Abstract
The conformational dynamics in the flaps of HIV-1 protease plays a crucial role in the mechanism of substrate binding. We develop a kinetic network model, constructed from detailed atomistic simulations, to determine the kinetic mechanisms of the conformational transitions in HIV-1 PR. To overcome the time scale limitation of conventional MD simulations, our method combines replica exchange molecular dynamics with transition path theory to study the diversity and temperature dependence of the pathways connecting functionally important states of the protease. At low temperature the large scale flap opening is dominated by a small number of paths; at elevated temperatures the transition occurs through many structurally heterogeneous routes. The expanded conformation in the crystal structure 1TW7 is found to closely mimic a key intermediate in the flap opening pathways at low temperature. We investigated the different transition mechanisms between the semi-open and closed forms. The calculated relaxation times reveal fast semi-open ↔ closed transitions and infrequently the flaps fully open. The ligand binding rate predicted from this kinetic model increases by 38-fold from 285 K to 309 K, which is in general agreement with experiments. To our knowledge, this is the first application of a network model constructed from atomistic simulations, together with transition path theory to analyze conformational changes between different functional states of a natively folded protein.
Introduction
HIV-1 protease is a major drug target in anti-viral treatment against AIDS.1,2 The efficacy of the existing active-site inhibitors is frequently reduced by the emergence of drug resistant mutations. Novel inhibition strategies against HIV-1 PR including targeting specific allosteric sites continue to be actively pursued.3–8 The active site of the protease is capped by two flexible β-hairpin flaps which control the substrate access to the binding cavity.3,9–11 An atomistic understanding of the conformational dynamics of HIV-1 PR flaps is important for elucidating the molecular basis of ligand binding and allosteric inhibition. Recently introduced protein kinetic network models offer a promising tool to explore the conformational dynamics of HIV-1 PR and other receptor drug targets.12–19
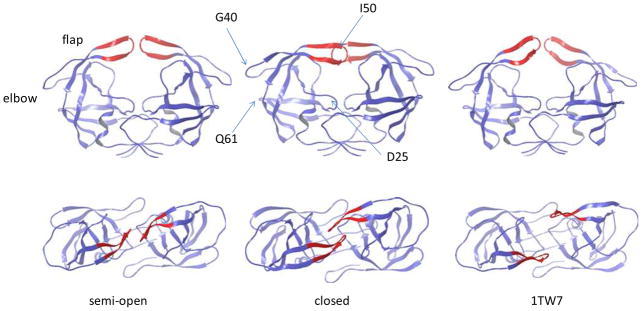
In HIV-1 PR, the β-hairpin flaps can exist in different conformational states (Fig. 1). Most apo structures adopt a semi-open conformation in which the flap tips shift away from the binding site. In ligand bound protein, the two flaps mostly form a closed conformation. The semi-open and closed forms show a characteristic difference in the relative orientation of the two hairpin flaps (the handedness, Fig. 1). Due to the intrinsic flexibility of the flaps, free enzyme can also exist in closed form,20 and conversely, ligand bound protease can also adopt the semi-open conformation.7 Although experimental structure containing fully open flaps has not been found, a partially open conformation has been reported for a multidrug-resistant mutant21 (PDB entry 1TW7, Fig. 1). However, the relevance of this structure in the conformational equilibrium of HIV-1 PR remains to be clarified, as subsequent MD simulations indicated that the 1TW7 structure is likely stabilized by crystal contacts.22,23 Solution NMR experiments suggest that the semi-open, closed and fully open conformations are in dynamic equilibrium on the microsecond timescale, while the flap tips exhibit fluctuations on the nanosecond timescale.24,25
Figure 1.
Three crystal structures of HIV-1 PR: the semi-open (PDB 1HHP), closed (PDB 1HVR, without ligand) and partially open (PDB 1TW7) conformations. Upper: side view. Lower: top view. The flap tips are shown in red.
Three simulation methods have been employed in the studies of HIV-1 PR dynamics: conventional MD,22,23,26–33 targeted MD/umbrella sampling,34,35 and coarse-grained simulations.10,36–38 Many insights into the dynamic equilibrium of the flaps have been obtained from these studies. In a pioneering early work35 using umbrella sampling, a transition path and free energy profile of the semi-open ↔ closed transition were revealed along a reaction coordinate determined by energy minimization. Reversible opening and closing of the flaps have been observed in recent unbiased MD simulations.29,32,33 Simulations with coarse grained potentials allowed numerous flap opening events to be observed, and provided estimates for the gated association rate constants.10,36–38 Despite this progress, our understanding of the flap dynamics and its impact on ligand binding remains incomplete. Several aspects regarding the conformational transitions in HIV-1 PR flaps need to be better understood, including descriptions of the flap opening pathways, the role of the curling of flap tips,30 the mechanisms of the semi-open ↔ closed and fully open transitions, and the relevance of the more expanded structure 1TW7.21 Although flap opening and closing were observed in long MD simulations, they do not give quantitative information about the transition rates or thermodynamic stabilities of the different states. Since only a very small number of the transition events can be observed by direct MD simulation, it is difficult to assess the statistical significance of the transition paths and mechanisms inferred from conventional MD simulations. While coarse-grained potentials greatly increased the time scale of simulation, simplified models may lack the atomic detail needed for understanding key features like modeling the flexibility of the β-hairpin structure of the flaps.3 Simulations using umbrella sampling require predefined reaction coordinates, which can be difficult to identify.
To address the unresolved questions about HIV-1 protease motions on longer time scales, we investigate its conformational dynamics using a new method based on a kinetic network model constructed from atomistic replica exchange simulations (REMD).14 The method allows pathways to emerge naturally from the statistical mechanical analysis of the network using the recently developed transition path theory (TPT).17,39,40 By combining the power of atomistic REMD simulation, TPT and stochastic simulations, we determine the full ensemble of transition paths and their fluxes in a statistically significant way. The calculated paths and associated fluxes give both qualitative and quantitative insights into the diversity and temperature dependence of the transitions among functionally important states of HIV-1 PR. The results presented here show how conformational changes can occur along a variety of different paths, rather than following a single route, which was the impression offered by conventional MD studies. Such information about the diversity of pathways is very difficult to obtain from conventional MD simulations. The statistical characterization of dynamics allowed us to answer questions such as: what is the probability for the semi-open ↔ closed transition to occur through fully-open states? How to quantify the role of curled flap tips in the flap opening pathway ensemble?
We also compute the potential of mean force (PMF) along principal component coordinates in order to provide a framework for visualizing the transitions in high dimensional conformational space. The results give new insights into the impact of flap dynamics on ligand binding. The present study demonstrated a new and powerful way to study conformational dynamics in complex biomolecular systems.
Theory and methods
Understanding the mechanisms by which transitions occur between functionally important states of proteins is one of the central goals of molecular biophysics.18 Although molecular dynamics simulation is well suited for studying conformational transitions, its utility has been limited by the long timescale of biomolecular transitions.41 Recently, a class of methods based on Markov state models (MSM, also called kinetic network models) has been developed for the calculation of transition paths. In this approach, the protein conformational space is discretized into a network of relevant substates. The dynamic transitions correspond to hopping between different nodes on the network, which can be modeled by a master equation assuming Markovian behavior.12–18,39,40,42–50 The network approach and its variants have been shown to produce plausible folding pathways for small proteins and peptides.12,14–16,19,47 Recently, Pietrucci et al.51 reported a novel application of a kinetic network model to simulate the binding of HIV-1 PR with a substrate ligand that illustrates the power of Markov state models. Their approach shares some important features with our present study, including the use of biased MD sampling schemes to sample state space in order to build the kinetic network, and the use of stochastic simulations on the constructed networks to study transition paths and mechanisms. While both studies used a kinetic network approach, the emphasis of their study is on the dynamics of protein-ligand binding; while our study is focused on the transitions among the functionally important states of the receptor in the absence of ligand. Also, the nature of the bias used to generate the transition paths is very different in the two approaches. While the metadynamics simulations employed in their work makes use of low dimensional predefined reaction coordinates,51 this is not needed with our biasing scheme which uses the temperature bias built into our replica exchange plus T-WHAM scheme to construct the nodes and their weights. Our method also facilitates the characterization of the temperature dependence of the transition paths.
Our approach to the construction of a kinetic network is based on replica exchange sampling (REMD) of states over a range of temperatures.12–14 Compared with MD simulations at a single temperature, our treatment has the advantage of including conformations in transition regions with high energy that are rarely visited at low temperatures. Sampling these transition regions using REMD greatly increases the likelihood of observing the full ensemble of transition pathways. The statistical weights of conformations sampled at different temperatures are reweighted at the target temperature using the T-WHAM method.52 The connectivity of the network is established by allowing transitions between structurally similar nodes,14,42 and the kinetic rates between connected nodes are assigned in a way which ensures that the detailed balance condition to preserve the equilibrium weights of all the states is satisfied.13,14 The network model thus constructed allows one to compute the reactive flux between any pair of nodes using a central result from transition path theory (TPT):15,17,39,40
| (1) |
Here Kji is the microscopic rate constant for transition from state i to state j, peq(i) is the equilibrium probability of state i and pfold is the committor probability. Using a recursive algorithm from TPT, the total flux obtained from Eq. (1) can be decomposed into unidirectional pathways ranked by their fluxes.17,39,40. Finally, the ensemble of these pathways is coarse-grained into a smaller number of essential (clustered) pathways to provide insights into the mechanism for the large structural rearrangement.
Stochastic simulations53 that satisfy the master equation of the network provide another powerful tool for probing the kinetics of the system that is complementary to TPT analysis.12,14 Because the simulations are carried out in discretized state space, they are much more efficient than conventional dynamics simulations, allowing numerous transition events to be observed for the calculation of fluxes and passage time distributions. Stochastic simulations on the network provide a means to directly test the validity of TPT for calculating fluxes. We have applied this approach to study the pathways for Trp-cage folding and obtained very good agreement between the TPT calculated pathways and kinetic MC stochastic trajectories.14 Here, we use this approach to characterize the conformational dynamics of HIV-1 PR. To our knowledge, this is the first study which applies a kinetic network model, TPT and stochastic simulations to analyze transitions among different states of a natively folded protein. Details about the REMD simulations, construction of the kinetic network, the TPT analysis and stochastic simulations are provided in the Supporting Information.
Results and Discussion
Conformational equilibrium and free energy surfaces
We first describe the thermodynamic states and free energy surfaces constructed using the data from REMD simulations with the OPLSAA forcefield54 in the AGBNP continuum solvent model.55 The snapshots from REMD were grouped into three macrostates: semi-open, closed and fully open. The semi-open and closed sets are defined by grouping together snapshots whose RMSD of the flaps are within 2.0 Å from the corresponding crystal structures (PDB entries 1HHP and 1HVR). The fully open states are defined as conformations with large flap-flap separations (I50Cα-I50’Cα distance ≥ 15 Å, see Fig. 2). The populations of the different states were calculated using T-WHAM (Table 1). At temperatures in the range 285 – 314 K, the semi-open (38.3% – 45.7%) and the closed forms (18.4% – 36.4%) are the two major components, and the fully open states (6 × 10−4 – 3.5%) account for a small fraction of the population. Although the fully open states are only transiently visited, they are functionally important since these are the only states that allow access for ligand binding. We note that a significant population (≥ 25.2%) is occupied by conformations other than the three aforementioned states (denoted as “other states” in Table 1). This is in agreement with previous long MD simulations in which the flaps were found to frequently visit conformations different from the three macrostates.29,32 These “other states” do not form major basins on the PMF. Instead, they consist of heterogeneous conformations lying between the three macrostates. The heterogeneity in the “other states” is largely responsible for the diverse pathways seen at elevated temperatures, as shown below.
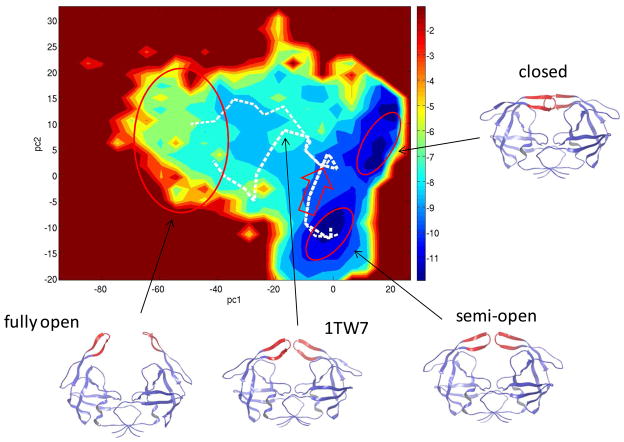
Figure 2.
The potential of mean force and the dominant pathways (white dotted lines) between the semi-open and fully open states projected onto the first two principal components PC1-PC2. The three macrostates are indicated by the red ovals. Representative structures are shown to illustrate the dominant motions associated with each principal component. The red arrow indicates the initial flap movements that change the flap handedness at the beginning of flap opening. T = 285 K.
Table 1.
Fractional populations of different conformational states from T-WHAM
| Temperature | Closed | Semi-open | Fully open | Other states |
|---|---|---|---|---|
| 285 K | 0.364 | 0.383 | 0.0006 | 0.252 |
| 314 K | 0.184 | 0.457 | 0.035 | 0.324 |
| 334 K | 0.080 | 0.326 | 0.0854 | 0.509 |
The calculated populations of different states are consistent with solution NMR data24,25 which suggest that the three conformations are in dynamic equilibrium, with the semi-open form being the most populated species. Based on the results of Table 1, the free energies of different states are estimated and the thermodynamic parameters are determined using and ΔG = ΔH − TΔS. For the semi-open → closed transition at T = 314 K, we found ΔG ≈ 0.6 kcal/mol, −TΔS ≈ 5.8 kcal/mol and 3H ≈ −5.2 kcal/mol. Since HIV-1 PR generally undergoes semi-open → closed transition upon ligand binding, ΔG(semi-open → closed) equals the protein reorganization contribution to binding free energy. Our calculation shows that this contribution is small compared with the typical binding free energies of ~ −10 kcal/mol for most HIV-1 PR inhibitors.56
Next, we examine the conformational fluctuations in the flaps region projected onto the PMF defined by the first few principal components. The PMF provides a convenient way for viewing the pathways between different states. Although pathways are computed without projection, the features of the PMF can be used as an aid to visualize pathways. We define a reaction zone consisting of flap residues 45–55 and the peripheral residues 79–82. The PMF is calculated using the principal component analysis (PCA) of 84 distances between nonadjacent Cα atoms of the reaction zone residues. This is based on the fact that the protein fluctuations are well represented by the flaps and peripheral residues. The PCA calculation indicates that the first six principal components account for more than 80 % of the fluctuations.
The PMF projected onto the first two principal components (PC) clearly separates the three macrostates: see Fig. 2. The first principal component (PC1) is dominated by flap separation: the fully open structures have highly negative PC1 < −40; the semi-open and closed structures show zero and positive values of PC1, respectively. The second principal component PC2 correlates with the relative lateral orientation of the flaps, and thus serves as an order parameter for distinguishing the closed and semi-open states.
Pathways between the semi-open and fully open states
To use TPT to compute flap opening pathways, we treat the semi-open and fully open states as the reactant and product, respectively. The remaining states are considered as the intermediate states. To study the temperature dependence of pathways, we perform the calculations at T = 285 K and 334 K.
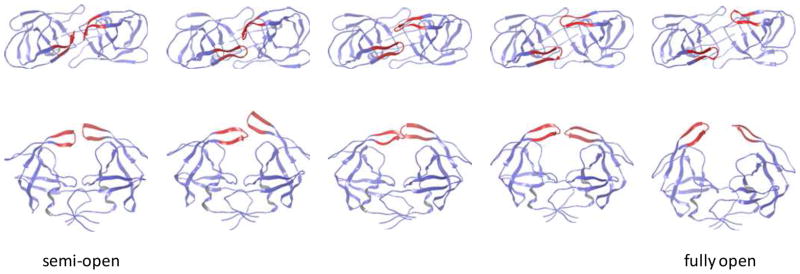
Fig. 2 shows the clustered pathways for flap opening at T = 285 K projected onto the PC1-PC2 PMF. A clustered pathway contains contributions from many similar unclustered pathways, which typically contain between 12 and 25 intermediate network nodes. The nodes along a pathway are ordered sequentially according to their Pfold values, which increase monotonically from Pfold = 0 (semi-open) to Pfold = 1 (fully open). The transition at T = 285 K is dominated by the top two clustered pathways which carry 42% and 30% of the total flux (Fig. 2). The remaining fluxes are distributed among a large number of smaller clustered pathways, which are omitted from Fig. 2. Starting from the semi-open form, the two dominant pathways initially follow the same route and only diverge during the second half of the transition. Fig. 3 shows the intermediate structures along the top pathway. The β-hairpin structure of the flaps and the intraflap hydrogen bonding are not disrupted in these pathways. Based on the intermediate structures and their projections onto the PMF, we find that the pathways go through two sequential phases: in the first phase, the flap tips change their handedness from the semi-open form to the orientation of the closed form. The binding pocket remains mostly closed during this initial phase. The handedness change is indicated in Fig. 2 by a red arrow. In the second phase, the two flaps begin to separate, resulting in exposure of the binding pocket. The flap separation occurs through a combination of upward rotations and lateral movements of the flaps. The two major pathways differ in the second phase: while both involve upward flap rotations, the second pathway shows larger lateral movements in the flap tips. We note that the non-flap residues experience little variation during the flap opening process: the RMSD of these residues remain below 2.1 Å from the semi-open structure; in contrast the flap residues move apart by > 14 Å. The large-scale movement in the non-flap region is not required for flap opening at 285 K.
Figure 3.
The intermediate conformations in the most dominant pathway for the semi-open → fully-open transition. T = 285 K. Upper: top view. Lower: side view.
With the availability of a large number of HIV-1 PR structures from X-ray and NMR, we examine whether the transition pathways involve known experimental structures. The clustering of 327 experimental structures reveals four conformational clusters.57 Two of them correspond to the semi-open and closed states; the other two include more expanded flaps. One of the more expanded clusters is represented by the crystal structure 1TW7 (Fig. 1),21 which is well sampled in the REMD simulations. More importantly, we found that the 1TW7 structure is visited by the dominant pathways from the semi-open to fully open states. This can be seen from Fig. S1 (Supporting Information) where 1TW7 is superimposed onto an intermediate node in the second dominant pathway. With a RMSD ≈ 1.7 Å, the two superimposed structures show the same flap handedness and a similarly expanded binding cavity. Superimposition of the 1TW7 and the structures along the top two pathways shows that the RMSD drops to below 2 Å in the middle of the flap opening. Note that this observation is obtained without the knowledge a priori of the experimental structure. The result suggests that, although 1TW7 has been shown to be only stable in a crystal environment,22,23 the structure actually represents an important on-path intermediate along the room-temperature pathways for large-scale flap opening.
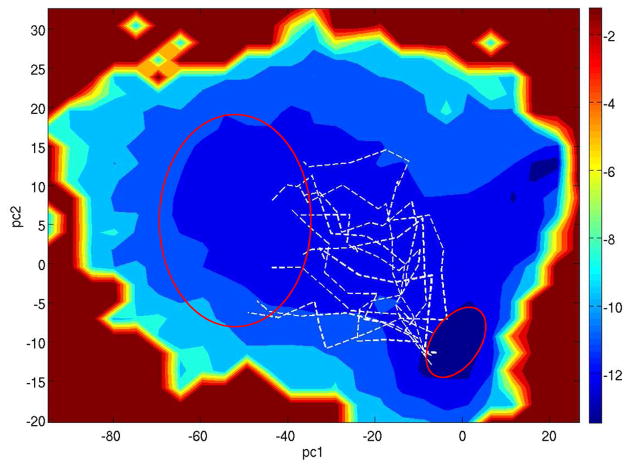
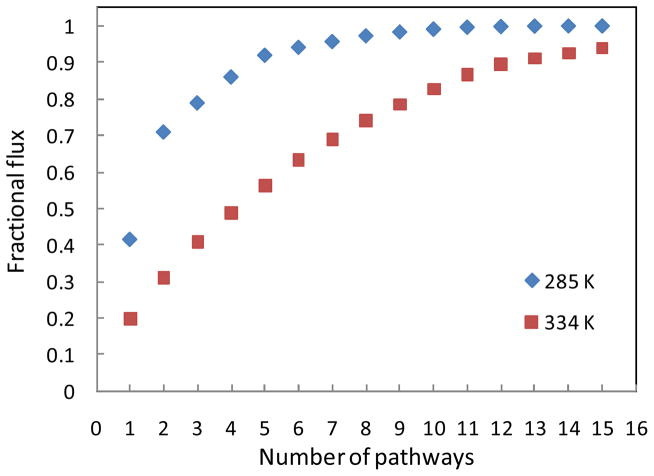
We now discuss the results of flap opening at T = 334 K. At this temperature, the PMF appears flatter compared with that at the lower temperature; many pathways are now accessible for flap opening: see Fig. 4. These paths follow structurally different routes: on average the Cα RMSD between two nodes located in the middle of two pathways is ≈ 3.7 Å. The greater diversity of the paths at T = 334 K is also apparent from Fig. 5, which shows the cumulative flux as a function of number of pathways. While at 285 K about 72% of the flux is carried by the top two pathways, at 334 K it requires 8 pathways to carry the same fraction of the flux. In contrast to the low temperature results shown in Fig. 2, here only a small number of the pathways (1.3% flux) go through the handedness change prior to flap separation. There exist pathways (4.5% flux) in which the fully open state is reached via upward flap rotations only, without substantial lateral movements of the flaps. Furthermore, many pathways visit intermediate structures with L-bent flap tips. Fig. S2 illustrates the difference between a bent flap tip and an uncurled one. At T = 285 K, a small fraction of the pathways (~10% of total flux) show flap bending; at T = 334 K over 50% of the flux come from paths containing bent flap tips. The curling of the flap tips was previously proposed to trigger large scale flap opening.30 The present results show that the flap curling primarily affects the high temperature pathways for flap opening.
Figure 4.
The pathways representing 83% of the total flux for the semi-open → fully-open transition projected onto the PMF of PC1-PC2. T = 334 K.
Figure 5.
The cumulative fluxes as a function of the number of pathways for the semi-open → fully-open transition. Blue: T = 285 K; Red: T = 334 K.
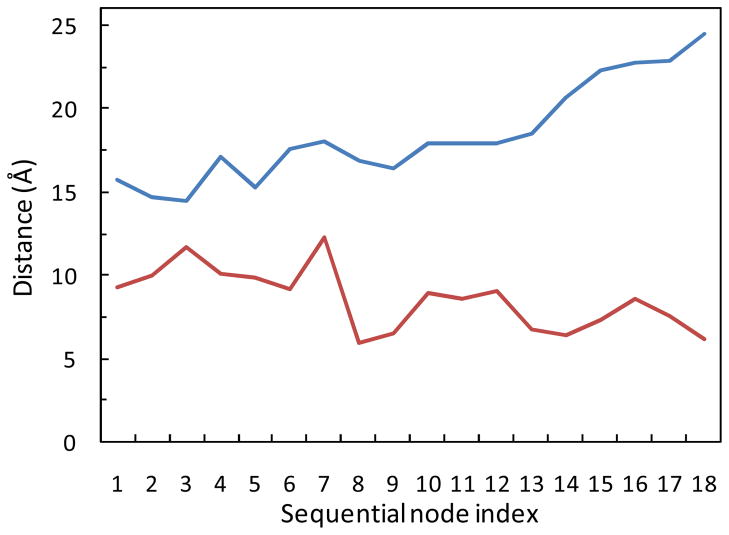
The computed flap opening pathways provide clarification for a potential allosteric site near the elbow region, located between residues Gly40 and Gln61 (see Fig. 1). The possibility for this region to serve as an allosteric binding site for the control of flap opening/closing has been suggested based on previous MD simulations.26,58 Perryman et al. observed anticorrelated motion between compression of the two elbows and increased flap tip motions in a 22-ns MD simulation.26 However, the flaps did not open in these simulations, even with the elbows compressed by using simulation restraints.58 Here, we investigate the possible correlation between the elbows and flap regions without restraining the elbows. The extent of flap opening (measured by the Ile50-Asp25 distance) and the size of the elbow region (defined by the Gly40-Gln61 distance) are monitored along the dominant flap opening pathways. Fig. 6 illustrates their anticorrelation seen in the top pathway at T = 285 K. Starting from the semi-open state, the Ile50-Asp25 distance increases from ≈ 15 Å to the fully open values of ≈ 24.5 Å as the Gly40-Gln61 distance decreases from ≈ 10 Å to ≈ 6 Å during the transition. Note that the extent of the flap opening is much larger than the flap motions observed in the previous MD simulations.26,58 The pathways at the higher temperatures show the same trend. The strong anticorrelation revealed by the dominant flap opening pathway provides more clear-cut evidence for the possibility of targeting the elbow region to allosterically interfere with flap opening/closing.
Figure 6.
The anticorrelation between the flap opening (I50-D25 distance, blue curve) and the size of elbow region (G40-Q61 distance, red curve) along the top pathway for the full opening of the flaps. T = 285 K.
Pathways between the semi-open and closed states
The semi-open and closed forms show a striking difference in the relative flap orientation, the positional difference of the flap tips can be as large as 7 Å (Fig. 1). The most direct path to reverse the flap handedness as occurs in the semi-open ↔ closed transition is via a lateral flap rotation in the plane of the lower graph of Fig. 1, but this route is blocked due to the clash of the main chain and side chain atoms involving flap tip residues G49, I50, G51 and G52. Using the kinetic network model and TPT, we determined the physical pathways leading to the reversal of the flap handedness at three temperatures: T = 285 K, 314 K and 334 K.
At the lowest temperature T = 285 K, the transitions are dominated by a single pathway carrying ≈ 92% of the total flux (Table 2). The conformations along the pathway show that the semi-open ↔ closed transitions involve relatively compact structures (Fig. S3), in which the β-hairpin structure of the flaps and the intraflap hydrogen bonds are intact, and the binding pocket remains covered throughout the rearrangement. During the early stage of the semi-open → closed transition, one flap tip moves up above the other (Fig. S3). The unsymmetrical upward movement creates space for the rearrangement of flap handedness. The observed sequential events leading to the handedness reversal is in good agreement with a recent 300 ns MD simulation in explicit solvent.32
Table 2.
Fractional fluxes of the top pathways for the semi-open ↔ closed transitions at different temperatures
| Pathway index | 285 K | 314 K | 334 K |
|---|---|---|---|
| 1 | 0.92 | 0.32 | 0.28 |
| 2 | - | 0.15 | 0.10 |
| 3 | - | 0.12 | 0.08 |
| 4 | - | 0.08 | 0.07 |
| 5 | - | 0.07 | 0.07 |
At the two higher temperatures T = 314 K and 334 K, the semi-open → closed transition pathways become significantly more diverse. The fluxes carried by the top five pathways at the three temperatures are shown in Table 2. The largest flux pathways at the two higher temperatures carry ≈ 32% and ≈ 28% of the total flux, which are much smaller than the 92% carried by the single dominant pathway at T = 285 K. Many higher temperature pathways between the semi-open and closed states contain conformations with bent-L flap tips. At higher temperatures, some pathways visit the fully open states in the middle of the transition, i.e. pathways which follow the routes: semi-open ↔ fully open ↔ closed. These paths account for about 10% of the total flux. The bent flaps and the open flaps provide two possible ways to create space for the rearrangement of the flap handedness. These conformations are higher in energy, due to the structural strain and loss of favorable interflap interactions; therefore they are seen only in the pathways at higher temperatures.
Stochastic simulations of the conformational transitions
We performed stochastic simulations on the network to more directly explore the kinetics of the semi-open → fully-open and semi-open → closed transitions, and to test the validity of the pathways calculated using transition path theory. Starting from randomly chosen nodes, 1000 reactive trajectories were collected from long equilibrium simulations, with each trajectory containing a single transition event. The total flux and mean first passage time (MFPT) obtained from these trajectories are shown in Table 3 and Table 4. The results are in good agreement with those calculated using TPT. The distributions of the first passage times can be fitted with single exponential functions. For the semi-open → fully-open transition with a MFPT of 375 ns, it would require hundreds of microseconds of conventional MD simulation to obtain comparable statistics provided by the 1000 reactive trajectories from stochastic simulations on the network.
Table 3.
Flux and relaxation time for semi-open → closed transition (T = 285 K)
| Net flux J (s−1) | Relaxation time τ (ns) | |
|---|---|---|
| Transition Path Theorya | 1.51×107 | 33.3 |
| Stochastic Simulationb | 1.52×107 | 29.1 |
In TPT, J is computed using Eq. (6) in the Supporting Information (SI), and τ is the inverse of the rate constant k calculated from Eq. (7) in SI.
In stochastic simulation, both J and τ are derived from 1000 reactive trajectories. J is obtained as the number of transition events divided by the total simulation time. τ is calculated from the mean first passage time for these trajectories.
Table 4.
Flux and relaxation time for semi-open → fully-open transition (T = 285 K)
| Net flux J (s−1) | Relaxation time τ (ns) | |
|---|---|---|
| Transition Path Theorya | 2.63×106 | 375 |
| Stochastic Simulationb | 2.63×106 | 345 |
In TPT, J is computed using Eq. (6) in the Supporting Information (SI), and τ is the inverse of the rate constant k calculated from Eq. (7) in SI.
In stochastic simulation, both J and τ are derived from 1000 reactive trajectories. J is obtained as the number of transition events divided by the total simulation time. τ is calculated from the mean first passage time for these trajectories.
We projected the stochastic trajectories onto the TPT pathways using the procedure described in the method section (SI). The percentage of the trajectories assigned to a pathway gives the fractional flux through that pathway. This allows us to compare the flux distributions along the pathways obtained from the TPT calculation and those from the stochastic simulations. The results for the semi-open → fully open and semi-open → closed transitions at different temperatures are shown in Fig. S4 and Fig. S5. There is a good correlation between the fluxes calculated from the two methods. The results from stochastic simulations therefore provide direct validation of the TPT calculation of fluxes and pathways.
In general, stochastic simulations not only serve to confirm the TPT results, but they can also provide additional kinetic information of the system under study; the two methods are complementary. For example, stochastic simulations provide passage time distributions and rate constants associated with each pathway, which are not directly available from TPT calculation. On the other hand, TPT analysis provides a natural framework for analyzing pathways ranked by flux from largest to smallest.
Implications for ligand binding kinetics
We analyze the implications of the conformational dynamics for the kinetics of ligand binding. The calculated relaxation times of the transitions between the three macrostates are given in Table 3 and Table 4. At T = 285 K, the relaxation time for the semi-open → closed transition was found to be τsc ≈ 33 ns. For the semi-open → fully-open transition, the relaxation time τso ≈ 375 ns. The results reveal a kinetic picture with fast internal rearrangements and infrequent openings. This is qualitatively consistent with the flap dynamics observed in solution NMR studies, which revealed motions on two time scales: a sub-nsec to nsec motion of flap tips and a μsec or longer time fluctuation attributed to flap opening.24,25 The time scales calculated in the present study are also roughly comparable to the results of recent MD simulations of length ranging from 40 ns to 300 ns at room temperature.29,32
For large ligands of HIV-1 PR, the two flaps serve as a gate that controls the accessibility of the binding site.10 On the basis of the flap opening relaxation time τso, we can estimate the magnitude and temperature dependence of ligand binding rate kon. In gated binding, the binding rate kon is governed by the ratio of the gate opening relaxation time τso and the characteristic diffusion relaxation time τd = R2/D.59,60 Here R is approximately the radius of the reactive surface area for binding. D is the ligand diffusion coefficient given by the Stokes-Einstein relation D = kBT/6πηr. Assuming R = 10 Å and a ligand radius r = 8 Å yields τd ≈ 4 ns, which is small compared with the relaxation time τso of 375 ns for gate opening. According to rate theories for gated ligand binding,59,60 in the limit τd < τso, the binding rate is the ungated rate ku multiplied by the fraction of the time the gate is open, i.e. kon = kuPopen. Popen is obtained from the equilibrium population of the fully open states. The ungated binding rate ku may be estimated by the diffusion controlled rate for reactive particles, which yields ku ≈ 4πRD ≈ 3×109 M−1s−1. At 298 K, the Popen is found to be ≈ 5×10−3. The binding rate is thus estimated to be kon = kuPopen ≈ 1.5 × 107 M−1s−1. Experimentally, kon for seven HIV-1 PR drugs are between 5.8×105 and 6.6×106 M−1s−1 at 25 °C.61 The estimated kon is within an order of magnitude of the experimental values. Here, we note that there is some arbitrariness in defining the fully open states as including conformations with I50Cα-I50’Cα distance ≥ 15 Å. Using a threshold of 13 Å would change the fully open population to Popen ≈ 1.06×10−2 from Popen ≈ 5×10−3. However, the distance threshold of 15 Å that we used is more consistent with the fact that many of the HIV-1 PR ligands have a dimension of ≥ 15 Å. Our estimate also assumes that the free energy barrier for the reaction process within the binding cavity is not rate limiting. We emphasize that our estimation only provides a semi-quantitative account of kon because of the assumptions summarized above.
Many HIV-1 PR ligands show strong temperature dependence in kon, which can increase 20-fold within the temperature range from 278 K to 308 K.61 This correlates with the calculated changes in the fully open population Popen, which increases by 38-fold from 285 K to 309 K. From the result for gated binding kon = kuPopen, the strong temperature dependence of kon is qualitatively reproduced. These analyses appear to suggest that the gating due to large flap opening is an important factor in the binding kinetics of HIV-1 PR ligands.
Conclusion
We have used a kinetic network model constructed from detailed atomistic simulations to compute pathways between different native forms of HIV-1 protease and determined conformational equilibrium and key features of kinetics without specifying the reaction coordinates in advance. The large-scale flap opening occurs through a combination of lateral flap movements and upward flap rotations. The mutual rotation of the dimer is not required for the transition. The crystal structure 1TW7 containing a more expanded cavity was shown to represent an important intermediate in the full opening of the flaps. The computed pathways reveal a strong correlation between the compression of the elbow region and the flap separation, providing new evidence that the elbow region may be useful for allosteric control of the flap opening and closing. The transitions were found to be temperature dependent: at low temperature, a small number of pathways dominate the transition, while at elevated temperatures the transition occurs through many parallel routes.
The calculated relaxation times between the three macrostates reveal a kinetic picture with relatively fast semi-open ↔ closed conversion and less frequent flap opening. The simulation and analysis provide reasonable estimates for the magnitude of the ligand binding rate and its temperature dependence, which appear to suggest that the gating due to flap opening plays an important role in the ligand binding kinetics.
The TPT analysis and stochastic simulations of the semi-open ↔ closed transition show that in the low temperature pathway, the two flaps remain in contact with each other, and the flap tips undergo local adjustments to change the handedness. More open conformations and curled flap tips are seen in the higher temperature pathways.
The results presented here demonstrate that the kinetic network approach of combining replica exchange molecular dynamics with TPT analysis provides a promising way to overcome the time scale limitation of conventional MD simulations for probing complex transition pathways and for obtaining atomistic insights into transitions among different conformational states of native proteins.
Supplementary Material
Acknowledgments
We thank Kristina Paris for providing the results of clustering experimental structures of HIV-1 PR. This work has been supported by a grant from the National Institute of Health (GM30580).
Footnotes
Supporting Information Available
Contains the details of the methodology, and more figures about the transition pathways in HIV-1 protease.
References
- 1.Emini E, Schleif W, Davis L, Heimbach J, Dixon R, Scolnick E, Sigal I, Kohl N. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner BG, Summers MF. Journal of Molecular Biology. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 3.Hornak V, Simmerling C. Drug Discovery Today. 2007;12:132–138. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee GM, Craik CS. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perryman AL, Zhang Q, Soutter HH, Rosenfeld R, McRee DE, Olson AJ, Elder JE, David Stout C. Chem Biol Drug Des. 2010;75:257–268. doi: 10.1111/j.1747-0285.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chellappan S, Kiran Kumar Reddy GS, Ali A, Nalam MNL, Anjum SG, Cao H, Kairys V, Fernandes MX, Altman MD, Tidor B, Rana TM, Schiffer CA, Gilson MK. Chem Biol Drug Design. 2007;69:298–313. doi: 10.1111/j.1747-0285.2007.00514.x. [DOI] [PubMed] [Google Scholar]
- 7.Böttcher J, Blum A, Dörr S, Heine A, Diederich WE, Klebe G. ChemMedChem. 2008;3:1337–1344. doi: 10.1002/cmdc.200800113. [DOI] [PubMed] [Google Scholar]
- 8.Damm KL, Ung P, Quintero JJ, Gestwicki JE, Carlson HA. Biopolymers. 2008;89:643–652. doi: 10.1002/bip.20993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markgren P-O, Schaal W, Hämäläinen M, Karlén A, Hallberg A, Samuelsson B, Danielson UH. J Med Chem. 2002;45:5430–5439. doi: 10.1021/jm0208370. [DOI] [PubMed] [Google Scholar]
- 10.Trylska J, Tozzini V, Chang C, McCammon JA. Biophysical Journal. 2007;92:4179–4187. doi: 10.1529/biophysj.106.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Liu MS, Ji B, Hwang K, Huang Y. J Chem Phys. 2009;130:215102. doi: 10.1063/1.3148022. [DOI] [PubMed] [Google Scholar]
- 12.Andrec M, Gallicchio E, Felts AK, Levy RM. Proceedings of the National Academy of Sciences. 2005;102:6801–6806. doi: 10.1073/pnas.0408970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W, Andrec M, Gallicchio E, Levy RM. J Phys Chem B. 2009;113:11702–11709. doi: 10.1021/jp900445t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng W, Gallicchio E, Deng N, Andrec M, Levy RM. J Phys Chem B. 2011;115:1512–1523. doi: 10.1021/jp1089596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhal N, Snow CD, Pande VS. J Chem Phys. 2004;121:415. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 16.Jayachandran G, Vishal V, Pande VS. J Chem Phys. 2006;124:164902. doi: 10.1063/1.2186317. [DOI] [PubMed] [Google Scholar]
- 17.Berezhkovskii A, Hummer G, Szabo A. J Chem Phys. 2009;130:205102. doi: 10.1063/1.3139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noe F, Fischer S. Current Opinion in Structural Biology. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Proceedings of the National Academy of Sciences. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins AH, Coman RM, Bracho-Sanchez E, Fernandez MA, Gilliland CT, Li M, Agbandje-McKenna M, Wlodawer A, Dunn BM, McKenna R. Acta Crystallogr D Biol Crystallogr. 2010;66:233–242. doi: 10.1107/S0907444909054298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Layten M, Hornak V, Simmerling C. J Am Chem Soc. 2006;128:13360–13361. doi: 10.1021/ja065133k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lexa KW, Damm KL, Quintero JJ, Gestwicki JE, Carlson HA. Proteins. 2009;74:872–880. doi: 10.1002/prot.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishima R, Freedberg DI, Wang Y-X, Louis JM, Torchia DA. Structure. 1999;7:1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 25.Freedberg DI, Ishima R, Jacob J, Wang Y-X, Kustanovich I, Louis JM, Torchia DA. Protein Science. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perryman AL, Lin J-H, McCammon JA. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornak V, Okur A, Rizzo RC, Simmerling C. J Am Chem Soc. 2006;128:2812–2813. doi: 10.1021/ja058211x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding F, Layten M, Simmerling C. J Am Chem Soc. 2008;130:7184–7185. doi: 10.1021/ja800893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornak V, Okur A, Rizzo RC, Simmerling C. Proceedings of the National Academy of Sciences. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott WR, Schiffer CA. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 31.Meagher KL, Carlson HA. Proteins. 2004;58:119–125. doi: 10.1002/prot.20274. [DOI] [PubMed] [Google Scholar]
- 32.Li D, Ji B, Hwang K, Huang Y. J Phys Chem B. 2010;114:3060–3069. doi: 10.1021/jp1005549. [DOI] [PubMed] [Google Scholar]
- 33.Sadiq SK, De Fabritiis G. Proteins. 2010;78:2873–2885. doi: 10.1002/prot.22806. [DOI] [PubMed] [Google Scholar]
- 34.Collins JR, Burt SK, Erickson JW. Nat Struct Mol Biol. 1995;2:334–338. doi: 10.1038/nsb0495-334. [DOI] [PubMed] [Google Scholar]
- 35.Rick SW, Erickson JW, Burt SK. Proteins. 1998;32:7–16. doi: 10.1002/(sici)1097-0134(19980701)32:1<7::aid-prot3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.Tozzini V, McCammon J. Chemical Physics Letters. 2005;413:123–128. [Google Scholar]
- 37.Tozzini V, Trylska J, Chang C, McCammon JA. Journal of Structural Biology. 2007;157:606–615. doi: 10.1016/j.jsb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Chang C, Shen T, Trylska J, Tozzini V, McCammon J. Biophysical Journal. 2006;90:3880–3885. doi: 10.1529/biophysj.105.074575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzner P, Schütte C, Vanden-Eijnden E. Multiscale Model Simul. 2009;7:1192. [Google Scholar]
- 40.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Proceedings of the National Academy of Sciences. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elber R. Current Opinion in Structural Biology. 2005;15:151–156. doi: 10.1016/j.sbi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Ozkan SB, Dill KA, Bahar I. Protein Sci. 2002;11:1958–1970. doi: 10.1110/ps.0207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swope WC, Pitera JW, Suits F. J Phys Chem B. 2004;108:6571–6581. [Google Scholar]
- 44.Chodera JD, Swope WC, Pitera JW, Dill KA. Multiscale Model Simul. 2006;5:1214. [Google Scholar]
- 45.Chekmarev DS, Ishida T, Levy RM. J Phys Chem B. 2004;108:19487–19495. [Google Scholar]
- 46.Sriraman S, Kevrekidis IG, Hummer G. J Phys Chem B. 2005;109:6479–6484. doi: 10.1021/jp046448u. [DOI] [PubMed] [Google Scholar]
- 47.Voelz VA, Bowman GR, Beauchamp K, Pande VS. J Am Chem Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao F, Caflisch A. Journal of Molecular Biology. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 49.Klimov D, Thirumalai D. Journal of Molecular Biology. 2005;353:1171–1186. doi: 10.1016/j.jmb.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 50.Carr JM, Wales DJ. J Phys Chem B. 2008;112:8760–8769. doi: 10.1021/jp801777p. [DOI] [PubMed] [Google Scholar]
- 51.Pietrucci F, Marinelli F, Carloni P, Laio A. J Am Chem Soc. 2009;131:11811–11818. doi: 10.1021/ja903045y. [DOI] [PubMed] [Google Scholar]
- 52.Gallicchio E, Andrec M, Felts AK, Levy RM. J Phys Chem B. 2005;109:6722–6731. doi: 10.1021/jp045294f. [DOI] [PubMed] [Google Scholar]
- 53.Gillespie DT. Markov processes: an introduction for physical scientists. Academic Press; Boston: 1992. [Google Scholar]
- 54.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 55.Gallicchio E, Levy RM. J Comput Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 56.Velazquez-Campoy A, Kisob Y, Freire E. Archives of Biochemistry and Biophysics. 2001;390:169–175. doi: 10.1006/abbi.2001.2333. [DOI] [PubMed] [Google Scholar]
- 57.Paris K. PhD Thesis. Rutgers University; Piscataway, New Jersey: 2010. Modeling receptor reorganization and strain in protein-ligand binding. [Google Scholar]
- 58.Perryman AL, Lin J-H, McCammon JA. Biopolymers. 2006;82:272–284. doi: 10.1002/bip.20497. [DOI] [PubMed] [Google Scholar]
- 59.Northrup SH, Zarrin F, McCammon JA. J Phys Chem. 1982;86:2314–2321. [Google Scholar]
- 60.Szabo A, Shoup D, Northrup SH, McCammon JA. J Chem Phys. 1982;77:4484. [Google Scholar]
- 61.Shuman CF, Hämäläinen MD, Danielson UH. J Mol Recognit. 2004;17:106–119. doi: 10.1002/jmr.655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.