Abstract
Background
Dyskerin, which is an important component of the telomerase complex and is needed for normal telomerase activity, is frequently overexpressed in neoplasia. Dyskerin also plays an essential role in ribosome biogenesis. Since protein synthesis increases during tumorigenesis, this led us to hypothesize that dyskerin expression would be upregulated independently of the cell immortalization mechanism.
Methods
Dyskerin and telomerase reverse transcriptase (TERT) expression were examined in oral squamous cell carcinomas (OSCC) and patient-matched controls, and in a panel of telomerase-positive and telomerase-negative cells. Antisense inhibition of TERT was used to test the effects of downregulation of telomerase on dyskerin expression.
Results
Dyskerin was frequently overexpressed in OSCC and in immortalized and transformed keratinocytes relative to primary cells, independently of TERT and telomerase activity. Instead, dyskerin expression strongly correlated with cell proliferation rates.
Conclusions
The role of dyskerin in tumorigenesis does not correlate with its function within the telomerase complex.
Keywords: TERT, ALT, telomere, ribosome biogenesis, oral carcinogenesis
INTRODUCTION
Telomeres are specialized nucleoprotein structures that cap and protect the ends of eukaryotic chromosomes, thereby preserving genomic integrity.1 To extend cellular replicative lifespan, human tumors use either one of two mechanisms for telomere maintenance. The most common is mediated by the telomerase ribonucleoprotein (RNP) complex which replicates telomeres through the addition of a variable number of hexanucleotide TTAGGG repeats to the chromosome ends.2-4 The other mechanism is the telomerase-independent, alternative lengthening of telomeres (ALT) pathway which mediates telomere replication by homologous DNA recombination; the exact mechanisms remain poorly understood.5
Telomerase activity is undetectable in most somatic cells except for those that are highly regenerative and undergo constant self-renewal, including oral keratinocytes. However, upregulation of telomerase activity is common to most forms of neoplasia, including 80-90% of oral squamous cell carcinomas (OSCC); telomerase levels increase early in oral carcinogenesis.3 In contrast, only a small subset of tumors, and only rarely OSCC and other head and neck squamous cell carcinomas, use the ALT mechanism for telomere homeostasis.6
The telomerase RNP has two core components, including telomerase reverse transcriptase (TERT), which is the catalytic subunit, and telomerase RNA (TERC), which is the template used to prime telomere replication.4 But several additional factors are also required for full activity of the complex, including dyskerin (encoded by the DKC1 gene), which directly binds to and stabilizes TERC within the complex.2,7,8
Dyskerin is a highly conserved and vital, 58 kDa nucleolar RNA binding protein that is required for the biogenesis of a subtype of RNP.7,9 Together with three other conserved factors, dyskerin forms a core complex that binds non-coding H/ACA RNA, including TERC and subsets of small nucleolar RNA (snoRNA) and Cajal body RNA.9 There are over 100 known human H/ACA RNA, and the specific function of each H/ACA RNP is dependent on the RNA that it incorporates. Loss of dyskerin function reduces steady-state levels of TERC, decreases telomerase activity and leads to premature telomere shortening.7,8 However, dyskerin has also been shown to play important roles in ribosome biogenesis and function. Through binding to H/ACA snoRNA, dyskerin is required for post-transcriptional processing of precursor rRNA.10,11 A recent report suggests that dyskerin also regulates translation of a subset of mRNAs via an internal ribosome entry site.12 Thus, dyskerin is a key component of two molecular pathways that are fundamentally important to the tumor cell phenotype; neoplasms require not only a mechanism for telomere homeostasis, but also have a high demand for protein synthesis.4,13 To that end, DKC1 mRNA is frequently and significantly overexpressed in a variety of human cancers, including lymphoma, melanoma, neuroblastoma, and adenocarcinomas of the breast, colon and ovary.14-21
We and others previously demonstrated that human dyskerin expression is increased in experimental conditions that promote cell growth and proliferation.22-24 Gu et al25 recently reported that a mutant form of murine dyskerin significantly impaired normal cell proliferation by a mechanism that was dependent upon the presence of a functional telomerase complex. They observed that in the absence of either Tert or Terc, the dyskerin-mutant and wild-type cells grew similarly. However, it is unknown if dyskerin also requires an active telomerase complex in order to exert its influence in human cells.
In this study, we investigated the dependency of the interaction between dyskerin and telomerase in human cells and during tumorigenesis. TERT expression is commonly used as a surrogate marker for telomerase.26,27 However, we show for the first time that dyskerin expression is frequently overexpressed in sporadic OSCC, and that upregulation of dyskerin does not require the presence of TERT or an active telomerase complex. Instead, dyskerin expression correlates with active cell proliferation.
Methods and Materials
Cell culture
Primary oral keratinocytes OKF4, OKF6, and their respective TERT-immortalized derivatives, OKF4-TERT1F and OKF6-TERT2 were cultured in Keratinocyte-Serum Free Media (K-SFM; Invitrogen, Carlsbad, CA) supplemented with 25 μg/ml bovine pituitary extract, 0.2 ng/ml epidermal growth factor, and 0.4 mM CaCl2. OKF4 and OKF6 cells and their TERT-derivatives have been well-characterized.28 These cells originate from the normal oral epithelium of two distinct individuals. K-SFM is not designed to grow cells to high density. Thus, to enable us to expand the cultures to densities high enough for our assays, cells were plated in rapid growth media composed of a 1:½:½ mix of K-SFM: calcium-free, glutamine-free Dulbecco's minimal essential mediun (DMEM; Invitrogen): Ham's F-12 (Invitrogen), supplemented with 0.2 ng/ml epidermal growth factor, 25 μg/ml bovine pituitary extract, 0.75 mM L-glutamine, and 0.2 mM CaCl2. OKF6-Δp53D1 cells were grown using Defined K-SFM (Invitrogen) with a pre-formulated growth supplement supplied by the manufacturer. OKF6-Δp53D1 is a telomerase-negative, ALT-immortalized derivative of OKF6 and has been previously described.29 The OSCC cell line, UM-SCC-1, and U2OS human osteosarcoma cells (purchased from American Type Culture Collection, Manassas, VA) were grown in D-MEM containing high glucose (4,500 μg/mL), 1 mmol/L of glutamine, and 10% fetal bovine serum. All media formulations were supplemented with 100 IU/mL penicillin and 100 IU/mL streptomycin.
mRNA analysis and quantitation
Thirteen distinct, fresh frozen OSCC and patient-matched normal oral mucosal tissue samples were obtained from the University of Pennsylvania Oral, Head and Neck Tumor, Tissue and Saliva Bank with appropriate informed consent and institutional regulatory approval. Total RNA was extracted from 20 to 110 mg of tissue using TRIzol Reagent (Invitrogen, Carlsbad, CA), as per the manufacturer's recommendations. For RNA extraction from cell lines, we used the Rneasy technique (Qiagen, Valencia, CA). Quantitative RT-PCR was performed as previously outlined.22 Quantitect Primer Assays (Qiagen) for DKC1, TERT, β-actin and Tata binding protein (TBP) were used, respectively. Relative DKC1 and TERT expression levels, respectively, were normalized to TBP or actin using the 2−ΔΔCT method. For DKC1 and TERT quantitation from the tissue samples, the reactions were performed three times. For analysis of cellular mRNA expression, three independent samples were used.
Western blot analysis
Whole cell lysates were extracted as described previously.22 Polyclonal antibodies against dyskerin, β-actin, GAPDH and secondary antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). All Western blot analyses were performed at least three times. NIH Image J was used to quantitate relative dyskerin expression.
Cell proliferation and cell cycle distribution assays
Equivalent numbers of cells were seeded and cell proliferation was assessed using the Cell Proliferation Reagent WST-1 (Roche Applied Science, Indianapolis, IN). Absorbance was measured using a Multiskan Ascent microplate photometer (ThermoScientific Labsystems, Hudson, NH). The relative absorbances were measured for up to four days after plating of the cells. For cell cycle analysis, cells were washed and fixed for at least 60 min with cold 80% ethanol. The cells were then resuspended in 1 mL cold PBS and stained with 10 μg/ml propidium iodide containing 1 mg/ml RNase A (Sigma-Aldrich) for 30 min. Samples were analyzed on a Becton-Dickinson FacstarPLUS flow cytometer (BD Biosciences). A minimum of 5,000 events were collected on each sample; cell cycle analysis was performed using Modfit (Verity Software House; Topsham, ME).
TERT inhibition and telomerase repeat amplification protocol (TRAP) assay
A TERT antisense DNA oligonucleotide, 5′-TTGAAGGCCTTGCGGACGTG-3′ (TERT-ANTI) and a control, non-specific oligonucleotide, 5′-TAAGCTGTTCTATGTGTT-3′ (TERT-CTRL), were synthesized by Invitrogen and prepared as 1 mM stocks in sterile water. Both molecules were protected by phosphorothioate linkages at their terminal ends (underlined). The TERT-ANTI and TERT-CTRL sequences were previously described in studies by Kraemer et al.30
OKF6-TERT2 cells were seeded in 24 well plates and grown for two days. On the 3rd day, cells were transfected in quadruplicate with either 10 μM TERT-ANTI or TERT-CTRL using Lipofectamine 2000 (Invitrogen) as the carrier. Twenty-four hours after transfection, the cells were pelleted, washed with PBS, and resuspended in ice-cold CHAPS lysis buffer (0.5% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate [CHAPS], 0.1 mM benzamidine, 10% glycerol, 10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM EGTA, and 5 mM 2-mercaptoethanol). Total protein concentrations were measured using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). Five micrograms of each of the respective protein extracts were then used to assess telomerase activity levels using the TRAPeze RT Telomerase Detection Kit (Millipore, Billerica, MA) as per the manufacturer's recommendations. This highly sensitive assay makes use of fluorimetric probes which allow for real time quantification of telomerase activity. In a parallel set of experiments, the cells were similarly transfected with the TERT-ANTI and TERT-CTRL oligonucleotides, except the cells were harvested for total RNA. All transfection experiments were performed twice. Where indicated, OKF-TERT2 and Δp53D1 cells were harvested in exponential growth phase and assayed for telomerase activity. However, for illustration purposes, the TRAP PCR products were resolved on a pre-cast 10% Novex TBE polyacrylamide gel (Invitrogen).
Mutation screening
Genomic DNA was extracted from each of the thirteen OSCC samples, as previously described.31 Using previously published primer sets32 with some modifications (Supplemental Table 1), the complete translated and untranslated regions of the DKC1 gene were amplified. This included amplification of all 15 exons and associated intron splice sites. The reaction conditions were: 40-100ng of genomic DNA, 10pmol of each primer; 4-8mM MgCl2, 2mM of each dNTP, 0.5 unit of AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA) and the manufacturer's buffer in a 50μl final volume. Amplifications were carried out as follows: an initial denaturation for 10min at 95°C followed by 35 cycles of 94°C for 30s, 55–63°C for 30s, 72°C for 1min, and a final extension for 10min at 72°C. The annealing temperatures and MgCl2 concentrations were optimized for each specific primer set. All PCR products were analyzed by agarose gel electrophoresis, purified using either the QIAQuick PCR Purification Kit or QIAQuick Gel Extraction Kit (Qiagen), and then sequenced on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Sequencing was done in the sense and anti-sense directions. All sequence analyses were performed using Sequence Analysis 3.7 and Sequencher 4.0.5 software (Gene Codes Corporation, Ann Arbor, MI).
Correlation of DKC1 and TERT expression in human cell lines
Relative DKC1 and TERT mRNA expression levels were correlated in 93 unique, transformed human cell lines using the NCI60 on U133A dataset available from the open access Gene Expression Atlas (http://symatlas.gnf.org/SymAtlas).33 As we previously described22, we first log transformed the average relative mRNA expression levels for both genes. Then, we performed a linear regression analysis of the log transformed values, characterizing DKC1 as the dependent variable and TERT expression as the independent variable. As controls, we also examined the relationship between DKC1 and MYC, NHP2, MKI67 and CASP3. All statistical analyses were performed using Sigma Plot 11.0 (SYSTAT Software, San Jose, CA).
Results
Inhibition of TERT and telomerase activity does not alter dyskerin expression
Similar to murine cells, it is possible that in normal human cells that exhibit appreciable levels of telomerase activity, dyskerin expression may be intimately linked to TERT and telomerase. Moreover, TERT promotes and sustains epithelial proliferation not only by telomere maintenance, but also by a telomere-independent mechanism through transcriptional modulation of various growth promoting genes.34,35 A recent study showed that DKC1 mRNA expression was significantly increased in association with TERT overexpression in normal human mammary epithelial cells.26 This prompted us to determine if dyskerin expression was dependent upon TERT in immortalized oral keratinocytes.
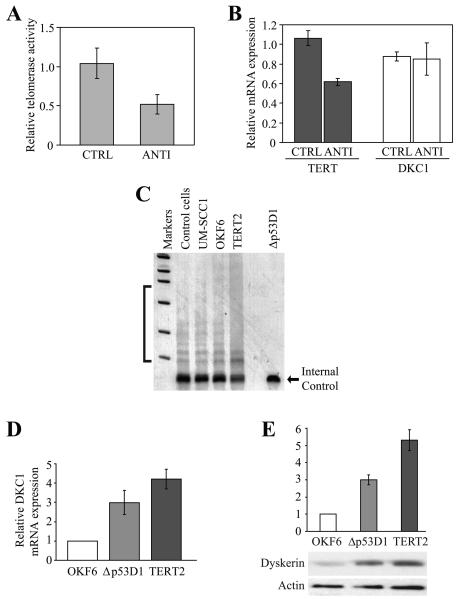
For these studies, TERT-immortalized human oral keratinocytes (OKF6-TERT2) cells were transfected with either 10 μM TERT antisense DNA oligonucleotide or a control, non-specific oligonucleotide. Twenty-four hours after transfection, the cells were harvested, and telomerase activity levels assayed by quantitative real-time RT-PCR. Due to technical difficulties, we were unable to reliably assess TERT expression by Western blot. However, while telomerase levels were decreased by almost 50%, and TERT mRNA expression reduced by more than 40% relative to the control cells, there was no effect on DKC1 mRNA expression (Figure 1A, B). Antisense inhibition of TERT in telomerase-positive H1299 lung adenocarcinoma cells also did not affect DKC1 expression (not shown). Thus, our findings reinforce those of a previous report which demonstrated no change in dyskerin expression following antisense inhibition of TERT and telomerase activity in oral cancer cells.2
Figure 1. Upregulation of dyskerin in immortalized oral keratinocytes is not dependent upon TERT.
A, OKF6-TERT2 cells were transfected with either 10 μM TERT antisense DNA oligonucleotide (ANTI) or a control, non-specific oligonucleotide (CTRL). Twenty-four hours later, relative telomerase activity levels were assessed by quantitative RT-PCR and normalized to telomerase levels in the untransfected OKF6-TERT2 cells. B, TERT / β-actin and DKC1 / β-actin expression was normalized to levels of the respective transcripts in the OKF6-TERT2 cells. The solid bars represent the mean values of four transfections conducted in one representative experiment; error bars denote the standard deviations. C, A qualitative TRAP assay was used to measure telomerase activity in exponentially-growing OKF6-TERT2 and OKF6-Δp53D1 cells. The characteristic laddering effect was observed in the OKF6-TERT2 and other telomerase-expressing cells, including UM-SCC1 oral cancer cells and a control cell pellet included with the kit, but not in the ALT-immortalized OKF6-Δp53D1 cells. The internal control served as a control for the PCR reaction. D, DKC1 mRNA expression was assessed by quantitative RT-PCR in OKF6, OKF6-Δp53D1 and OKF6-TERT2 cells. E, Dyskerin protein levels were quantitated relative to β-actin using NIH Image J.
Dyskerin is upregulated in immortalized keratinocytes independently of telomerase activity
Telomerase-positive OKF6-TERT2 and ALT-immortalized OKF6-Δ53D1 cells are derived from the same parental OKF6 primary oral keratinocyte strain.28,29 OKF6-Δ53D1 cells do not exhibit telomerase activity and basal TERT mRNA levels are decreased relative to the parental OKF6 keratinocytes (Figure 1C and not shown; see Ref. 29). However, quantitative RT-PCR revealed DKC1 mRNA expression to be elevated at least 3-fold in proliferating OKF6-Δ53D1 and more than 4-fold in OKF6-TERT2 cells relative to OKF6 cells (Figure 1D). Protein expression was also increased approximately 3-fold in the OKF6-Δ53D1 cells and at least 5-fold in OKF6-TERT2 relative to the primary cells (Figure 1E). We cannot exclude the possibility that TERT may at least partially contribute to regulation of DKC1 expression in human cells. Nonetheless, these data indicate that upregulation of dyskerin in immortalized oral keratinocytes is not dependent upon TERT expression or the presence of telomerase activity.
Dyskerin expression correlates with the rate of active cell proliferation
We and others previously demonstrated that human dyskerin expression is increased in experimental conditions that promote cell growth and proliferation.22-24 Since dyskerin contributes to other important cellular processes that are typically upregulated during active cell growth and proliferation9,11, we next asked if dyskerin expression correlates with the rate of cell proliferation. For this analysis, we examined two distinct, exponentially-growing primary oral keratinocyte strains, OKF4 and OKF6, their respective immortalized derivatives, OKF4-TERT-1F, OKF6-TERT2 and OKF6-Δ53D1, and telomerase-positive UM-SCC1 oral cancer cells. To our knowledge, there is no ALT-immortalized derivative of OKF4 cells, and we are not aware of any telomerase-negative oral cancer cell lines. Thus, we also included the ALT-immortalized U2OS human osteosarcoma cell line in the assay. U2OS cells do not express either TERT or TERC and do not exhibit any telomerase activity (not shown and see Ref. 36).
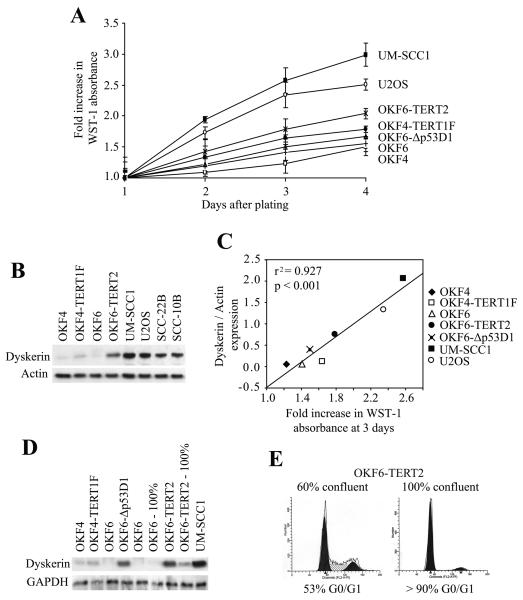
The immortalized cells proliferated more rapidly than their primary counterparts, while the transformed cells were the fastest growing (Figure 2A). Relative dyskerin expression strongly correlated with cell proliferation rates (r2 = 0.927, p < 0.001) insofar as there was low basal expression in the slow-growing primary keratinocytes, significantly increased levels in the immortalized cells and even more so in the transformed cell lines (Figures 2B-D). We note that dyskerin expression was greater in OKF6-Δ53D1 cells compared to the faster-growing OKF4-TERT1F cells (Figure 2D). Moreover, the observation that dyskerin expression was significantly decreased following contact inhibition of OKF6 and OKF6-TERT2 cells reinforces the notion the dyskerin expression is closely associated with active cell proliferation and not telomerase status (Figures 2D, E and not shown).
Figure 2. Dyskerin expression correlates with the rate of cell proliferation.
A, Fold increase in cell proliferation rates were determined by normalizing the relative WST-1 absorbance for each of the respective cell lines to those measured on the first day after plating. Error bars denote standard deviation from triplicate wells for each time point. Experiments were repeated twice with similar results. B, Cells were harvested in log growth phase (three days after initial plating) and total cell lysates were subjected to Western blot. Dyskerin was increased in the TERT-immortalized cells relative to their parental cells, and even more so in the transformed cells. For comparison purposes, dyskerin expression was also examined in the transformed squamous epithelial cell lines, SCC-10B and SCC-22B. C, Dyskerin expression was normalized to actin, and then compared to the fold increase in WST-1 absorbance three days after initial plating. A linear regression analysis showed strong correlation (r2 = 0.927, p < 0.001) between relative dyskerin levels and cell proliferation. D, Dyskerin expression was reduced in contact-inhibited OKF6 and OKF6-TERT2 cells (100%) relative to the exponentially growing cells (50%-60% confluent). Except where indicated, all protein lysates were obtained from actively proliferating cells. E, Cell cycle distribution of OKF6-TERT2 cells at 60% and 100% confluency. OKF6 cells showed a similar profile (not shown).
DKC1 is upregulated and not mutated in patient-derived OSCC
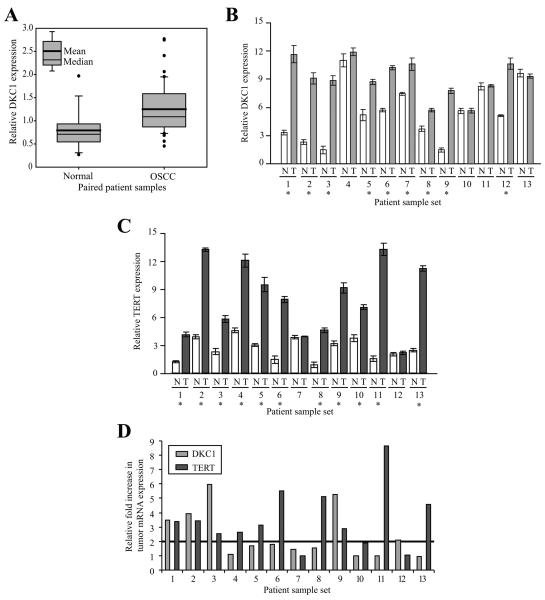
Our in vitro studies indicated that dyskerin expression was upregulated in actively proliferating oral keratinocytes, irrespective of the cell immortalization mechanism. However, while DKC1 mRNA is significantly overexpressed in several cancer types, its status in OSCC had not been previously established. Thus, we initially analyzed OSCC microarray data, a portion of which was previously published in a study aimed at identifying gene signatures that are predictive of regionally metastatic OSCC.31 In culling the dataset, we found that DKC1 mRNA expression was significantly upregulated (p < 0.023) in OSCC (N=49) relative to normal, patient-matched mucosal controls (N=16; Figure 3A).
Figure 3. DKC1 and TERT mRNA expression do not correlate in patient-derived OSCC.
A, Box plot illustrating DKC1 upregulation in OSCC (N=49) relative to matched normal controls (N=16; p<0.023). Data represent the average relative signals for DKC1 from two independent arrays.31 B, DKC1 mRNA levels were significantly (p<0.03) increased (*) in 9/13 OSCC (light grey bars) relative to patient-matched normal mucosal controls (white bars). C, TERT mRNA levels were significantly (p<0.001) increased (*) in 11/13 OSCC (dark grey bars) relative to the controls (white bars). D, There was no correlation between the relative fold increase in TERT and DKC1 mRNA expression in the tumors. Using an arbitrary threshold of a 2-fold increase in tumor mRNA expression relative to the matched normal control (horizontal line), 10/11 high TERT expressing tumors surpassed the threshold, while only 4/9 high DKC1 expressing tumors were increased beyond this threshold. DKC1 and TERT mRNA levels were measured relative to TBP) mRNA by quantitative RT-PCR using the 2−ΔΔCt method. N = normal, T = tumor.
We then validated DKC1 expression by quantitative RT-PCR in a subset of the tumors and controls. Of the samples tested, DKC1 mRNA levels were variably increased in 9/13 tumors (69%, p<0.03) relative to the patient-matched controls (Figure 3B); the transcript was ubiquitously expressed in all paired samples. Since our sample set was limited, we were unable to reliably correlate DKC1 expression with any demographic or prognostic variables.
Germline mutations in DKC1 give rise to the X-linked recessive form of dyskeratosis congenita.32,37 This is an unusual disorder associated with a wide-ranging phenotype, including cancer susceptibility; OSCC is among the most common cancers that these individuals develop.38 To our knowledge, none of the tumor samples were derived from individuals with this disorder; most of the patients had classic histories of long-standing smoking and/or alcohol intake. Nonetheless, we proceeded to analyze the thirteen tumors for possible DKC1 mutations. Using genomic DNA extracted from each of the tumors, we bi-directionally sequenced all fifteen DKC1 exons and their associated intron splice sites. In comparing the sequences of the amplicons to those of the known wild-type DKC1 exon sequences, mutations were not observed in any of the tumors (not shown). However, a synonymous single nucleotide polymorphism (SNP) was identified in one tumor. This SNP, Ex14+93G>A, L477L, was confirmed by sequence analysis of the patient-matched normal tissue sample. While a matched blood sample was not available for further confirmation, this SNP has been identified in healthy individuals and is not thought to be of any functional or clinical significance.37,39
Neither protein nor a sufficient amount of fixed tissues were available from any of the patient samples described above. Thus we used immunohistochemistry to evaluate dyskerin protein expression from a different series of OSCC and preneoplastic oral epithelial lesions obtained from formalin-fixed, paraffin-embedded oral biopsy samples. There was at least some evidence of dyskerin expression in all of the tissue samples tested (benign and malignant), and there was wide intra- and inter-specimen variability with respect to the intensity and extent of nucleolar staining (data not shown). Thus, differences in dyskerin expression between normal, dysplastic, and transformed epithelium could not be reliably quantitated. Similarly, other investigators were also unable to reliably compare dyskerin expression in tumor tissues relative to controls through use of immunohistochemical methods.17,20 This suggests that evaluation of dyskerin expression by immunohistochemistry may not be of any practical use.
DKC1 and TERT mRNA expression do not correlate in OSCC or in transformed cell lines
Telomerase levels are increased in the vast majority of human cancers, including OSCC.2,3 However, while TERT and DKC1 mRNA are significantly upregulated in many of the same cancer types, the interrelationship between DKC1 and TERT in tumor tissues is not known.
Using the same thirteen patient-derived OSCC samples and matched normal mucosal controls described above, we found that TERT mRNA was significantly increased in 11/13 tumors relative to their respective controls (p<0.001; Figure 3C). However, there was no correlation between the relative increase in tumor expression levels of TERT and DKC1 (r2 = 0.07, p=0.18; Figure 3D). When normalized to the expression of Tata binding protein (TBP) mRNA, there was also no correlation between the expression of TERT and DKC1 in the tumor tissues (r2 = 0.0089, p>>0.5; data not shown). TBP mRNA levels remained relatively constant in all the tissue samples tested.
Since our cohort was limited, we proceeded to evaluate TERT and DKC1 expression levels in a panel of 93 unique human cell lines derived from a variety of tumor types. To do this, we analyzed one of the gene microarray datasets (NCI60 on U133A, gcRMA) available from the open access Gene Expression Atlas.33 To our knowledge, at least five of the cell lines in this dataset are ALT-immortalized (Supplemental Table 2). These include U2OS, SAOS2 (osteosarcoma), Hs578T (breast adenocarcinoma), NCI H226 (non-small cell lung carcinoma) and IOSE80 (ovarian adenocarcinoma). The vast majority of the remaining lines are known to be telomerase-positive.
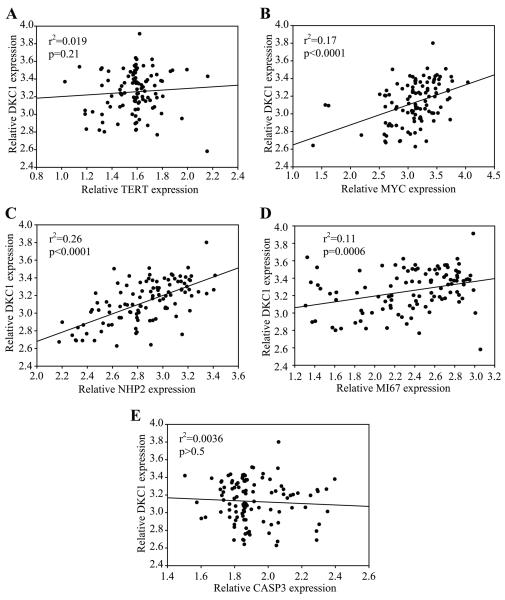
A linear regression analysis of the log-transformed expression values revealed no correlation (r2 = 0.0048, p>0.05) between TERT and DKC1 mRNA expression (Figure 4A). A similar result was obtained using the Human Gene Atlas GNF1H, gcRMA dataset which is derived from a collection of 78 normal and transformed human tissues and cell lines (not shown). As a positive control for the in silico analysis, we also examined the relationship between DKC1 and MYC expression. MYC promotes cell proliferation and oncogenesis, and its amplification is common to many different tumor types.22,23 As we have previously shown22, DKC1 and MYC expression showed significant correlation (r2=0.17, p<0.0001; Figure 4B). Similarly, the relative levels of DKC1 and its H/ACA RNP binding partner, NHP2/NOLA2, also strongly correlated (r2=0.26, p<0.0001; Figure 4C). There was also significant correlation between DKC1 and MKI67 (Ki-67) expression (r2=0.11, p=0.0006; Figure 4D). Ki-67 is required for normal ribosomal RNA synthesis and is widely used as a marker of cell proliferation; high tumor levels of Ki-67 may be associated with poor prognosis in some cancer types.40 There was no correlation between DKC1 and CASP3 expression (r2=0.0036, p>0.5; Figure 4E); CASP3 was randomly chosen as a factor that does not have any known relationship to DKC1. Together, these cumulative findings suggest that there is no overt relationship between DKC1 and TERT gene expression in human cancer. Instead, as suggested by our in vitro data, DKC1 expression correlates with other factors that are known to be upregulated in actively proliferating cells irrespective of telomerase status.
Figure 4. DKC1 and TERT mRNA expression do not correlate in transformed human cell lines.
A, A linear regression analysis of the log-transformed expression values revealed no correlation (r2 = 0.019, p=0.21) between TERT and DKC1 mRNA expression. B, DKC1 and MYC expression showed significant correlation (r2=0.17, p<0.0001). C, Relative levels of DKC1 and its H/ACA snoRNP binding partner, NHP2, also strongly correlated (r2=0.26, p<0.0001). D, DKC1 and the cell proliferation marker MKI67 also showed correlation (r2=0.11 p=0.0006). E, There was no correlation between DKC1 and CASP3 expression (r2=0.0036, p>0.5); CASP3 was randomly chosen as a factor that does not have any known relationship to DKC1. The raw data and corresponding log transformations for each of the genes are listed in Supplemental Table 2.
Discussion
The mechanisms by which DKC1 mRNA levels are increased in neoplasia as well as during active cell proliferation remain to be clarified. Dyskerin binds to NHP2 in H/ACA RNPs and to TERT within the telomerase complex.9,41 However, while DKC1 and NHP2 expression showed strong correlation in the NCI60 dataset, there was no relationship between DKC1 and TERT. We have previously shown that DKC1 is a direct and conserved transcriptional target of the MYC oncoprotein.22 MYC has also been shown to directly regulate the transcription of TERT41 and NHP2.43 Yet, while MYC and DKC1 expression strongly correlated, there was only weak correlation between MYC and TERT and MYC and NHP2, respectively (not shown). We note that the in silico analysis doesn't account for post-transcriptional or post-translational modifications that may influence the expression and/or function of either dyskerin or TERT. Nonetheless, our findings suggest that DKC1 and TERT mRNA may be differentially regulated in tumor cells, and likely by different factors. To that end, we have observed overexpression of DKC1 in the absence of any appreciable changes in MYC expression (unpublished data), and TERT is also known to be regulated by a variety of different factors.44
TERT expression is regarded as a reliable marker for telomerase. Although recent reports suggest TERC levels may be a rate-limiting determinant of telomerase activity under certain conditions, amplification of TERT is the primary limiting factor for telomerase activity.26,27,45 To that end, similar to the findings described in our current report, Cao et al26 demonstrated that DKC1 mRNA expression was upregulated 2- to 4-fold following TERT-immortalization of mammary epithelial cells, but there was no consistent correlation between DKC1 mRNA expression and either TERT expression or telomerase activity levels. While it remains possible that TERT may exert at least a partial influence on dyskerin expression in telomerase-positive cells, telomerase activity is not sufficient for immortalization of human oral keratinocytes or mammary epithelial cells; these cells also require loss of pRB/p16INK4a cell cycle regulatory control.28,46
Defects in the pRB/p16INK4a molecular pathway are common in oral and breast carcinogenesis28,46, and presumably are relevant in the pathogenesis of both telomerase-positive and telomerase-negative tumors. Under normal conditions, pRB binds to and sequesters the transcription factor E2F1 thereby preventing G1 to S phase transition. However, loss of pRB/p16INK4a regulatory control results in dysregulation of E2F1 leading to its transcriptional overactivity47; this could be a contributing factor in dyskerin upregulation in cancer. In support of this notion, a recent high-throughput microarray study suggests that E2F1 may directly regulate DKC1 expression.48 Moreover, analysis of the Gene Expression Atlas NCI60 on U133A dataset33 indicates that E2F1 and DKC1 expression do weakly correlate in transformed cells (not shown). Thus, future studies will be needed to determine if E2F1 and/or other components of the pRB/p16INK4a pathway regulate dyskerin.
As an individual variable, high DKC1 mRNA levels are associated with increased tumor aggressivity and poor prognosis in various cancer types.14-19 In addition, DKC1 expression is upregulated in high-grade lymphomas but downregulated relative to normal controls in indolent lymphomas.14,49,50 Moreover, a recent high-throughput study identified DKC1 as one of only seventy genes that, collectively, constitute a gene expression profile that strongly correlates with the development of aneuploidy and poor clinical prognosis.51 Although our sample cohort was too limited to assess statistical significance, further study is needed to determine if the same is true for OSCC. Moreover, more detailed studies will be needed to determine the exact mechanisms by which dyskerin contributes to tumorigenesis. Nonetheless, our cumulative findings indicate that dyskerin expression is not dependent upon TERT in either immortalized or transformed cells, or in patient-derived OSCCs. However, it is apparent that mutations, targeted inhibition, or low endogenous levels of dyskerin, respectively, do have negative impacts on normal function of the telomerase RNP, including in tumors.2,7,8 Yet, the effect is one of only partial functional abrogation. Conversely, Wong et al44 demonstrated that re-introduction of wild-type dyskerin into dyskerin-mutant cells did not revert telomerase activity to normal levels. Moreover, we have also observed that ectopic expression of dyskerin into cells with wild-type dyskerin does not confer additional increases in telomerase activity (unpublished data). Gu et al25 showed that murine dyskerin could contribute to telomere maintenance through a mechanism that was independent of telomere shortening. Thus, we cannot exclude the possibility that human dyskerin may also play a role in telomere homeostasis in TERT- and ALT-immortalized cells that can be uncoupled from the role of the protein within the telomerase complex. Alternatively, it is possible that dysregulation of dyskerin may also perturb other vital cellular functions.
Emerging evidence implicates aberrant ribosome biogenesis and function in neoplastic transformation and progression.13 Whether upregulation of dyskerin contributes to alterations in ribosome synthesis and/or activity is currently unknown. It has also been suggested that high levels of dyskerin may be broadly disruptive to the assembly of H/ACA snoRNPs.45 To that end, one of dyskerin's obligate binding partners, NHP2, is also upregulated in neoplasia; high levels of this factor are associated with poor prognosis in lung squamous cell carcinoma and colorectal adenocarcinoma.19,43 Like dyskerin, NHP2 is also essential for the assembly and maturation of H/ACA RNPs, and is needed for normal rRNA processing and telomere maintenance; NHP2 is also a telomerase component.9 We have shown that DKC1 and NHP2 mRNA expression strongly correlate in transformed cell lines (Figure 4C). Therefore, it is possible that dysregulation of H/ACA RNP biogenesis, irrespective of the mechanism, may be broadly disruptive to normal cellular homeostasis and that such alterations may contribute to tumorigenesis. This will require more detailed investigation.
Finally, it is possible that dyskerin may contribute to the regulation of other important molecular processes beyond those for which the protein has already been implicated in. To that end, we have recently shown that acute loss of dyskerin function impairs the accumulation of a specific subset of microRNAs.52 MicroRNAs are a class of small non-coding RNAs that directly regulate post-transcriptional gene expression.53 MicroRNAs regulate a wide array of cellular functions including cell proliferation; a number of microRNAs have been implicated in tumorigenesis.53 Thus, future studies will be needed to determine if dyskerin contributes to normal and tumor cell growth through regulation of miRNA-mediated post-transcriptional gene expression.
Supplementary Material
Acknowledgements
We are very grateful to Drs. James Rheinwald, Anil Rustgi, and Thomas Carey for providing us with the cell lines described here. This work was supported in part by NIH grants DE018416 (FA) and DE015856 (BZ), University of Pennsylvania Research Foundation, University of Pennsylvania's Abramson Cancer Center, and by the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analysis, interpretations or conclusions.
Abbreviations used
- ALT
Alternative lengthening of telomeres
- OSCC
oral squamous cell carcinoma
- TERC
telomerase RNA
- TERT
telomerase reverse transcriptase
- RNP
ribonucleoprotein
References
- 1.Artandi SE, Chang S, Lee SL, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 2.Chang JT, Chen YL, Yang HT, Chen CY, Cheng AJ. Differential regulation of telomerase activity by six telomerase subunits. Eur J Biochem. 2002;269:3442–3450. doi: 10.1046/j.1432-1033.2002.03025.x. [DOI] [PubMed] [Google Scholar]
- 3.Patel MM, Parekh LJ, Jha FP, et al. Clinical usefulness of telomerase activation and telomere length in head and neck cancer. Head Neck. 2002;24:1060–1067. doi: 10.1002/hed.10169. [DOI] [PubMed] [Google Scholar]
- 4.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 5.Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- 6.Reddel RR. An alternative lifestyle for immortalized oral keratinocytes. J Clin Invest. 2001;108:665–667. doi: 10.1172/JCI13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 8.Montanaro L, Calienni M, Ceccarelli C, et al. Relationship between dyskerin expression and telomerase activity in human breast cancer. Cell Oncol. 2008;30:483–490. doi: 10.3233/CLO-2008-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 11.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc Natl Acad Sci U S A. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 13.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 14.Piva R, Pellegrino E, Mattioli M, et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J Clin Invest. 2006;116:3171–3182. doi: 10.1172/JCI29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westermann F, Henrich KO, Wei JS, et al. High Skp2 expression characterizes high-risk neuroblastomas independent of MYCN status. Clin Cancer Res. 2007;13:4695–4703. doi: 10.1158/1078-0432.CCR-06-2818. [DOI] [PubMed] [Google Scholar]
- 16.Montanaro L, Brigotti M, Clohessy J, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- 17.Schaner ME, Ross DT, Ciaravino G, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sieron P, Hader C, Hatina J, et al. DKC1 overexpression associated with prostate cancer progression. Br J Cancer. 2009;101:1410–1416. doi: 10.1038/sj.bjc.6605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkowska A, Gumprecht J, Glogowska-Ligus J, et al. Expression profile of significant immortalization genes in colon cancer. Int J Mol Med. 2010;25:321–329. doi: 10.3892/ijmm_00000348. [DOI] [PubMed] [Google Scholar]
- 20.McDonald SL, Edington HD, Kirkwood JM, Becker D. Expression analysis of genes identified by molecular profiling of VGP melanomas and MGP melanoma-positive lymph nodes. Cancer Biol Ther. 2004;3:110–120. doi: 10.4161/cbt.3.1.662. [DOI] [PubMed] [Google Scholar]
- 21.Turano M, Angrisani A, De RM, Izzo P, Furia M. Real-time PCR quantification of human DKC1 expression in colorectal cancer. Acta Oncol. 2008;47:1598–1599. doi: 10.1080/02841860801898616. [DOI] [PubMed] [Google Scholar]
- 22.Alawi F, Lee MN. DKC1 is a direct and conserved transcriptional target of c-MYC. Biochem Biophys Res Commun. 2007;362:893–898. doi: 10.1016/j.bbrc.2007.08.071. [DOI] [PubMed] [Google Scholar]
- 23.Schuhmacher M, Kohlhuber F, Holzel M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaatinen T, Hemmoranta H, Hautaniemi S, et al. Global gene expression profile of human cord blood-derived CD133+ cells. Stem Cells. 2006;24:631–641. doi: 10.1634/stemcells.2005-0185. [DOI] [PubMed] [Google Scholar]
- 25.Gu BW, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci U S A. 2008;105:10173–10178. doi: 10.1073/pnas.0803559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Huschtscha LI, Nouwens AS, et al. Amplification of telomerase reverse transcriptase gene in human mammary epithelial cells with limiting telomerase RNA expression levels. Cancer Res. 2008;68:3115–3123. doi: 10.1158/0008-5472.CAN-07-6377. [DOI] [PubMed] [Google Scholar]
- 27.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 28.Dickson MA, Hahn WC, Ino Y, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opitz OG, Suliman Y, Hahn WC, Harada H, Blum HE, Rustgi AK. Cyclin D1 overexpression and p53 inactivation immortalize primary oral keratinocytes by a telomerase-independent mechanism. J Clin Invest. 2001;108:725–732. doi: 10.1172/JCI11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraemer K, Fuessel S, Schmidt U, et al. Antisense-mediated hTERT inhibition specifically reduces the growth of human bladder cancer cells. Clin Cancer Res. 2003;9:3794–800. [PubMed] [Google Scholar]
- 31.Ziober AF, Patel KR, Alawi F, et al. Identification of a gene signature for rapid screening of oral squamous cell carcinoma. Clin Cancer Res. 2006;12:5960–5971. doi: 10.1158/1078-0432.CCR-06-0535. [DOI] [PubMed] [Google Scholar]
- 32.Knight SW, Heiss NS, Vulliamy TJ, et al. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J, Southworth LK, Sarin KY, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 36.Tilman G, Loriot A, Van Beneden A, et al. Subtelomeric DNA hypomethylation is not required for telomeric sister chromatid exchanges in ALT cells. Oncogene. 2009;28:1682–1693. doi: 10.1038/onc.2009.23. [DOI] [PubMed] [Google Scholar]
- 37.Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 38.Knight S, Vulliamy T, Copplestone A, Gluckman E, Mason P, Dokal I. Dyskeratosis Congenita (DC) Registry: identification of new features of DC. Br J Haematol. 1998;103:990–996. doi: 10.1046/j.1365-2141.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 39.Packer BR, Yeager M, Burdett L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullwinkel J, Baron-Lühr B, Lüdemann A, Wohlenberg C, Gerdes J, Scholzen T. Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol. 2006;206:624–635. doi: 10.1002/jcp.20494. [DOI] [PubMed] [Google Scholar]
- 41.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu KJ, Grandori C, Amacker M, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 43.Wu CH, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW. Combined analysis of murine and human microarrays and ChIP analysis reveals genes associated with the ability of MYC to maintain tumorigenesis. PLoS Genet. 2008;4:e1000090. doi: 10.1371/journal.pgen.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 47.Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–11620. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- 48.Zeller KI, Zhao X, Lee CW, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 50.Poncet D, Belleville A, t'kint de Roodenbeke C, et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–2391. doi: 10.1182/blood-2007-09-111245. [DOI] [PubMed] [Google Scholar]
- 51.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 52.Alawi F, Lin P. Loss of dyskerin reduces the accumulation of a subset of H/ACA snoRNA-derived miRNA. Cell Cycle. doi: 10.4161/cc.9.12.11922. Manuscript in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.