchromatin) and is placed in an opposing chromatin context (e.g. heterochromatin) via chromosomal rearrangement or transgene insertion. A classic example of PEV in Drosophila is the expression of the white eye color gene in the chromosomal inversion In(1)wm4 (Muller 1930), in which the euchromatic white gene is placed immediately adjacent to X chromosome centric heterochromatin. The proximity to heterochromatin causes mosaic white repression, resulting in a mottled eye color phenotype. Multiple genetic screens for modifiers of PEV [Mod(var)s] have identified a large number of mutations that dominantly enhance [E(var)s] or suppress [Su(var)s] heterochromatin-mediated gene silencing (Reuter and Wolff 1981; Locke et al. 1988; Dorn et al. 1993).
In Schizosaccharomyces pombe, mutations that alleviate centromere-mediated silencing also induce elevated rates of minichromosome loss and missegregation (Allshire et al. 1995; Grewal et al. 1998; Freeman-Cook et al. 1999). In Drosophila melanogaster, mutations in the Su(var)2–5 locus, which encodes Heterochromatin Protein 1 (HP1), exhibit recessive mitotic defects, including telomere fusions (Fanti et al. 1998) and chromosome undercondensation (Kellum and Alberts 1995). Other Mod(var) mutations — specifically Su(var)2–8, Su(var) 2–10 and Su(var)3–6 — cause chromosome loss, undercondensation and overcondensation, respectively (Axton et al. 1990; Wines and Henikoff 1992; Hari et al. 2001). Mammalian HP1 can be delocalized as a consequence of prolonged exposure to deacetylase inhibitors or by mutations in the histone H3 methyltransferase SUV39, and this is accompanied by defects in chromosome segregation (Rea et al. 2000; Lachner et al. 2001; Taddei et al. 2001). Chromosome segregation defects associated with mutations in the S. pombe HP1 homolog, Swi6, appear to be caused by defects in cohesin recruitment to centromeric regions (Bernard et al. 2001; Nonaka et al. 2002). In Drosophila, a recent study has demonstrated that some Mod(var) mutations affect meiotic recombination (Westphal and Reuter 2002). These results indicate that Mod(var) mutations can impact chromosome inheritance and behavior; however, no comprehensive study of the inheritance roles of Mod(var) genes in metazoans has been reported.
The fully functional 1.3 Mb Drosophila minichromosome Dp(1;f)1187 (Dp1187) has been a useful substrate for studies of centromere structure and function and the molecular biology of heterochromatin (Le et al. 1995; Murphy and Karpen 1995b; Karpen et al. 1996; Sun et al. 1997, 2003). Previous studies using this minichromosome and its radiation-induced deletion derivatives localized the functional centromere to a 420 kb region within the heterochromatin of Dp1187. Derivatives of Dp1187 with deletions of the centromere region show reduced transmission. The derivative J21A is missing approximately one-third of the genetically defined centromere, and is transmitted to progeny half as well as intact minichromosomes (Sun et al. 1997). Its intermediate level of transmission makes it an ideal candidate to look for dominant interactions. Indeed, known inheritance mutations dominantly enhance or suppress the transmission of J21A and other ‘sensitized’ minichromosomes (Murphy and Karpen 1995a; Cook et al. 1997). Seventy-eight P element-induced mutations that dominantly affect minichromosome inheritance were isolated by screening for increased or decreased transmission of J21A (Dobie et al. 2001). P element insertions were recovered in genes previously characterized to play roles in inheritance and many of the novel homozygous lethal mutations affected the function of the normal Drosophila chromosome complement, further substantiating a role for these mutations in inheritance.
Here, we present evidence that suggests that a high proportion of Mod(var)s are required for normal chromosome inheritance. We describe the results of a genetic screen in which 59 Mod(var) mutations, representing 49 complementation groups, were tested for dominant effects on the transmission of J21A. Nearly half of the Mod(var) mutations dominantly increased or decreased J21A transmission to progeny. Cytological analysis demonstrated that endogenous Drosophila chromosomes display visible mitotic chromosome defects in homozygous lethal mutants. We conclude that a large percentage of modifiers of PEV have essential, diverse roles in chromosome inheritance. Our findings suggest that a primary function of heterochromatin is to promote chromosome inheritance, and that some properties of heterochromatin, such as gene silencing, may be the consequence of the DNA and protein structures required to ensure proper inheritance of chromosomes from parent to progeny.
Materials and methods
Fly stocks and crosses
J21A is the result of a terminal deletion that removed approximately one-third of the genetically defined centromere and the rest of the flanking heterochromatin from an inversion derivative (Dp238)of Dp(1;f)1187. It is 580 kb in size. J21A and modifiers of terminal deficiency associated PEV [Mod( TDA-PEV)] are described in more detail elsewhere (Murphy and Karpen 1995b; Donaldson et al. 2002). Previously identified Su(var)s (Reuter and Wolff 1981; Sinclair et al. 1992; Eissenberg et al. 1992) were obtained from B. Wakimoto, G. Reuter, and J. Eissenberg. Balancer-GFP (green fluorescent protein) stocks came from the Bloomington Stock Center. All mutations were crossed into a standard y1; ry506 background prior to use.
J21A monosome transmission assay
J21A is a Dp1187 minichromosome deletion derivative marked with two insertions of a ry+-marked P element construct (Murphy and Karpen 1995b). Transmission tests for dominant effects on minichromosome inheritance were performed as described previously (Murphy and Karpen 1995a; Cook et al. 1997). Single, heterozygous mutant females bearing one copy of J21A were crossed to several C(1;Y)1, y1; ry506 (X^Y) males; transmission frequency was determined by counting ry+ ( J21A present) and ry− (J21A absent) female progeny. The attached X–Y (X^Y) is utilized to produce X / X^Y female progeny, such that the presence of an extra Y suppresses the partial silencing of the ry+ marker. Only vials containing at least 30 progeny were included in this analysis. Standard Student’s t-tests were used to compare mutant results with y1;ry506 controls; P values of ≤0.05 were considered statistically significant.
Cytological analysis
Females and males of the genotype mutation/GFP-marked balancer were crossed to produce homozygous mutant, non-GFP larvae for cytological analysis of neuroblast mitoses. The oldest developing homozygous mutant larvae produced from each line were subsequently selected for dissection. Larval brains were dissected in 0.7% NaCl, fixed with 45% acetic acid for 30–60 s, squashed onto slides, and frozen in liquid nitrogen. After coverslip removal, chromosomes were stained with 1 μg/ml 4′,6-diamidino-2-phenylindole and scored on a Zeiss Axiophot Epifluorescence Microscope. Images were captured with a cooled CCD camera (Princeton Instruments) and IP Lab Spectrum software (Scanalytics). In addition to the above method, a subset of neuroblasts from some lines were also prepared with a 5 min soak in 0.8% sodium citrate after the dissection. The hypotonic swell is known to cause artificial chromatid arm separation (Gatti et al. 1994) and this phenotype was ignored in these neuroblasts. Mitotic figures from mutant larvae were compared with y1; ry506 control larvae. Since early instar larvae have fewer mitotic figures than later stage larvae, random sampling can lead to sample bias. Therefore, the entire brain was analyzed for mitotic and anaphase figures. The mitotic index here is defined as the total number of mitotic and anaphase figures divided by an approximation of the total number of cells (calculated by multiplying the average number of cells from five randomly selected fields by the total number of fields scored). Standard Student’s t-tests were used to determine the significance of differences between mutants and controls. The fields were scored at 100× magnification with a 1.25× optivar.
Results
A high proportion of previously identified Su(var)s dominantly affect J21A transmission
The minichromosome deletion derivative J21A is transmitted to only 28% of progeny as a monosome, compared with 50% transmission for a full-length Dp1187 minichromosome (Murphy and Karpen 1995b; Cook et al. 1997). The partial function of J21A makes it an excellent substrate for identifying genes that play a role in chromosome inheritance, since a reduced level of a protein required for inheritance may not affect the normal complement of chromosomes, but may affect J21A transmission. Consequently, potential inheritance loci can be identified by observing dominant effects of heterozygous mutations on J21A transmission. This approach also circumvents the recessive lethality associated with many loss-of-function mutations in inheritance genes. Known inheritance mutations dominantly affect J21A transmission, but do not dominantly alter the transmission of minichromosomes with the entire genetically defined centromere, or the normal complement of chromosomes, demonstrating the sensitivity of J21A (Murphy and Karpen 1995a; Cook et al. 1997). The J21A assay has also been used successfully to identify new chromosome inheritance genes (Dobie et al. 2001).
We tested nine previously identified Su(var) mutations representing five different loci (Reuter and Wolff 1981; Wustmann et al. 1989; Eissenberg et al. 1992) for dominant effects on J21A minichromosome transmission (see Materials and methods). We also tested the effect of an extra Y chromosome, known to be a strong suppressor of many types of PEV (Spofford 1976). Three of the nine Su(var) mutations and the extra Y chromosome reduced transmission significantly (P ≤0.05, Table 1), while one mutation increased transmission. Su(var)2051 and the extra Y chromosome displayed the most dramatic reductions of J21A transmission, to 5% and 8%, respectively. Su(var)2–10 reduced transmission to 15%, and in-depth analysis demonstrated that Su(var)2–10 affects chromosome condensation and nuclear organization (Hari et al. 2001). Thus, three out of five loci tested had at least one allele that altered J21A transmission significantly, suggesting a general role for Su(var)s in chromosome inheritance.
Table 1.
The effects of previously identified Su(var)s on J21A transmission. (N, number of vials with >30 progeny)
| Gene |
J21A transmission |
P value | |
|---|---|---|---|
| (%±SD) | N | ||
| Su(var)2051 | 5±3 | 13 | <0.01 |
| Extra Y | 8±5 | 10 | <0.01 |
| Su(var)2–102 | 15±10 | 20 | <0.01 |
| Su(var)2–11 | 14±13 | 16 | <0.01 |
| Su(var)2–101 | 20±14 | 14 | 0.12 |
| Su(var)2–571 | 23±8 | 16 | 0.13 |
| Su(var)2–12–13 | 23±12 | 9 | 0.43 |
| Su(var)2–481 | 25±14 | 15 | 0.62 |
| Su(var)2–19 | 29±13 | 17 | 0.47 |
| Su(var)2–55 | 34±8 | 14 | 0.01 |
Nearly half of the Mod(var)s induced in an isogenic background significantly affect J21A transmission
Data from screening the previously identified Su(var)s suggested that this class of mutations would provide a rich source of genes involved in chromosome transmission. However, genetic background in the form of additional mutations induced on the parental chromosomes or accumulated in the balanced stocks could be responsible for the transmission phenotype. A large group of ethyl methanesulfonate (EMS)-induced modifiers of TDA-PEV (Donaldson and Karpen 1997; Donaldson et al. 2002) was generated in the same isogenic y; ry genetic background used for previous J21A transmission screens (Cook et al. 1997). Most of the mutations affected multiple examples of PEV (Donaldson et al. 2002). These mutations were used to test the generality of Mod(var) effects on inheritance.
Fifty modifiers of TDA-PEV, representing maximally 45 complementation groups (Donaldson et al. 2002), were tested for dominant effects on J21A transmission (Table 2). The original y1; ry506 stock used for the EMS screen was used as a control, and showed 27% transmission, as expected from previous studies. Twenty-three of the 50 (46%) lines tested altered J21A transmission significantly. A majority of the modifiers analyzed were Su(var)s. In total, 20 out of 45 Su(var) lines significantly altered J21A transmission. Seventeen of the 20 lines reduced transmission to 7–21%, representing decreases of 22–74% from the control transmission value of 27%. Three lines increased transmission to 37–41%, showing an increase of at least 50% from the control. Five enhancers of TDA-PEV were also tested. Three of five enhancers affected J21A transmission significantly. One line (E1060) reduced transmission weakly to 20%, while two lines (E1178 and E1377) increased transmission to 39–40%. In total, 21 of 45 complementation groups tested (47%) altered J21A transmission significantly.
Table 2.
Results of screening modifiers of position effect variegation (PEV) in an isogenic background for J21A transmission effects. (E, Enhancer of variegation; N, number of vials with >30 progeny; S, Suppressor of variegation)
| Line |
J21A transmission |
P value | |
|---|---|---|---|
| (%±SD) | N | ||
| Decreased transmission (18) | |||
| 1038 | 7±4 | 14 | <0.01 |
| 224 | 10±6 | 14 | <0.01 |
| 1116 | 11±9 | 14 | <0.01 |
| 1126 | 12±7 | 7 | <0.01 |
| 1025 | 13±6 | 14 | <0.01 |
| 1009 a | 13±8 | 12 | <0.01 |
| 1474 | 14±8 | 12 | <0.01 |
| 1453 | 14±6 | 13 | <0.01 |
| 1447 | 14±8 | 11 | <0.01 |
| 1539 | 16±10 | 13 | <0.01 |
| 1315 | 16±9 | 10 | <0.01 |
| 234 c | 16±10 | 14 | <0.01 |
| 1260 | 17±6 | 9 | <0.01 |
| 600 | 17±8 | 13 | <0.01 |
| 1207 a | 19±7 | 15 | <0.01 |
| E1060 b | 20±5 | 10 | <0.01 |
| 1200 | 19±7 | 11 | 0.01 |
| 242 | 21±7 | 15 | 0.02 |
| Increased transmission (5) | |||
| 1094 | 37±8 | 11 | <0.01 |
| 1173 | 39±9 | 9 | <0.01 |
| 1144 | 41±12 | 19 | <0.01 |
| E1178 b | 40±16 | 15 | 0.01 |
| E1377 | 39±13 | 9 | 0.03 |
| No effect (27) | |||
| 25S, 2E Control |
25±10 | 322 | |
| y1;ry506 | 27±7 | 19 | |
Complementation groups:
1227 and 1545 (no effect), 1009 and 1207
E1060, E1178
234, 1551 (no effect)
Combining the results from the analyses of previously identified Su(var)s and the isogenic collection of modifiers of TDA-PEV, 27 of 59 Mod(var) mutations affected J21A transmission significantly (Table 3). Twenty-four of the 54 Su(var)s significantly altered J21A transmission; the vast majority of these mutations (20/23, or 87%) decreased transmission. Of the three E(var) lines that altered J21A transmission, two increased transmission while the other decreased it. One complementation group [Su(var)2–5] is represented by both previously identified and newly induced Mod(var) mutations, thus lowering the total number of loci that affect J21A transmission to 23 of 49. These results show that a large proportion of Mod(var)s (23/49 loci, 47%) affect the inheritance of the J21A ‘sensitized’ minichromosome.
Table 3.
Summary of effects of Su(var) and E(var) mutations on J21A transmission
| Lines tested |
Complementation groups tested |
|||||
|---|---|---|---|---|---|---|
| Altered transmission | Total | % with effects | Altered transmission | Total | % with effects | |
| Previously isolated | 4 | 9 | 44 | 3 | 5 | 60 |
| Su(var)s | ||||||
| New Su(var)s | 20 | 45 | 44 | 19 | 41 | 46 |
| New E(var)s | 3 | 5 | 60 | 2 | 4 | 50 |
| Total | 27 | 59 | 46 | 24(23)a | 50(49)a | 48(47)a |
Alleles of Su(var)2–5 are present among the previously isolated and new Su(var)s. Therefore, the total of 24/50 is reduced to 23/49
Mitotic defects are observed in homozygous lethal Mod(var) mutants
In previous studies, mutations in inheritance genes did not have dominant effects on the transmission of minichromosomes containing the entire genetically defined centromere (Cook et al. 1997); therefore, the Mod(var)s that have dominant effects on J21A transmission are unlikely to have dominant effects on the transmission of the normal complement of chromosomes. Nevertheless, the Mod(var)s may have recessive effects on the inheritance of normal, full-length chromosomes, which could lead to aneuploidy and lethality. Therefore, we examined larval neuroblasts from nine homozygous lethal Mod(var) mutations (six complementation groups) that altered J21A transmission for cytological defects in chromosome behavior.
Homozygous mutants for many of the lines had either small brains and/or diminutive or missing imaginal discs, phenotypes previously demonstrated to correlate with mitotic defects (Gatti and Baker 1989). Larval brains from lines 234, 1097, 1227, 1545, and 1552 displayed significantly lower frequencies of cells in metaphase and anaphase, in comparison with y1;ry506 controls and line 1259, which is homozygous viable and had no effect on J21A transmission (Table 4). The low mitotic indices may be due to cell cycle arrest in interphase, a prolonged interphase, or decreased length of mitosis. The rare metaphase nuclei in these lines had no obvious chromosomal abnormalities.
Table 4.
Homozygous lethal mutations affecting J21A transmission display reduced mitotic index. (N, number of brains scored. See Materials and methods for how mitotic index was calculated)
| Line | Mitotic Index (×10−4) | N | P value | Larval stage |
|---|---|---|---|---|
| y1;ry506 | 2±2 | 7 | N/A | 1st |
| y1;ry506 | 30±10 | 7 | N/A | 2nd |
| 234 | 6±4 | 5 | 0.03 | 2nd |
| 1097 | 3±2 | 5 | 0.01 | 2nd |
| 1227 | 4±4 | 5 | 0.03 | 2nd |
| 1545 | 0 | 6 | 0.03 | 1st |
| 1552 | 5±3 | 5 | 0.01 | 2nd |
| 1259 | 40±40 | 5 | 0.23 | 2nd |
| 1126 | 3±3 | 4 | 0.67 | 1st |
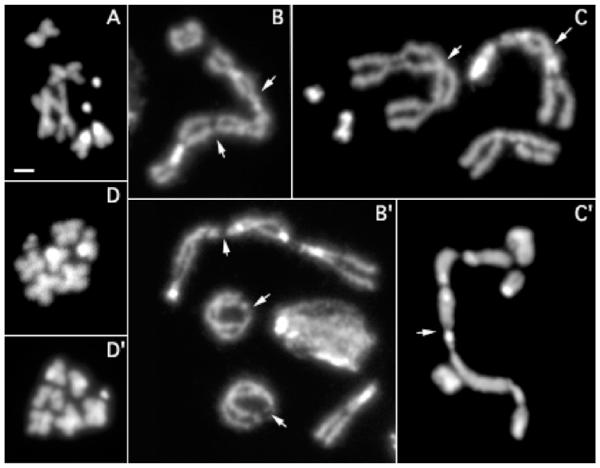
Mitotic chromosomes from two other complementation groups displayed striking cytological defects (Fig. 1). Telomere fusions were seen in second and third instar larval neuroblasts from Su(var)1207, including interchromosomal fusions of two or more chromosomes to form large rings, intrachromosomal fusions to form small rings, and multiple chromosomes fused to form linear ‘trains’. Such fusions have been observed in homozygous mutant larvae for Su(var)2–5 (Fanti et al. 1998), and for UbcD (also known as effete, Cenci et al. 1997). 1207 and 1009 fail to complement Su(var)2–502 for viability and contain nonsense mutations in the Su(var)2–5 open reading frame, indicating that they are new 2–5 alleles (Donaldson et al. 2002). Analysis of third instar larval brains from homozygotes of the E(var) line E1060 revealed that mitotic chromosomes were substantially shorter and more compact than control chromosomes (Fig. 1). This overcondensed chromosome phenotype correlates well with the ‘tighter’ chromatin structure of genes silenced by PEV (Wallrath and Elgin 1995), which is likely further condensed by E(var) mutations.
Fig. 1.
A–D Metaphase figures from homozygous lethal mutants demonstrate effects on chromosome behavior and morphology. A Wild type. B, B’ Chromosome fusion phenotypes associated with line 1009. Note chromosome ‘trains’ (B) and ‘rings’ (B’). C, C’ Chromosome fusion phenotypes associated with line 1207. Arrows point to telomere fusions. D, D’ Chromosome overcondensation phenotype associated with line E1060. Bar represents 2 μm
Line 1126, which decreased J21A transmission to 12% and is homozygous lethal, displayed a normal mitotic index and no obvious cytological defects. This mutation may not affect the activity of the gene enough to cause abnormal behavior of full-length chromosomes, or the defect may be too subtle to observe in cytological analysis.
In summary, cytological examination of homozygous lethal lines that disrupted J21A transmission revealed that mutations in five out of six complementation groups caused visible chromosome defects represented by three different phenotypes: low mitotic index, telomere fusions, and overcondensed chromosomes. These results demonstrate that mutations identified through dominant effects on J21A transmission also have recessive effects on the inheritance of normal Drosophila chromosomes.
Discussion
Here, we have analyzed 59 Mod(var) mutations for dominant effects on chromosome inheritance, and discovered that 47% of the loci tested had at least one allele that significantly affected the transmission of the J21A ‘sensitized’ minichromosome. The demonstration that a particular locus or protein is responsible for the transmission phenotype must await cloning of the gene and phenotypic rescue by P element constructs. However, the general conclusion that Mod(var)s are highly enriched for genes involved in chromosome inheritance is sound. Strong support for this comes from the observation that only 2.6% of approximately 3000 randomly selected P element insertions affected J21A transmission (Dobie et al. 2001); thus, Mod(var) s are enriched, perhaps as much as 20-fold, for factors involved in inheritance. In addition to pulling out known inheritance genes, cytological characterization of the Dobie et al. mutants revealed that they had mitotic defects, which further substantiates J21A as a legimate tool for identifying chromosome inheritance factors. Finally, rescue experiments with the first of these loci to be cloned, Su(var)2–10/dPIAS, demonstrated that the dominant effect on J21A transmission and condensation defects map to that locus (Hari et al. 2001).
The 59 mutations investigated here correspond maximally to 49 loci, based on lethal complementation analysis (Donaldson et al. 2002). This is likely to be an overestimate of the number of loci, since many mutations are not homozygous lethal and cannot be definitively tested for complementation. On the other hand, we may be underestimating the proportion of Mod(var)s with effects on inheritance, since some mutations in inheritance genes may not reduce or alter protein function enough to observe dominant effects on J21A transmission. For example, the Su(var)3–9 protein (Schotta et al. 2002) is homologous to the histone H3 K9 methyltransferases that play key roles in heterochromatin structure and function (for review see Jenuwein and Allis 2001). Line 1699 contains a mutation in Su(var)3–9 and is the strongest Su(var) isolated in our screen (Donaldson et al. 2002), yet only reduced J21A transmission to 22%. It is likely that the absence of significant effects on J21A transmission result from redundancy in 3–9 function; 1699 and all other 3–9 alleles are homozygous viable. Nonetheless, our results demonstrate that proteins affecting heterochromatin function in Drosophila play a more important role in inheritance than previously appreciated.
Cytological analyses revealed that neuroblasts from homozygous mutant larvae displayed visible mitotic defects in five out of six Mod(var) complementation groups tested — specifically telomere fusions, overcondensation of full-length chromosomes and low mitotic indices. These results suggest that these Mod(var)s are bona fide chromosome inheritance mutations, and that their effects are not specific to J21A.
Several of the Mod(var) lines (234, 1097, 1227, 1545, 1552) had low mitotic indices, suggesting that the genes may affect cell cycle progression directly or indirectly. Defective chromosome inheritance may be the result of altered cell cycle timing or inactivation of a cell cycle checkpoint (for review see Clarke and Gimenez-Abian 2000). If mitosis is abbreviated, there may not be enough time for the functionally impaired J21A to assemble the required segregation machinery and attach securely to the spindle, although normal centromeres would be able to. In a similar fashion, if the spindle attachment checkpoint is defective (for review see Burke 2000), mitosis may not be delayed enough to allow time for the J21A to compensate for its partial inheritance defects. Further analysis is necessary to determine exactly how these mutations affect cell cycle progression or timing, and the roles that heterochromatin proteins play in these functions.
The telomere fusions observed in homozygotes of the Su(var)2–5 mutant alleles 1009 and 1207 could cause chromosome loss. Telomeres are thought to play a role in nuclear organization (Gasser 2001; Hari et al. 2001) and are important for chromosome inheritance in many organisms (Nimmo et al. 1998; Cooper 2000). Fusions of telomeres can result in dicentric chromosomes, which are broken during anaphase (McClintock 1938). No abnormal chromosomal fragments were seen in anaphases from 1009 and 1207 homozygotes, suggesting that these telomere fusions are weak enough to be resolved, as observed previously for other HP1 alleles (Fanti et al. 1998). However, the fact that the telomeres undergo end-to-end fusions indicates that the telomeres are not functioning properly, and that defects in telomere function may cause lethality through mechanisms other than chromosome breakage. For example, Su(var)2–10/dPIAS mutations affect telomere-telomere clustering and telomere-lamina interactions, which may be responsible for the undercondensation and gene expression phenotypes (Hari et al. 2001).
We also observed that some Mod(var)s affect general chromosome structure. The overcondensation of whole chromosomes observed in homozygous E1060 mutants is consistent with the enhancement of PEV seen in E1060 heterozygotes. Gene silencing of euchromatic genes placed in or near heterochromatin may be due to the genes adopting a ‘closed’ or compacted, heterochromatin-like structure, and E(var) loci may promote further such ‘heterochromatinization’. Su(var)2–10/dPIAS, which also affects J21A transmission (Hari et al. 2001 and Table 1), has the opposite effect: homozygous mutant neuroblasts display undercondensed chromosomes, which also supports a correlation between effects on gene expression and chromosome condensation. However, there was not a strict correlation between the direction of the effects on PEV and J21A transmission; both E(var)s and Su(var)s were observed to cause decreases and increases in J21A transmission. This is not surprising, since in most cases we do not know what protein product and processes are affected. In some cases, effects on gene expression could be regulated differently from effects on inheritance.
The results presented here demonstrate that modifiers of PEV are highly enriched for factors that affect the inheritance of a minichromosome. Cytological evidence along with previously published Mod(var) data suggest that inheritance of the normal complement of chromosomes is also compromised. This further bolsters the idea that these genes play a crucial role in a variety of inheritance functions in a metazoan, as previously demonstrated for S. pombe (Allshire et al. 1995; Bernard et al. 2001). It is quite likely that other Mod(var)s with effects on chromosome inheritance will disrupt other chromosomal processes, such as meiotic pairing (Dernburg et al. 1996; Karpen et al. 1996) and NOD-mediated antipoleward forces (Murphy and Karpen 1995a). Indeed, a study has recently been published demonstrating effects of Mod(var)s on meiotic recombination (Westphal and Reuter 2002). While some Mod(var) loci will likely encode protein components of heterochromatin, others will undoubtedly have indirect effects on heterochromatin function and chromosome transmission. For example, some Mod(var) mutations may alter the expression, function or localization of chromosome inheritance proteins. Still others may be involved in general aspects of nuclear organization and function critical for normal inheritance, as recently demonstrated for Su(var)2–10 (Hari et al. 2001). The unusually high percentage of Mod(var)s that affect inheritance intriguingly suggests that the primary function of heterochromatin is to regulate chromosome inheritance and nuclear organization by determining chromosome structure, and that PEV is a secondary consequence of the chromatin structures required to carry out these functions. Further characterization of these Mod(var)s will provide important insights into inheritance mechanisms that involve heterochromatin.
Acknowledgements
We would like to thank Cameron Kennedy and Beth Sullivan for technical assistance, Barbara Wakimoto, Gunther Reuter, and Joel Eissenberg for Su(var) stocks used in the initial pilot screen and M. Blower, K. Hari, K. Maggert, B. Sullivan, R. Truelove, and C. Yan for critical comments. This work contributed to partial fulfillment of the requirements for a doctorate of philosophy in Biology at the University of California San Diego for K.D. and was funded by NIH R01 GM 54549 to G.H.K., and by NIH Institutional National Research Service Award CA-09370 and an American Cancer Society Postdoctoral Fellowship to K.R.C.
References
- Afshar K, Barton NR, Hawley RS, Goldstein LS. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995;81:129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Axton JM, Dombradi V, Cohen PT, Glover DM. One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell. 1990;63:33–46. doi: 10.1016/0092-8674(90)90286-n. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Orr-Weaver TL. Holding chromatids together to ensure they go their separate ways. Bioessays. 1996;18:293–300. doi: 10.1002/bies.950180407. [DOI] [PubMed] [Google Scholar]
- Burke DJ. Complexity in the spindle checkpoint. Curr Opin Genet Dev. 2000;10:26–31. doi: 10.1016/s0959-437x(99)00040-4. [DOI] [PubMed] [Google Scholar]
- Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 1997;11:863–875. doi: 10.1101/gad.11.7.863. [DOI] [PubMed] [Google Scholar]
- Clarke DJ, Gimenez-Abian JF. Checkpoints controlling mitosis. Bioessays. 2000;22:351–363. doi: 10.1002/(SICI)1521-1878(200004)22:4<351::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Cook KR, Murphy TD, Nguyen TC, Karpen GH. Identification of trans-acting genes necessary for centromere function in Drosophila melanogaster using centromere-defective minichromosomes. Genetics. 1997;145:737–747. doi: 10.1093/genetics/145.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP. Telomere transitions in yeast: the end of the chromosome as we know it. Curr Opin Genet Dev. 2000;10:169–177. doi: 10.1016/s0959-437x(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Dej KJ, Orr-Weaver TL. Separation anxiety at the centromere. Trends Cell Biol. 2000;10:392–399. doi: 10.1016/s0962-8924(00)01821-3. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Dobie KW, Kennedy CD, Velasco VM, McGrath TL, Weko J, Patterson RW, Karpen GH. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics. 2001;157:1623–1637. doi: 10.1093/genetics/157.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Karpen GH. Trans-suppression of terminal deficiency-associated position effect variegation in a Drosophila minichromosome. Genetics. 1997;145:325–337. doi: 10.1093/genetics/145.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Lui A, Karpen GH. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics. 2002;160:995–1009. doi: 10.1093/genetics/160.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Szidonya J, Korge G, Sehnert M, Taubert H, Archoukieh E, Tschiersch B, Morawietz H, Wustmann G, Hoffmann G, et al. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics. 1993;133:279–290. doi: 10.1093/genetics/133.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, Pillus L. The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM. Positions of potential: nuclear organization and gene expression. Cell. 2001;104:639–642. doi: 10.1016/s0092-8674(01)00259-8. [DOI] [PubMed] [Google Scholar]
- Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 1994;44:371–379. doi: 10.1016/s0091-679x(08)60924-3. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2–10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. Heterochromatin. Chromosoma. 1999;108:1–9. doi: 10.1007/s004120050346. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Le MH, Duricka D, Karpen GH. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics. 1995;141:283–303. doi: 10.1093/genetics/141.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, Kotarski MA, Tartof KD. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Missouri Agric Exp Stn Bull. 1938;290:1–48. [Google Scholar]
- Muller HJ. Types of invisible variations induced by X-rays in Drosophila. Genetics. 1930;22:299–334. [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995a;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995b;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reuter G, Wolff I. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet. 1981;182:516–519. doi: 10.1007/BF00293947. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Ruddell AA, Brock JK, Clegg NJ, Lloyd VK, Grigliatti TA. A cytogenetic and genetic characterization of a group of closely linked second chromosome mutations that suppress position-effect variegation in Drosophila melanogaster. Genetics. 1992;130:333–344. doi: 10.1093/genetics/130.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford JB. Position-effect variegation in Drosophila. In: Ashburner M, editor. The genetics and biology of Drosophila. Academic Press; London: 1976. pp. 955–1018. [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen G. Molecular structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines DR, Henikoff S. Somatic instability of a Drosophila chromosome. Genetics. 1992;131:683–691. doi: 10.1093/genetics/131.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Szidonya J, Taubert H, Reuter G. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet. 1989;217:520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]