Abstract
Sol-gel synthesized nickel oxide (NiO) film deposited onto indium tin oxide (ITO) coated glass plate has been utilized for the development of sensitive and stable DNA biosensor and demonstrated for diagnosis of Visceral Leishmaniasis also known as Kala-azar. Leishmania specific sensor is developed by immobilizing 23mer DNA sequence (oligonucleotide) identified from 18S rRNA gene sequences from Leishmania donovani. Characterization studies like X-ray diffraction and Scanning Electron Microscopy revealed the formation of nano-structured NiO, while immobilization of single strand (ss)-DNA of Leishmania was supported by UV-visible, Fourier Transform Infrared Spectroscopy and Scanning Electron Microscopy techniques. Response studies of ss-DNA/NiO/ITO bioelectrode are carried out using differential pulsed voltammetry in presence of methylene blue redox dye as a redox mediator. A linear response is obtained in the wide concentration range of 2pg/ml to 2μg/ml of complementary target genomic DNA (parasite DNA) within the variation of 10% for 5 sets of studies. The observed results holds promise for not only diagnosis of Kala-azar patients but also enormous potential of the nano NiO based probe for development of stable and sensitive biosensors.
Keywords: Visceral Leishmaniasis, Kala-azar, Leishmania donovani, Nickel oxide, DNA biosensor, Electrochemical sensor
1. Introduction
The protozoan parasites of the genus Leishmania are the causative agents of a group of diseases called Leishmaniasis or Kala-azar, endemic in more than 98 countries worldwide 1. Visceral Leishmaniasis (VL) is a debilitating protozoan disease, next only to malaria and TB in terms of annual incidences and severity. It is a potentially fatal vector-borne parasitic disease due to infection by L. donovani in the Indian subcontinent and Eastern Africa2. Proper diagnosis is necessary for suitable treatment strategies because the clinical symptoms are not specific enough to allow diagnosis of a VL infection and evaluating prognosis. The disease is generally confirmed by microscopic demonstration of the parasite in bone marrow or spleen aspirates which is considered to be the “gold standard” for the diagnosis of VL but requires invasive sampling techniques which are occasionally fatal 1, 2. As clinical features of VL mimics several other common diseases, accurate diagnosis of VL becomes an important issue as the treatment is associated with significant toxicity. Serological tests using rK39 antigen is most promising tool for diagnosis of VL using ELISA or strip3. Despite of high sensitivity of rK39 antigen it has no prognostic value 4. Other serological tests like IFAT and immunoblotting have shown high sensitivity and specificity but are restricted to research laboratories only due to complex procedures and again cannot discriminate between past and current infections 5-8. Traditional laboratory diagnosis of this infection is carried out by microscopy, culture and PCR techniques 9-12. Efforts are being made towards the development of sensitive, noninvasive and cost effective and specific VL sensing techniques. The basic element of a DNA sensor is single strand oligonucleotide (ss-DNA) probe, immobilized on the transducer surface (here NiO coated ITO electrode). The development of sensing probes immobilized with single strand (ss-DNA) is found to be the most crucial part which decides sensitivity of response, stability and accuracy of the biosensor. The surface property variations, after hybridization with complementary ss-DNA (target strand) are subsequently measured by various analytical techniques including optical methods via fluorescence labeled oligonucleotides, quartz crystal microbalance, surface plasmon resonance and electrochemical analysis 13-15.
Electrochemical DNA biosensors based on nucleic acid hybridization have received considerable attention due to their potential application for the diagnosis of various diseases 16–22. They offer great advantages due to simplicity, speed, miniaturization, sensitivity, selectivity and low cost for detection of desired DNA sequence or mutant genes 23–25. Immobilization of the ss-DNA has been studied over a variety of substrates/electrodes using various methods such as adsorption, covalent coupling and entrapment 26. However, various issues were raised like poor compatibility of immobilizing materials with the transducer, poor binding of DNA, mismatching of conductivity, creation of inactive or poor conducting layers, less surface area and poor stability. Very recently semi conducting nano metal oxide materials have shown promising role for electrochemical transduction of DNA-sensing events because of having high ionic conductivity, capacitive action, catalytic properties, large surface area and high isoelectric point (Ip) 27-28.
Recently, Choi et al. have fabricated a nano-porous niobium oxide film for label free detection of DNA hybridization events 29. In another approach, a nucleic acid sensor has been fabricated using a single-stranded DNA-immobilized multiwall carbon nanotube–nano-ZrO2–chitosan-modified glassy carbon electrode 22. Solanki et al. have more recently reported application of a nano-structured ZrO2 film modified electrode for detection of DNA hybridization 30. Various methods like electrochemical deposition, sol-gel technique, CVD, sputtering etc. have been adopted to form the nano oxide material coating over the transducers. The sol–gel method is considered particularly more attractive for the development of desired metal oxide electrodes because it is simple and fast. Moreover, sol–gel materials can be prepared under ambient conditions and exhibit tunable porosity, high retention capacity, high surface area, biocompatibility, excellent thermal stability, chemical inertness and negligible swelling in aqueous and non-aqueous solutions 31-34. Besides this, a sol-gel derived nano-porous film can retain its bioactivity at different environmental and experimental conditions such as pH and temperature and can be used for direct electron transfer between bio-molecules and the electrode.
Past research on nano-sized NiO has demonstrated its excellent catalytic, magnetic, electrochromic, optical and electrochemical properties 35-38. Nano structured NiO has also been used for the immobilization of various enzymes and direct voltammetry. Moghaddam et al. and Salimi et al. have demonstrated excellent biocompatibility, direct and enhanced electron transfer and electrochemical properties of NiO nano-particles which can be put to biosensor applications 36, 37. In a later study by Salimi et al. the electrochemical oxidation of guanine on the guanine/NiOx-modified glassy carbon electrode showed high sensitivity for the pM detection of insulin 38.
The Isoelectric Point of NiO which is 10.8 is comparatively higher than the other metal oxides like ZnO, ZrO2, TiO2 etc. which have been recently used for the biosensors applications. Therefore, NiO can serve as an excellent matrix for the better immobilization of the biomolecules. In addition, NiO is stable and has much better electro-catalytic and electron transfer properties thus enhancing the intensity of the electrochemical signal viz. sensing current suitable for detection of the analyte at very low concentration levels 37-39.
In this work, successful attempts have been made towards the fabrication of voltammetry based VL (Kala-azar) biosensor by immobilization of single-stranded DNA on the surface of the nano structured NiO film deposited on ITO conducting glass plate. Methylene blue (MB), a well-known hybridization indicator of DNA due to its association with the free guanine bases of single-strand DNA 40, 41 is used to generate a marked electrochemical signal for ss-DNA and hybridized DNA. Electrochemical reduction of MB (using a trace amount) is used to get indication of presence of single strand of DNA and hybridization process. A linear response is obtained in the wide concentration range of 2pg/ml to 2μg/ml of complementary target genomic DNA (parasite DNA) within the variation of 10% for 5 sets of studies.
2. Experimental
2.1 Materials
All chemicals were purchased from Sigma–Aldrich. Leishmania specific probes (23 bases) were identified from the 18S rRNA gene sequences from L. donovani (Gen Bank Accession ID X0773). The 50 mM phosphate buffer saline of pH 7 having 0.7% NaCl is used for experiment and mentioned as phosphate buffer saline in the text.
2.2 Probe design
Since DNA probes are of crucial importance to the success of any genosensor based assay, a rigorous bioinformatics analysis was done to design the probe (in the Institute of Medical Sciences, BHU). The 23mer ss-DNA probe was synthesized by Metabion, Germany. The sequence of the probe (5′GCCGAATAGAAAAGATACGTAAG3′) was taken from elsewhere 42. 18S rRNA gene was chosen as the target region. 18S rRNA gene sequences from L. donovani (Gen Bank Accession ID X0773), T b. gambiense (AJ009141) and T. cruzi (AF303660) were aligned with using ClustalW software version 2 (http://www.ebi.ac.uk/Tools/clustalw2) and probe was designed using the most stringent conditions allowed in the software package against the conserved regions by using OLIGO4 primer analysis software (Molecular Biology Insights, Inc., Cascade, CO, USA). Specificity of the probe was examined using the BLAST program at NCBI site (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3 Cultured Isolates and Genomic DNA Extraction
Splenic aspirate containing Leishmania donovani parasites was collected in Novy Mac-Neal-Nicolle media containing biphasic media (blood-agar and RPMI) in sterile condition and cultured at 260C in BOD incubator to get promastigote form of parasite (in the Institute of Medical Sciences, BHU). Approximately 107 (number of parasites in logarithmic phase of their growth curve) phase promastigotes of isolates were harvested from culture by centrifugation. The pellets were washed 3 to 4 times with chilled 50 mM phosphate buffer saline (pH 7.0 and 0.7% NaCl) and suspended in 750μl of NET buffer (5M NaCl, 0.5M EDTA, 1M Tris Buffer pH 8.0). Cells were lysed with Proteinase K (100 μg /ml) and 1% Sodium dodecyl sulphate for 3-4 hrs at 56°C. Total DNA was extracted by standard Phenol-chloroform method as described earlier.43 The parasitic DNA was then diluted in 10 fold serial dilutions starting from 20ng up to 0.002pg.
Genomic DNA Extraction from healthy individual
Total genomic DNA was extracted from 2 ml of blood obtained from healthy individual by using standard Phenol-chloroform method as described earlier. 43
2.4 Preparation of NiO nanoparticles and modified ITO electrode
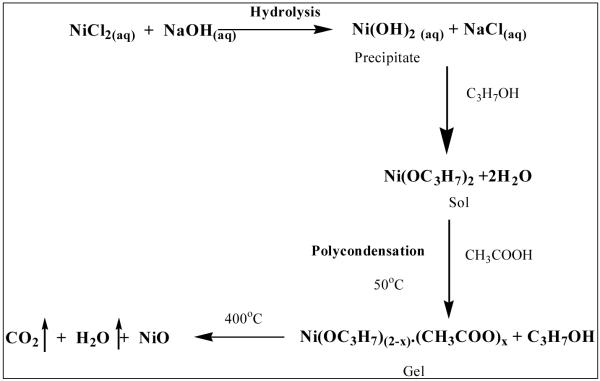
2.37g of NiCl2.6H2O (0.2 M) was dissolved in 50 ml of deionised water. 25ml of 1M NaOH solution was added to the nickel chloride solution drop by drop under constant stirring till pH 7.5. A green precipitate of Ni(OH)2 was formed after some time. The solution was allowed to settle overnight and the Ni(OH)2 precipitate was separated using centrifugation followed by 2-3 times washing with water. The precipitate was further dissolved in 2-propanol (minimum amount) with constant stirring followed by addition of 10 drops of acetic acid (1.5 M) as chelating agent. The sol thus obtained was continuously stirred for an hour at ambient temperature to get gel. A green colored gel was obtained due to the reactions involved as in the scheme given below 44.
Scheme 1.
Scheme for the formation of NiO nanoparticles by sol-gel method.
The obtained sol-gel solution was used to fabricate film onto ITO coated glass plates. Gel was coated over ITO by dip coating method and calcined at 400°C in a furnace for an hour to obtain Nickel Oxide (NiO) thin film over ITO glass plate (ITO/NiO).
2.5 Immobilization of DNA
ITO/NiO surface was washed with phosphate buffer saline and dried in nitrogen atmosphere. 10μl of 23mer oligonucleotide (ss-DNA, 26.7 nM) specific to L. donovani was immobilized onto ITO/NiO electrode and allowed to 10-15 minutes incubation to physically adsorb over the surface of NiO (in a humid chamber at 25°C). The ITO/NiO/ss-DNA electrode was then subsequently washed with phosphate buffer saline to remove weakly adsorbed ss-DNA. It was stored at 4°C in a sealed container after drying in nitrogen environment when not in use.
2.6 Hybridization over ss-DNA probe
ss-DNA immobilized probe as discussed above was used for hybridization studies using full genomic DNA of L. donovani or healthy human genome. Hybridization process was achieved in 5 ml vial by taking full genomic DNA followed by dehybridization (denaturation) of the target ds-DNA. Ds-DNA solution was first sonicated and the heat treatment of the genomic ds-DNA at 92-94°C was carried out for 3 minutes followed by cooling for 3 minutes at 75°C. During cooling step ss-DNA immobilized probe was dipped in the solution for hybridization. Hybridization was studied by incubation of probe in MB solution after washing.
2.7 Measurement and Instruments
The ITO/NiO and ITO/NiO/ss-DNA electrodes were characterized using UV-visible spectrophotometer (UV-vis), Fourier transform infrared spectroscopy (FT-IR), Scanning Electron Microscopy (SEM) techniques, differential pulse voltammetry (DPV) and X-ray diffraction (XRD) techniques.
DPV measurements were carried out by electrochemical workstation 17041C CH instruments Inc. (USA), using a three-electrode cell configuration with Ag/AgCl electrode as a reference electrode and platinum foil as a counter electrode in 10ml of phosphate buffer saline. UV-vis and FT-IR analysis were carried out with Lambda-25 UV-visible Spectrophotometer, Perkin-Elmer, Germany and 8400 FT-IR, Schimadzu, Japan respectively. SEM images were taken using ZEISS Supra 40 SEM. X-ray diffraction (XRD) patterns were recorded using 18 kW rotating anode (Cu) based (Rigaku, Japan) powder diffractometer operating in the Bragg-Brentano geometry and fitted with a graphite monochromator in the diffracted beam. Data were recorded at a scanning rate of 4°/min at 6 kW energy.
3. Result and Discussion
3.1 X-Ray diffraction
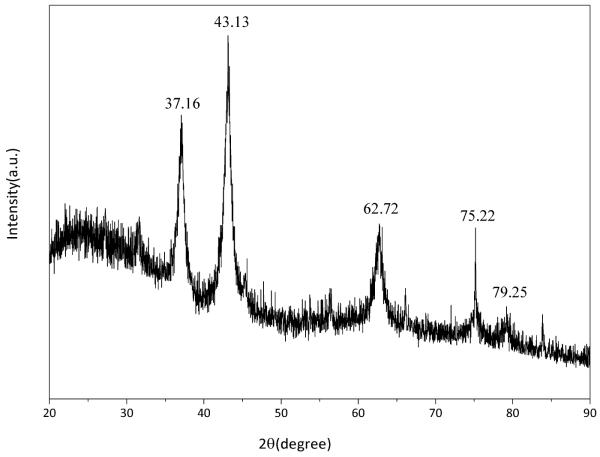
X-ray diffraction pattern was recorded for NiO film annealed at 400°C for 1h to confirm the formation of NiO, purity and particle size of the NiO. The observed peaks were matched with the corresponding pure cubic-structured crystalline NiO 35.
Characteristic peaks were observed at 2θ=37.16°, 43.13°, 62.72°, 75.2°, and 79.25° assigned to the (111), (200), (220), (311) and (222) crystal planes, respectively as shown in Fig. 1. The indexed peaks were fully consistent with the cubic-structured crystalline NiO (JCPDS 47-1049) 35. The cell parameter was obtained as 4.1862 A°. No impurity peaks such as Ni(OH)2, Ni2O3, or other phases were observed in the pattern which further confirmed the pure phase formation of the cubic NiO using sol-gel method. The NiO crystallite size was calculated using following Scherer formula. 45
| (1) |
Where k is a constant which is taken as 0.9, λ is wavelength of X-rays used (15.406 nm), ß is full width at half maximum obtained as 0.74 radians for 2θ= 43.13 and θ is the angle of diffraction. The average diameter of NiO nano particles was found to be about 40 nm.
Fig. 1.
X-Ray diffraction pattern of sol–gel derived nano-structured NiO film annealed at 400°C for 1h.
3.2 UV-vis studies
A strong absorption was observed at wavelength about 345 nm as shown in Fig. 2 can be attributed to intra-3d transition of Ni2+ in the cubic structure of NiO 35, 46. The characteristic peak of NiO at wavelength about 345 nm is shifted at 339 nm after immobilization of ss-DNA along with the presence of new absorption peak at 270 nm which appeared due to the DNA bases 47. The presence of characteristic peak of DNA and shift in the peak of NiO support the immobilization of ss-DNA onto the surface of NiO oxide matrix and formation of ss-DNA coated NiO film.
Fig. 2.
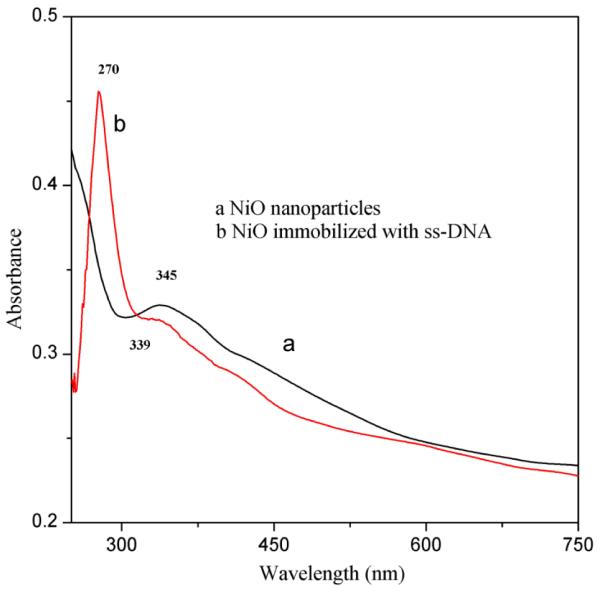
UV-vis absorption spectrum of (a) nano-structured ITO/NiO film (b) ITO/NiO/ss-DNA film
3.3 FT-IR studies
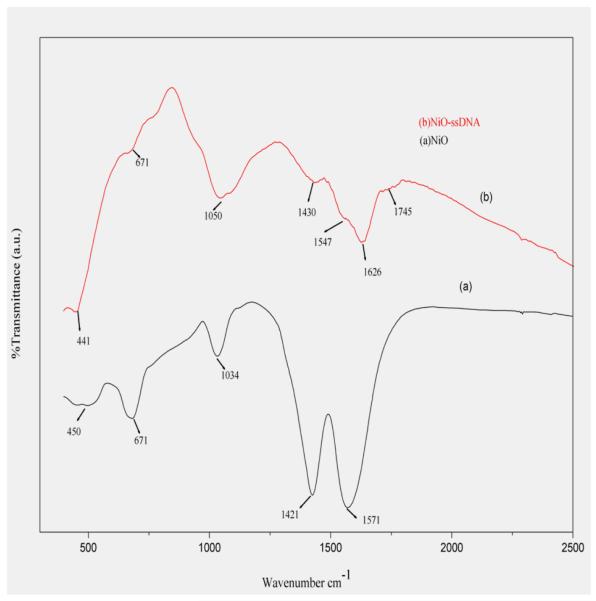
The Fourier transformed infrared spectroscopy of the as-synthesized sample was performed as shown in Fig. 3. The peak at 450 cm−1 in spectrum, (curve a), showing Ni–O bond 35, 48. This peak shifted to 441 cm−1 on immobilization with ss-DNA. The peaks at 671, 1034, 1421 and 1571 cm−1 in both the curves (a) and (b) are due to the various stretching and bending vibration modes of Ni-O bond 49. Also the two new peaks at 1626 cm−1 and 1745 cm−1 (curve-b) due to characteristic cytosine and guanine bases of the DNA molecule confirmed the stable immobilization of ss-DNA. 50 Intensity of NiO peaks decreased after immobilizing ss-DNA over NiO.
Fig. 3.
FT-IR spectrum of the (a) nanostructured NiO/ ITO thin film (b) ss-DNA/ NiO/ ITO sol-gel film.
3.4 Scanning Electron Microscopy Studies
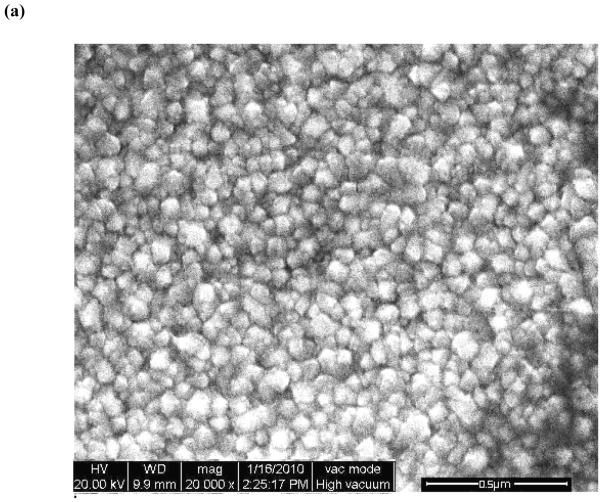
The surface morphology of the as-prepared NiO nanoparticle film and annealed at 400 °C for 1 hour as well as NiO film immobilized with ss-DNA, has been studied by scanning electron microscopy. It is clearly observed in SEM that the nanoparticles are extra-fine, and uniformly distributed on the ITO coated glass substrate (cf. Fig. 4-a). The morphology of nano NiO changes into dense morphology consisting of large globules like thick clusters as can be seen in Fig. 4-b, after immobilization of the ss-DNA, which revealed presence of ss-DNA onto NiO/ITO electrode. This is probably adsorption of the ss-DNA over nanoparticles and agglomeration of the particles in presence of DNA.
Fig.4.
SEM images of (a) pure NiO (b) NiO nanoparticles after immobilization with ss-DNA.
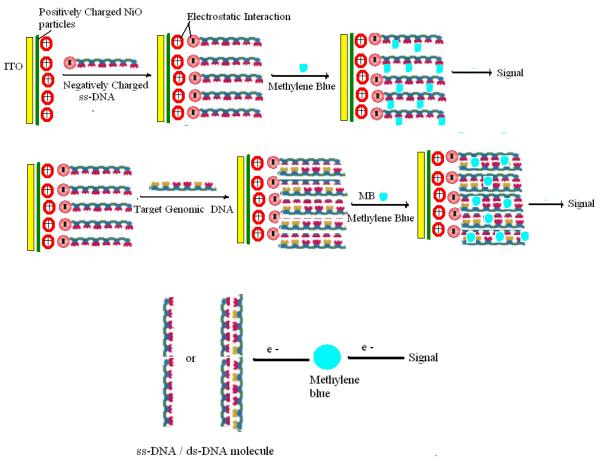
The mechanism of immobilization of DNA onto NiO/ITO surface occurs due to the electrostatic interactions between NiO surface and DNA molecules as shown in the Fig. 5. At the physiological pH 7.5, NiO having a high isoelectric point (IEP) of 10.8 behaves as a positively charged matrix which is suitable for adsorption of negatively charged DNA molecules due to its low IEP of 4.2. Hence the positively charged NiO nano-particles not only provide a biocompatible environment for immobilizing negatively charged DNA molecules, but also promote electron transfer between DNA and the electrode 30, 39, 51.
Fig. 5.
Schematic representation of sensing mechanism over the bioelectrode (ordered arrangement of ss-DNA is just assumption for easy presentation).
3.5 DPV studies
Electrochemical studies for nucleic acid hybridization
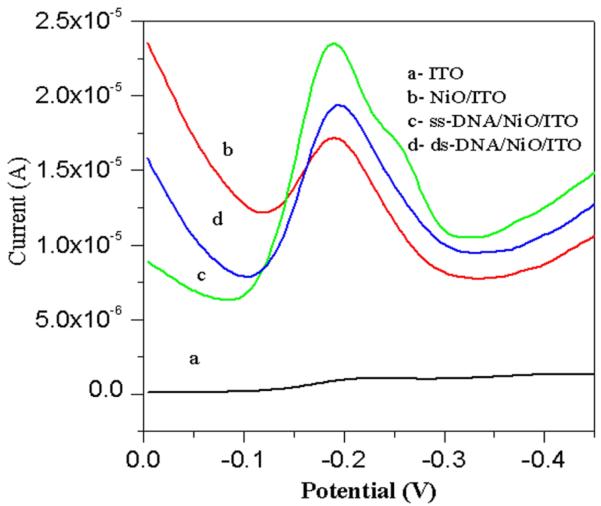
DPV studies (Fig. 6.) of ITO electrode, NiO/ITO electrode, ss-DNA/NiO/ITO, ds-DNA/NiO/ITO bioelectrodes in phosphate buffer saline (at pulse height of 50 mV) with methylene blue (MB, 20 mM) as an indicator have been conducted to distinguish presence of ss-DNA and double stranded ds-DNA onto electrode surface.
Fig.6.
Differential pulse voltammograms (at a pulse height of 50 mV) of (a) ITO electrode, (b) NiO/ITO electrode, (c) ss-DNA/NiO/ITO bioelectrode, (d) ds-DNA/NiO/ITO bioelectrode in phosphate buffer saline and methylene blue (20 mM).
Immobilization of the ss-DNA for development of sensing probe was done as described above. The hybridization event was carried out first by dehybridizing (denaturation) of the target ds-DNA51, 52 (as discussed under experimental) followed by incubation in MB solution. A peak was observed at −0.19V due to the reduction of MB. The increased magnitude of electrochemical current response observed for NiO/ITO electrode (Fig. 6.b) is higher than that of ITO indicating the large surface area of NiO for electro-reduction process. MB binds with the guanine moieties of the DNA molecules. The magnitude of current response further increases for ss-DNA/NiO/ITO bioelectrode (Fig. 6.c), due to strong affinity of MB with free guanine bases and hence greatest amount of MB accumulation occurs at the electrode surface. However, a significant reduction in current is obtained on incubating with the complementary sequence (Fig. 6.d) due to decreased MB accumulation on the ds-DNA/NiO/ITO bioelectrode since the guanine bases become inaccessible to MB after hybridization takes place.
Detection of VL (Kala-azar) from clinical isolates
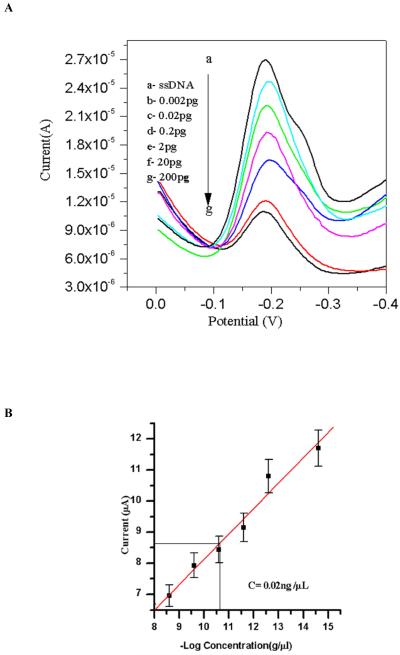
Nucleic acid (oligonucleotide) functionalized NiO/ITO based sensor has been used for the detection of L. donovani in cultured isolates. Fig. 8, which shows response of ss-DNA/NiO/ITO bioelectrode to detect the presence of complementary target sequence of L. donovani in cultured isolates. Electrochemical response of ss-DNA/NiO/ITO bioelectrode after hybridization with different concentrations of 2fg/μl, 0.02pg/μl, 0.2pg/μl, 2pg/μl, 0.02ng/μl, 0.2ng/μl, 2ng/μl of target genomic DNA (L. donovani) has been studied by DPV technique in phosphate buffer saline using methylene blue as an indicator (Fig. 8. A. b-g). It has been observed that magnitude of the current with respect to methylene blue decreases with the increase in target genomic DNA concentration (Fig. 8. B). This may be explained due to the enhanced number of double stranded DNA molecules at the surface of the bioelectrode with the increase in target genomic DNA concentration which results in the steric inhibition to methylene blue dye and also due to the blocking of guanine sites due to the double helix formation at the electrode surface. Wide concentration range linearity was observed with detection level of 2fg/μl concentration. The sensor showed reproducibility as the deviation was within 10% from the mean value of the readings of 5 sets.
Fig.8.
(A) Differential pulse voltammograms (at a pulse height of 50 mV) of (a) ss-DNA/NiO/ITO bioelectrode and (b-d) showing response of the ss-DNA/NiO/ITO bioelectrode after hybridization (60 s hybridization time) with complementary target probe concentration 0.002pg -200pg in phosphate buffer saline and methylene blue (20 mM). (B) Showing the calibration graph of the same concentration range in which the unknown sample detected (detection limit is 0.02 ±0.002ng/μl).
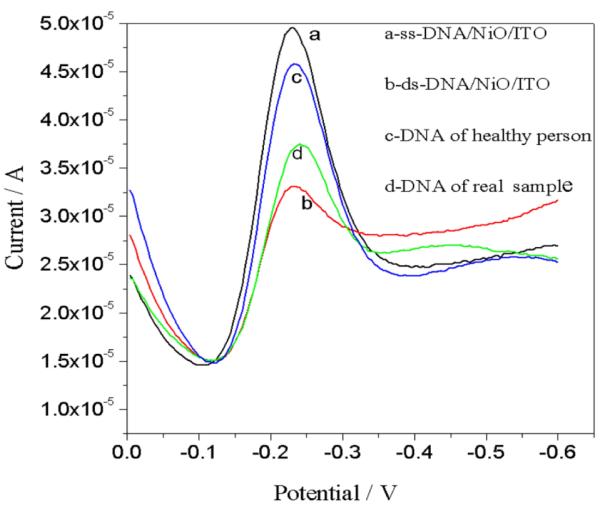
Detection of mismatch in target DNA
Our system was efficient enough to detect mismatch target DNA obtained from healthy person. Figure 9 shows the differentiation between DNA of healthy person with the DNA of the VL affected patient. The peak current of ss-DNA/NiO/ITO electrode obtained is 4.58*10−5 A which decreases after hybridization with the DNA of the VL affected patient to 1.88*10−5 A, while showed 4.56*10−5 A after hybridization with target DNA of healthy person.
Fig.9.
Differential pulse voltammograms of (a) ss-DNA/NiO/ITO bioelectrode (b) ds-DNA/NiO/ITO (after hybridization with complementary target DNA) (c) DNA of healthy person (d) DNA of diseased person in phosphate buffer saline and methylene blue (20 mM).
Conclusion
Sol-gel synthesized nano structured nickel oxide (NiO) film deposited onto indium tin oxide (ITO) coated glass electrode has been used for the development of a genosensor for the precise diagnosis of VL (Kala-azar). It was characterized using various techniques and found that the sol-gel derived nano-structured NiO film is an excellent matrix for the immobilization of the DNA and development of genosensors. Further it was demonstrated using 23mer DNA sequence (oligonucleotide) of L. donovani species for DNA hybridization detection. The ss-DNA–NiO/ITO bioelectrode was used for detection in clinical isolates, which exhibits linearity in the wide concentration range of 2pg/ml to 2μg/ml of complementary target genomic DNA (parasite DNA) within the variation of 10% from the mean value for 5 sets of studies. Besides this, the bioelectrode is found to be highly specific to distinguish between DNA from L. donovani and genomic DNA from healthy person with the help of DPV electrochemical technique. We are in process of investigating the performance of our kala-azar sensor in clinical samples since we got encouraging results in this Phase I study on cultured isolates. Our assay could be a promising new tool for molecular detection of kala-azar and other leishmanial diseases.
ACKNOWLEDGEMENT
Authors (Pankaj Srivastava and Shyam Sundar) are thankful to NIAID-NIH (grant number-1P50AI074321-01) and Rajiv Prakash is thankful to DST, India for financial supports.
References
- 1.Salotra P, Sreenivas G, Pogue GP, N.L., Nakhasi HL, Ramesh V, Negi NS. J. Clin. Microbiol. 2001;39:849–854. doi: 10.1128/JCM.39.3.849-854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deborggraeve S, Boelaert M, Rijal S, De Doncker S, Dujardin JC, Herdewijn P, Buscher P. Trop. Med. Int. Health. 2008;13:1378–1383. doi: 10.1111/j.1365-3156.2008.02154.x. [DOI] [PubMed] [Google Scholar]
- 3.Salam MA, Mondal D, Kabir M, Ekram ARMS, Haque R. Acta Trop. 2010;113:52–55. doi: 10.1016/j.actatropica.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Sundar S, Singh RK, Maurya R, Kumar B, Chhabra A, Singh V, Rai M. Trans. R. Soc. Trop. Med. Hyg. 2006;100:533–537. doi: 10.1016/j.trstmh.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Kar K. Crit. Rev. Microbiol. 1995;21:123–125. doi: 10.3109/10408419509113537. [DOI] [PubMed] [Google Scholar]
- 6.Sundar S, Rai M. Clin. Diag. Lab. Immunol. 2002;9:951–958. doi: 10.1128/CDLI.9.5.951-958.2002. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zijlstra EE, Nur Y, Desjeux P, Khalil EA, El-Hassan AM, Groen J. Trop. Med. Int. Health. 2001;6:108–113. doi: 10.1046/j.1365-3156.2001.00680.x. [DOI] [PubMed] [Google Scholar]
- 8.Oskam L, Nieuwenhuijs JL, Hailu A. Trans. R. Soc. Trop. Med. Hyg. 1999;93:275–277. doi: 10.1016/s0035-9203(99)90021-4. [DOI] [PubMed] [Google Scholar]
- 9.Chulay JD, Bryceson AD. Am. J. Trop. Med. Hyg. 1983;32:475–479. doi: 10.4269/ajtmh.1983.32.475. [DOI] [PubMed] [Google Scholar]
- 10.Siddig M, Ghalib H, Shillington DC, Peterson EA. Trans. R. Soc. Trop. Med. Hyg. 1988;82:66–68. [PubMed] [Google Scholar]
- 11.Osman OF, Oskam L, Zijlstra EE, Kroon NC, Schoone GJ, Khalil ET, El-Hassan AM, Kager PA. J. Clin. Microbiol. 1997;35:2454–2457. doi: 10.1128/jcm.35.10.2454-2457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reithinger R, Dujardin JC. J. Clin. Microbiol. 2007;45:21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taton TA, Lu G, Mirkin CA. J. Am. Chem. Soc. 2001;123:5164. doi: 10.1021/ja0102639. [DOI] [PubMed] [Google Scholar]
- 14.Feng K, Li J, Jiang JH, Shen GL, Yu RQ. Biosens. & Bioelectron. 2007;22:1651–1657. doi: 10.1016/j.bios.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Gong P, Lee CY, Gamble LJ, Castner DG, Grainger DW. Anal. Chem. 2006;78:3326–3334. doi: 10.1021/ac052137j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, Chen Z, Li Y, He S, Xie S, Xu X, Liang Z, Meng X, Li Q, Zhu Z, Li M, Le XC, Shao Y. Anal. Chem. 2008;80:6323–6328. doi: 10.1021/ac800601y. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Tian S, Nielsen PE, Knoll W. Chem. Commun. 2005:2969–2971. doi: 10.1039/b419425j. [DOI] [PubMed] [Google Scholar]
- 18.Odenthal KJ, Gooding JJ. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- 19.Shrestha S, Yeung CMY, Mills CE, Lewington J, Tsang SC. Angew. Chem., Int. Ed. Engl. 2007;119:1–6. doi: 10.1002/anie.200604827. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Xu D, Polsky R. J. Am. Chem. Soc. 2002;124:4208–4209. doi: 10.1021/ja0255709. [DOI] [PubMed] [Google Scholar]
- 21.Marrazza G, Chianella I, Mascini M. Biosens. & Bioelectron. 1999;14:43–51. doi: 10.1016/s0956-5663(98)00102-x. [DOI] [PubMed] [Google Scholar]
- 22.Zhu N, Zhang A, Wang Q, He P, Fang Y. Anal. Chim. Acta. 2004;510:163–168. [Google Scholar]
- 23.Zhang J, Song SP, Zhang LY, Wang LH, Wu HP, Pan D, Fan C. J. Am. Chem. Soc. 2006;128:8575–8580. doi: 10.1021/ja061521a. [DOI] [PubMed] [Google Scholar]
- 24.Zuo SH, Zhang LF, Yuan HH, Lan MB, Lawrance GA, Wei G. Bioelectrochemistry. 2009;74:223–226. doi: 10.1016/j.bioelechem.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Cai X, Rivas G, Shiraishi H, Farias PAM, Dontha N. Anal. Chem. 1996;68:2629–2634. doi: 10.1021/ac9602433. [DOI] [PubMed] [Google Scholar]
- 26.Ansari AA, Singh R, Sumana G, Malhotra BD. Analyst. 2009;134:997–1002. doi: 10.1039/b817562d. [DOI] [PubMed] [Google Scholar]
- 27.Wei A, Sun XW, Wang JX, Lei Y, Cai XP, Li CM, Dong ZL, Huang W. Appl. Phys. Lett. 2006;89:123902–123903. [Google Scholar]
- 28.Kouassi GK, Irudayaraj J, McCarty G. J. Nanobiotechnol. 2005;3:1–9. doi: 10.1186/1477-3155-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi J, Lim JH, Rho S, Jahng D, Lee J, Kim KJ. Talanta. 2008;74:1056–1059. doi: 10.1016/j.talanta.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Solanki PR, Kaushik A, Chavhan PM, Maheshwari SN, Malhotra BD. Electrochem. Commun. 2009;11:2272–2277. [Google Scholar]
- 31.Avnir D, Braun S, Ovadia L, Ottolengthi M. Chem. Mat. 1994;6:1605–1614. [Google Scholar]
- 32.Dave BC, Dunn B, Valentine JS, Zink JI. Anal. Chem . 1994;66:1120A–1126A. [Google Scholar]
- 33.Collinson MM. TrAC, Trends Anal. Chem. 2002;21:31–39. [Google Scholar]
- 34.Jin W, Brenan JD. Anal. Chim. Acta. 2002;461:1–36. [Google Scholar]
- 35.Niasari MS, Mir N, Davar F. Polyhedron. 2009;28:1111–1114. [Google Scholar]
- 36.Moghaddam AB, Ganjali MR, Dinarvand R, Ahadi S, Saboury AA. Biophys. Chem. 2008;134:25–33. doi: 10.1016/j.bpc.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Salimi A, Sharifi E, Noorbakhsh A, Soltanian S. Electrochem. Commun. 2006;8:1499–1508. [Google Scholar]
- 38.Salimi A, Noorbakhash A, Sharifi E, Semnani A. Biosens. & Bioelectron. 2008;24:792–798. doi: 10.1016/j.bios.2008.06.046. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez N, Moreno R, Sanchez-Herencia AJ, Fierro Jose L. G. J. Phys. Chem. B. 2005;109:4470–4474. doi: 10.1021/jp0448954. [DOI] [PubMed] [Google Scholar]
- 40.Erdem A, Kerman K, Meric B, Ozsoz M. Electroanal. 2001;13:219–223. [Google Scholar]
- 41.Ozkan D, Kara P, Kerman K, Meric B, Erdem A, Jelen F, Nielsen PE, Ozsoz M. Bioelectrochemistry. 2002;58:119–126. doi: 10.1016/s1567-5394(02)00131-7. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava P, Mehrotra S, Tiwary P, Chakravarty J, Sundar S. PloS ONE. 2011;6:e19304. doi: 10.1371/journal.pone.0019304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. pp. A.10–A.11. [Google Scholar]
- 44.Boschloo G, Hagfeldt A. J. Phys. Chem. B. 2001;105:3039–3044. [Google Scholar]
- 45.Sasi B, Gopchandran KG. Sol. Energy Mater. Sol. Cells. 2007;91:1505–1509. [Google Scholar]
- 46.Al-Ghamdi AA, Mahmoud WE, Yaghmour SJ, Al-Marzouki FM. J. Alloys Compd. 2009;486:9–13. [Google Scholar]
- 47.Sutherland JC, Griffin KP. Radiat. Res. 1981;86:399–409. [PubMed] [Google Scholar]
- 48.Dharmaraj N, Prabu P, Nagarajan S, Kim CH, Park JH, Kim HY. Mater. Sci. Eng. B. 128:111–114. b. [Google Scholar]
- 49.Korošec RC, Bukovec P. Acta Chim. Slov. 2006;53:136–147. [Google Scholar]
- 50.Taniguchi H, Saito M. J. Phys.: Condens. Matter. 2009;21:064242–064244. doi: 10.1088/0953-8984/21/6/064242. [DOI] [PubMed] [Google Scholar]
- 51.Mohan S, Nigam P, Kundu S, Prakash R. Analyst. 2010;135:2887–2893. doi: 10.1039/c0an00258e. [DOI] [PubMed] [Google Scholar]
- 52.Vermeeren V, Bijnens N, Wenmackers S, Daenen M, Haenen K, Williams OA, Ameloot M, vandeVen M, Wagner P, Michiels L. Langmuir. 2007;23:13193–202. doi: 10.1021/la702143d. [DOI] [PubMed] [Google Scholar]