Abstract
Background
Depressive symptoms are common among individuals with alcohol use disorders and impact treatment outcome. Substantial overlap exists among the neurobiological systems proposed in the pathophysiology of depressive and alcohol use disorders; however, specific genetic effects contributing to risk for depressive comorbidity remain poorly understood.
Methods
This study examines the association of depressive symptom scores for lifetime depression (the sum of DSM-IV MD co-endorsed criteria for lifetime depression) with markers in 120 candidate genes in 554 alcohol dependent individuals. The candidate genes code for molecules involved in dopamine, serotonin, glutamate, GABA, and opioid neurotransmission, cell signaling, pharmacokinetics, stress biology and behavioral control. Analyses were conducted at the single marker level with experimentwise permutation to control for multiple testing.
Results
Results revealed nominal associations for markers in 20 genes. Following experimentwise permutation, markers in the corticotropin releasing hormone binding protein (CRHBP) the μ-opioid receptor (OPRM1) and the β1 subunit of gamma-aminobutyric acid A (GABAA) receptors (GABRB1) met or exceeded the significance threshold. None of the markers associated with depressive symptom scores were significantly associated with alcohol dependence symptom scores.
Conclusion
These findings suggest potential risk genes for depressive symptoms in alcohol dependent individuals.
Keywords: major depression, alcohol dependence, genetic association
Introduction
Vulnerability to stress-related psychiatric disorders such as alcohol dependence (AD) is influenced by environmental factors; however, family, twin, and adoption studies consistently demonstrate that AD aggregates in families due partly to genetic risks (McGue, 1999). Biological vulnerability to addiction is supported by the observation that disturbances in neuroendocrine activity occur among offspring of AD individuals, which in turn predicts future experimentation with illicit substances (Lovallo, 2006). Despite the strong familial component to AD, complex interactions among neurobiological systems involved in addiction and high rates of comorbid clinical diagnoses complicate efforts to identify genetic vulnerabilities.
Genomewide association studies (GWAS) have had modest success in identifying genes associated with AD (e.g., Edenberg and Foroud, 2006). Because they are empirically rather than hypothesis driven, GWA studies require conservative statistical correction that renders detection of common vulnerability alleles difficult. An alternate strategy is to select candidate genes in a focused array based on the neurobiology of addiction. Whereas a neurobiology-guided approach may have its own limitations, this strategy has identified some important AD-linked genes such as the alcohol dehydrogenase cluster (Edenberg, 2007). In addition to ethanol metabolism, however, a strong motivational component involving multiple neurobiological systems is also important for the development and maintenance of addiction.
An influential conceptual framework provided by Koob and colleagues conceptualizes AD, and addiction generally, as a cycle of increasing dysregulation of brain reward/anti-reward mechanisms, resulting in a negative emotional state contributing to the compulsive use of drugs. According to this model, the reward mechanisms disturbed in AD are mediated by activation of dopamine, serotonin, opioid peptides, and γ-aminobutyric acid (GABA) systems, as well as receptor transduction mechanisms such as adenylate cyclase, protein kinase A, CREB, and deltaFosB. Activation of brain stress systems, most notably corticotropin-releasing hormone (CRH) but also noradrenaline, dynorphin, and the anti-stress neuropeptide Y system in the extended amygdala, are considered the anti-reward mechanisms associated with the emotional dysregulation observed in addiction (Koob and Le Moal, 2008). Koob argues that CRH and other brain stress systems play a role in the transition to dependence, its maintenance, and relapse during abstinence in the face of life stress by contributing to a “residual negative emotional state” in the cycle of addiction (Koob and Le Moal, 2008). The negative affective component in the cycle of AD consists of motivational changes including chronic irritability, emotional pain, malaise, dysphoria, alexithemia, sensitivity to stress and pain, sleep disturbance, and loss of interest in natural rewards (Valdez and Koob, 2004).
Although negative affect that occurs in the context of heavy drinking does not necessarily warrant an independent diagnosis of major depression (MD), clinical data show a large proportion of AD individuals meet criteria for concurrent or lifetime history of MD. Moreover, disturbances in the CRH system, the brain stress system central to Koob's model, is believed to play a central role in the pathogenesis of MD via similar feedback/feedforward processes (de Rijk and de Kloet, 2005). Epidemiological studies of AD and MD comorbidity show there is up to a 4-fold increased lifetime risk for developing one disorder if the other disorder has occurred (Hasin and Grant, 2002; Lynskey, 1998). Comorbidity rates are even higher among treatment-seeking populations (Grant et al., 2004). Research conducted by trained clinicians using standard diagnostic interviews has shown that over two-thirds of severely affected (i.e. hospitalized) AD patients meet criteria for lifetime history of MD (Grant et al., 1989). Among individuals with AD, symptoms of MD predict treatment outcomes, in particular relapse to drinking (Greenfield et al., 1998). These clinical implications further underscore the importance of understanding factors contributing to depressive symptoms in individuals with AD.
The present study examines genetic association of symptom scores from a lifetime history of depression in AD individuals. We conducted a candidate gene association study using a single nucleotide polymorphism (SNP) array designed by the group of D. Goldman at the National Institute on Alcohol Abuse and Alcoholism (NIAAA). The rationale for using this array was twofold: First, genes on the array are selected on the basis of their roles in functional domains important in the related phenotypes of addiction, depression and anxiety (Goldman et al., 2005). In some cases, specific genes and molecules have been identified by GWAS or candidate-gene focused linkage results; other genes are selected solely on the basis of animal studies or for their role in relevant biological pathways (Goldman et al., 2005). Second, along with high clinical comorbidity there is considerable overlap among neurobiological pathways hypothesized to be involved in pathogenesis of alcohol dependence, mood and anxiety disorders including signaling networks, neuroendocrine stress response, and neurotransmitter systems (Belmaker and Agam, 2008; Pacher and Kecskemati, 2004; Vengeliene et al., 2008). Neurotransmitter/neuromodulator systems with potential overlap in AD and MD include serotonin, acetylcholine, dopamine, GABA, opioid, glutamate, CRH and neuropeptide Y systems. With the exception of genes directly involved in drug metabolism, whose role in AD is well documented, we opted to conduct association analysis on the entire array. This includes glutamate, GABA, dopamine, serotonin, and opioid neural system genes, signaling genes, pharmacokinetic genes, and genes modulating stress biology and behavioral control.
Materials and Methods
Participants and Phenotypes
Participants in this study were recruited in Ireland and Northern Ireland. A detailed account of the study design, sample ascertainment and consent procedure is described elsewhere (Prescott et al., 2006). All procedures were approved by the Institutional Review Board at Virginia Commonwealth University, the ethics committees at the Irish Health Research Board and Belfast Health and Social Care Trust, and all the ethics boards at facilities referring participants. Briefly, ascertainment of probands was conducted primarily in public and private hospitals and community alcoholism treatment facilities. Probands were eligible for study inclusion if they met DSM-IV (American Psychiatric Association, 1994) criteria for AD and if all four grandparents had been born in Ireland, Northern Ireland, Scotland, Wales or England. After a prospective family was identified through probands, parents and potentially affected siblings whom the probands provided permission to contact were recruited.
Interviews were conducted by clinically trained research interviewers. The assessment included demographic and ancestral information as well as related traits and lifetime history of symptoms of AD and MD. Depressive symptoms were assessed using the Major Depression module from the SCID-R. Participants were asked about lifetime history of depressed mood or anhedonia lasting at least 2 weeks. Individuals responding positively to either item were assessed for presence of the remaining 7 criteria for a DSM-IV defined MD episode during that period (or during their worst episode if there were multiple periods of 2 weeks of symptoms; Spitzer and Williams, 1985). Co-occurrence of symptoms was required to be considered a positive endorsement, as per standard diagnostic interviews. To maximize comparability to prior literature on depressive symptoms, a depressive symptom score was created by summing the 9 co-occurring DSM-IV symptom criteria. Prior work comparing a continuum of depressive symptoms and MD diagnosis indicates that a continuum of depressive symptoms has greater predictive power (Kendler and Gardner, 1998). Lifetime AD was assessed using the modified SSAGA (Semi-Structured Assessment for the Genetics of Alcoholism) interview and included items to assess the 7 DSM-IV AD criteria (version 11 [Bucholz et al., 1994]). As with the depressive symptom score, an AD symptom score was created by summing the number of co-occurring DSM-IV AD symptom areas endorsed. Participants were also asked whether their alcohol problems occurred always, sometimes or never in conjunction with their depressive symptoms. Of 425 individuals with clinically significant levels of depressive symptoms (those meeting criteria for MD diagnosis), few participants reported either complete separation (3.5%) or complete overlap (4.9%) with regards to co-occurrence of depressive and alcohol dependence symptoms.
All participants provided informed consent. There were 1238 individuals who completed the SSAGA interview and met criteria for DSM-IV AD diagnosis, including 591 probands and 647 affected siblings. In families where more than one AD individual provided a DNA sample, one participant was randomly drawn for inclusion in the study. Genotyping was performed on 575 independent cases; of which 13 were dropped for poor DNA quality (see Data QC) and 8 for insufficient information regarding depression, yielding a final sample of 554 (354 male, 200 female). Demographics, drinking history and treatment use are shown in Table 1. This was a severely affected AD sample, with 84% of the sample endorsing at least 6 of 7 possible AD criteria, 73% of the sample having undergone 2+ different types of treatment (e.g. self-help group, inpatient, outpatient, hospital, other), with the median number of maximum drinks consumed in a 24-hour period equaling 36 drinks.
Table. 1. Demographic and clinical characteristics of the sample.
| Variable | Statistic |
|---|---|
| Age at interview M(SD) | 41.8 (9.6) |
| Sex (% female) | 36.1 |
| Marital status | |
| Married % | 39.9 |
| Divorced/Separated % | 27.8 |
| Widowed % | 2.7 |
| Never married % | 29.1 |
| Education | |
| < Secondary % | 11.3 |
| Some secondary % | 34.3 |
| Completed secondary % | 39.0 |
| Some postsecondary % | 15.4 |
| Employment | |
| >20 hours % | 44.8 |
| Unemployed % | 36.6 |
| Other (student, homemaker, retired) % | 18.6 |
| MD diagnosis % | 75.1 |
| Drinking history | |
| Age at first drink M(SD) | 15.7 (4.5) |
| Age at first intoxication M(SD) | 17.4 (6.0) |
| Age began drinking regularly M(SD) | 18.0 (5.3) |
| Age began drinking most heavily M(SD) | 31.8 (9.4) |
| Max drinks in 24 hours M(SD) | 39.2 (19.2) |
| Age meeting AD criteria M(SD) | 26.2 (8.9) |
| Treatment | |
| Any treatment % | 79.6 |
| Alcoholics Anonymous % | 72.2 |
| Inpatient alcohol treatment facility % | 64.8 |
| Outpatient alcohol treatment facility % | 49.6 |
| Medical facility % | 21.3 |
| Other treatment % | 9.7 |
Single Nucleotide Polymorphisms
A detailed description of the NIAAA “addictions array” has been published previously (Hodgkinson et al., 2008). Briefly, candidate genes were selected based on molecular pathways or prior evidence from human or animal studies suggesting involvement in the related phenotypes of addiction, depression and anxiety. Genes assessed in this study are indicated in Table 2. Of particular interest for an examination of depressive symptoms among alcohol dependent individuals are genes in systems potentially implicated in studies of both AD and MD, including serotonin, acetylcholine, dopamine, GABA, opioid, NMDA (glutamate), and stress (HPA axis and neuropeptide Y). Genomic regions containing 5kb upstream and 1 kb downstream of each candidate gene were screened for haplotype tagging single nucleotide polymorphisms (SNPs) selected to reconstruct all haplotypes with frequency greater than 0.5% observed in HAPMAP data from Africans, the most genetically diverse of human population groups. Performance of initially selected SNPs was validated by the manufacturer and replacements made when necessary. A total of 1536 loci on a custom Illumina array were genotyped, including 1350 SNPs and 186 ancestry informative markers (AIMs). AIMs were previously developed for and tested on the Illumina platform (Enoch et al., 2006) and were analyzed using structure 2.1 (Pritchard et al., 2000) to generate population assignments for all individuals.
Table 2. Genes analyzed.
| Adrenergic | GABA | Stress | Signal Transduction | Serotonin |
| ADRA1A | GABRA2 | CRH | ADCY7 | HTR1A |
| ADRA2A | GABRA3 | CRHBP | AVPR1A | HTR1B |
| ADRA2B | GABRA4 | CRHR1 | AVPR1B | HTR2A |
| ADRA2C | GABRA6 | CRHR2 | CDK51R | HTR2B |
| ADRB2 | GABRB1 | FKBP5 | CREB1 | HTR2C |
| ARRB2 | GABRB2 | GAL | CSNK1E | HTR3A |
| SLC6A2 | GABRB3 | NPY | FEV | HTR3B |
| DBH | GABRD | NPY1R | FOS | MAOA |
| GABRE | NPY2R | FOSL1 | MAOB | |
| Cholinergic | GABRG2 | NPY5R | FOSL2 | SLC6A4 |
| CHRM1 | GABRG3 | NR3C1 | GSK3B | TPH1 |
| CHRM2 | GABRQ | JUN | TPH2 | |
| CHRM3 | SLC6A7 | Dopamine | MAPK1 | |
| CHRM4 | SLC6A11 | COMT | MAPK3 | Other |
| CHRM5 | SLC6A13 | DDC | MAPK14 | BDNF |
| CHRNA4 | SLC32A1 | DRD1 | MPDZ | CART |
| CHRNB2 | GAD1 | DRD2 | NGFB | CCK |
| GAD2 | DRD3 | NTRK2 | CCKAR | |
| Opioid | DBI | DRD4 | NTSR1 | CCKBR |
| OPRM1 | DRD5 | NTSR2 | CLOCK | |
| OPRD1 | NMDA | SLC1BA2 | PPP1R1B | HCRT |
| OPRK1 | GRIK1 | SLC6A3 | PRKCE | LEP |
| OPRL1 | GRIN1 | TH | OXT | |
| PDYN | GRIN2A | Glycine | SLC29A1 | |
| PENK | GRIN2B | Cannabinoid | GLRA1 | TAC1 |
| PNOC | GRIN2C | CNR1 | GLRA2 | |
| POMC | GRM1 | FAAH | GLRB | |
| GPHN |
An additional 10 SNPs were selected in-house to tag FKBP5, which was not included on the array, and AVPR1B, for which there were no SNPs informative for Caucasian populations on the array. These SNPs were genotyped using an automated liquid handling system EP 5075 (Eppendorf) and standard lab procedures for Taqman genotyping (Applied Biosystems Inc).
Data analysis
Association analyses were conducted at the single marker level using SAS 9.1. Mantel-Haenszel χ2 for linear association was computed to test the association of depressive symptom scores and genotype of each marker while controlling for potential confounds of age and sex. X-linked markers were assessed for males and females separately. As expected with a clinically ascertained sample, depressive symptom scores were high (M=6.02, SD=3.4), with wide variability. One hundred and nineteen participants did not report any lifetime history of depression. Because the scores showed a bimodal distribution, normality assumptions required for linear regression analyses would have been violated. We therefore treated the depressive symptom score as an ordinal scale. To maximize generalizability to clinical populations, we grouped individuals with scores below the diagnostic cut-off of DSM-IV criteria at one level (0-4 criteria N=134) and the remaining individuals in 4 additional levels reflecting increasing numbers of depression criteria above the DSM-IV threshold for caseness (5-6 criteria N=70; 7 criteria N=65; 8 criteria N= 128; 9 criteria N=157). The ordinalized depressive symptom measure was highly correlated with the original score (raw total count of depressive symptom areas endorsed; Pearson's r = .93, p<.001). Power calculations for the depressive symptom measure are reported in Supplementary Table 1.
Because alcohol and depressive symptoms tend to be highly comorbid, we conducted a parallel Mantel-Haenszel χ2 analysis testing association of allelic frequencies with the alcohol symptom scores to examine whether associations were unique to endorsement of symptoms of depression or common to depression and alcohol dependence. Because the sample was clinically ascertained, the total alcohol dependence criteria endorsed was high (M=6.42, SD=0.96) and not normally distributed. For this reason, and to compare this scale with the depressive symptom scale, the alcohol dependence symptom scale was also treated as an ordinal variable (3-4 criteria N=36, 5 criteria N=50, 6 criteria N= 101, 7 criteria N=367). The number of individuals at each level of the depressive symptom and alcohol symptom scales are provided in Supplementary Table 2. There was a significant but modest correlation in the depressive symptom and alcohol dependence scores, (ρ=0.26, p<.01).
Determining appropriate threshold for significance was a challenge for this study. Candidate genes were selected on the basis of biological pathways purported to be involved in a limited set of related phenotypes, and thus many tests have higher a priori probabilities of yielding significant results than random genomic regions. Here, we consider results based on experimentwise permutation analysis, a conservative approach to interpreting significant results. We use the term observed p values to refer to nominal significance where alpha is set at p<.05. Empirical P values refer to p values generated through experimentwise permutation analysis.
Experimentwise permutation was conducted by randomly permuting depressive symptom scores across the sample while holding genotype fixed and retaining the correlational structure of all SNPs. Observed p values were compared to permuted p value distributions under the null hypotheses to generate empirical P values. Inference was based on 1000 permutations. To interpret the results of experimentwise permutation, we utilized the method introduced by Lander and Kruglyak (1995) used extensively in linkage analyses. Under this approach we consider a P value threshold that occurs by chance once per experiment as significant. Experimentwise thresholds can be easily calculated if all the tests performed are independent. In this study many tested SNPs were in high LD, thus analyses were conducted on empirical P values where LD structure was preserved. We determine experimentwise thresholds as Nth position where N=number of permutations.
Results
Data QC and stratification analysis
Among the 1360 genotyped SNPs, 4.7% were excluded on the basis of low genotyping rate (<0.5) and 25.2% on the basis of minor allele frequency (MAF) <0.05 and/or violation of Hardy-Weinberg equilibrium (<.001). Because markers on the array were originally selected for tagging in a different ethnic group, many markers would have been redundant in our sample. Thus we used genetic data from a sample of 530 Irish community controls previously collected for a study of AD (Prescott et al., 2006) to determine LD among SNPs on the Addictions Array. Non-independence was defined as r2 values exceeding .80. Where r2>.80 one SNP was randomly selected using the program Haploview (Barrett et al., 2005) for inclusion in analysis, resulting in a final SNP set of 644 markers. Thirteen individuals were excluded on the basis of low genotyping rate (<50%). The average genotyping rate of the remaining individuals was 92%. Of 186 AIMs, 4.8% were excluded for low genotyping rate.
Analyses of population structure testing two or three maximum populations were consistent in showing evidence for one ethnic cluster. For the two population test, individuals' assignments to population 1 was Mean=0.50, sigma =0.02 (Min p=0.45 max p=0.55). Discrepancies from the mean were very small, indicating there was only one population. Assessments of AIM data showed no evidence for stratification, as expected in this single-ethnicity sample.
Associations with age and sex
A small but significant correlation was observed for number of depressive symptom areas/criteria endorsed and age at time of interview (r=-.14, p<.01) with older participants on average endorsing a greater number of criteria. The number of depressive symptom areas endorsed was not correlated with sex (M=5.92 in men and 6.21 in women, t=0.97, ns). The lack of a sex difference in depressive symptoms is consistent with epidemiological data by Van de Velde et al. (2010) showing no sex difference for MD diagnoses in Ireland. That report also showed that the difference in prevalence of MD between men and women in Ireland is the smallest among 23 European countries. However, because sex differences are commonly observed in other cultures and may relate to biological or demographic factors (Van de Velde et al., 2010), both age and sex covariates were preserved for possible measured and unmeasured effects.
Single marker association
Results of single marker tests are indicated in Table 3. Mantel-Haenszel χ2 for linear association was computed to examine the association of depressive symptom scores with genotype after controlling for possible confounding effects of sex and age, where age was binned into 5-year intervals to accommodate the Mantel-Haenszel χ2 test. To examine whether associations were being driven by individuals whose depressive symptoms occurred only in the context of drinking, we ran associations on a subset of individuals excluding the 21 individuals who reported symptoms of MD and AD always occurred together. None of the significant effects differed from those of the full data set, and so data from the full sample are reported. Nominal associations were observed for 31 SNPs (observed p's<.05) in multiple molecular pathways and functional domains. They include the adrenergic, cholinergic, dopaminergic, opioidergic and serotonergic systems, GABA, NMDA receptors, as well as molecules involved in signal transduction and the neuroendocrine stress response.
Table 3. Single-marker association with depressive symptom score.
| Marker* | Gene | Chr. | Bp* | Position* | MAF | χ2 | Observed p |
|---|---|---|---|---|---|---|---|
| rs11688 | JUN | 1 | 59247993 | Exonic synon | 0.05 | 5.24 | 0.0221 |
| rs10495447 | CHRM3 | 1 | 240051340 | Intronic | 0.26 | 3.92 | 0.0478 |
| rs4688043 | GSK3B | 3 | 119591248 | Intronic | 0.08 | 5.51 | 0.0189 |
| rs1442060 | GABRA2 | 4 | 46366067 | Intronic | 0.45 | 5.34 | 0.0208 |
| rs2236781 | GABRB1 | 4 | 47035087 | Intronic | 0.49 | 5.76 | 0.0164 |
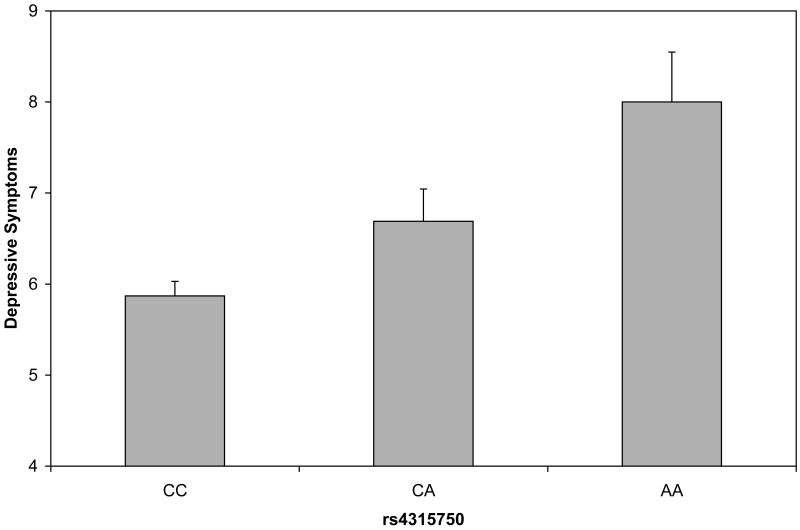
| rs4315750 | GABRB1 | 4 | 47051185 | Intronic | 0.09 | 9.64 | 0.0019 |
| rs971353 | GABRB1 | 4 | 47102894 | Intronic | 0.28 | 3.95 | 0.0469 |
| rs9996854 | GABRB1 | 4 | 47340895 | Intronic | 0.36 | 5.97 | 0.0145 |
| rs6350 | SLC6A3 | 5 | 1443199 | Exonic synon | 0.08 | 5.63 | 0.0176 |
| rs10474485 | CRHBP | 5 | 76270853 | 3′ | 0.16 | 5.83 | 0.0157 |
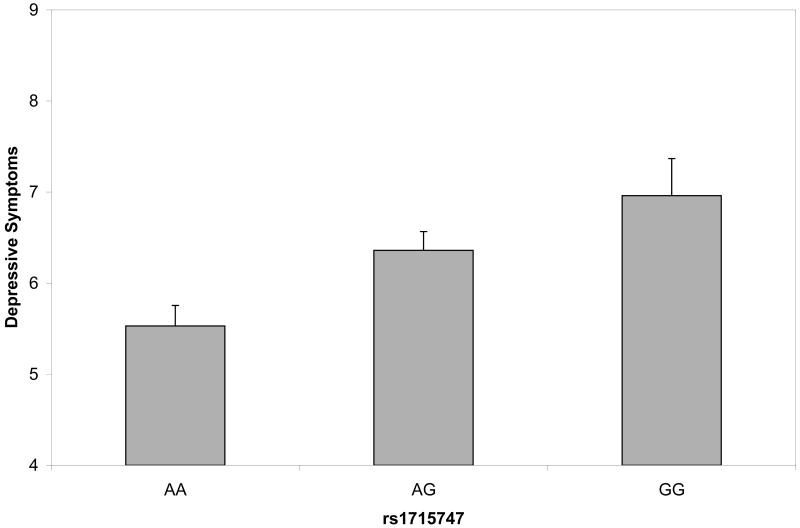
| rs1715747 | CRHBP | 5 | 76274537 | 3′ | 0.31 | 10.61 | 0.0011 |
| rs1042718 | ADRB2 | 5 | 148206917 | Exonic synon | 0.14 | 4.03 | 0.0446 |
| rs7724086 | GABRB2 | 5 | 160724214 | Intronic | 0.09 | 5.82 | 0.0158 |
| rs1799971 | OPRM1 | 6 | 154360797 | Exonic missense | 0.10 | 4.80 | 0.0285 |
| rs3778151 | OPRM1 | 6 | 154393680 | Intronic | 0.17 | 5.12 | 0.0237 |
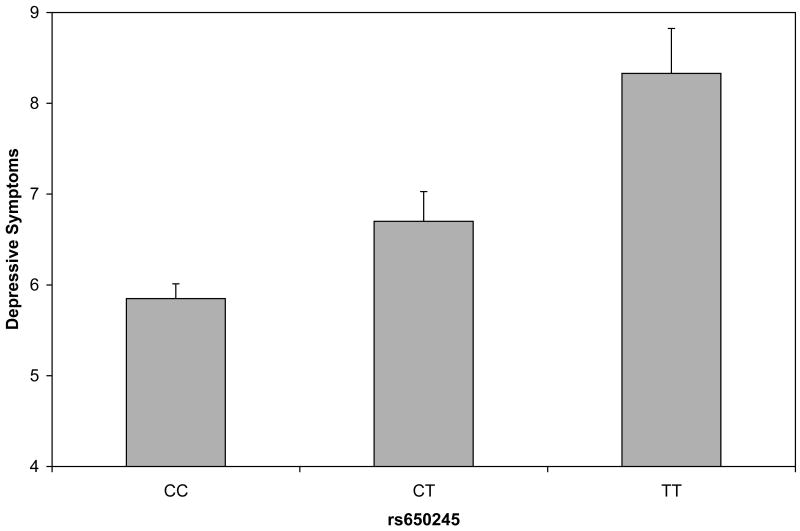
| rs650245 | OPRM1 | 6 | 154428702 | Intronic† | 0.10 | 9.86 | 0.0017 |
| rs548339 | OPRM1 | 6 | 154460799 | Intronic | 0.33 | 3.95 | 0.0468 |
| rs10488599 | CHRM2 | 7 | 136589094 | Intronic | 0.15 | 7.06 | 0.0079 |
| rs10102186 | ADRA1A | 8 | 26624651 | Intronic | 0.31 | 4.26 | 0.0390 |
| rs6356 | TH | 11 | 2190951 | Exonic missense | 0.37 | 5.30 | 0.0213 |
| rs2587548 | DRD2 | 11 | 113292212 | Intronic | 0.40 | 4.08 | 0.0434 |
| rs1125394 | DRD2 | 11 | 113297185 | Intronic | 0.16 | 5.55 | 0.0185 |
| rs4274224 | DRD2 | 11 | 113319452 | Intronic | 0.46 | 5.07 | 0.0244 |
| rs1352252 | TPH2 | 12 | 72452041 | 3′ | 0.37 | 4.43 | 0.0352 |
| rs11179071 | TPH2 | 12 | 72455185 | 3′ | 0.14 | 6.18 | 0.0129 |
| rs6582086 | TPH2 | 12 | 72498191 | 3′ | 0.18 | 5.68 | 0.0172 |
| rs12324292 | GABRB3 | 15 | 27003081 | Intronic | 0.31 | 5.27 | 0.0217 |
| rs4611457 | ADCY7 | 16 | 50295994 | 5′ | 0.47 | 6.74 | 0.0094 |
| rs7219247 | GRIN2C | 17 | 72847205 | Intronic | 0.27 | 5.85 | 0.0156 |
| rs2235751 | PDYN | 20 | 1969934 | Intronic | 0.20 | 4.10 | 0.0429 |
| rs460401 | GRIK1 | 21 | 31176765 | Intronic | 0.10 | 3.95 | 0.0469 |
As identified in dpSNP Build 129
NCBI lists this marker as intronic. Recent work identifies it as exonic or in the 3′-UTR of some isoforms (Shabalina et al., 2009)
Analysis of experimentwise permutation
Depressive symptom scores were permuted across participants 1000 times while retaining the correlational structure of SNPs to generate empirical P values. Using the Lander and Kruglyak (1995) approach to interpretation of experimentwise permutation, the threshold for significance was set at p=0.0017. Empirical P values below the threshold included the CRHBP marker 1715747 and the OPRM1 marker rs650245 (empirical P's=0.0010, 0.0014). The GABRB1 marker rs4315750 was at the permutation threshold (empirical P=.0017). Figure 1 shows the relation of the genotypes to depressive symptom scores. In each case, the minor allele was additively associated with higher depressive symptom scores. Associated allele frequencies for each depressive symptom score are indicated in Table 4. Notably, none of the markers that were significantly associated with depressive symptom scores were even nominally associated with alcohol dependence symptom scores (see Supplementary Table 3).
Figure 1.
Depressive symptom scores plotted by variants in CRHBP (fig. 1a), OPRM1 (fig 1b) and GABRB1 (fig 1c). Analyses were conduced treating the depressive symptom score as an ordinal measure statistically correcting for age and sex; data are shown on the original scale for ease of interpretation. Error bars denote standard error of the mean.
Table 4. Associated allele frequency for each level of depressive symptoms scale.
| Gene | Marker | 0-4 Symptoms† | 5-6 Symptoms | 7 Symptoms | 8 Symptoms | 9 Symptoms |
|---|---|---|---|---|---|---|
| CRHBP | rs1715747 | .233 | .286 | .315 | .372 | .334 |
| GABRB1 | rs4315750 | .059 | .057 | .069 | .098 | .127 |
| OPRM1 | rs650245 | .059 | .071 | .069 | .118 | .127 |
Individuals below clinical threshold for MD diagnosis
Discussion
The purpose of this study was to examine the association of potential candidate genes with lifetime history of depressive symptoms in alcohol-dependent individuals. Initial results revealed significant associations for markers in 20 genes. Because there is not yet a clear consensus as to the most appropriate method of multiple testing correction for genetic association analysis, we used experimentwise permutation, a conservative approach to multiple testing. Of the 20 genes reaching nominal significance, SNPs in CRHBP, OPRM1, and GABRB1 were at or below the multiple test correction threshold.
CRHBP codes for the corticotropin releasing hormone binding protein that regulates the bioavailability of CRH to act at its receptors, inhibits CRH activation of the HPA axis, and may be involved in HPA axis feedback and feedforward regulation via effects in limbic system structures (Behan et al., 1995; Van Den Eede et al., 2005). The CRH system is central to both the neuroendocrine hypothesis of depression (DeRijk and de Kloet, 2005) and the proposed anti-reward mechanism promoting the transition to, maintenance of, and relapse in addiction (Koob and Le Moal, 2008). Moreover, HPA axis feedback influences reward pathways and GABAA receptor function involved in ethanol sensitivity (Morrow et al., 2006; Oswald and Wand, 2004). Family studies show altered HPA axis activity in relatives of depressed patients and HPA response to ethanol in sons of alcoholics (DeRijk and de Kloet, 2005; Schuckit et al., 1987; Zimmerman et al., 2004). Two CRHBP SNPs were nominally significant in our study, with one passing threshold for experimentwise correction. The CRHBP SNP significant in our study was associated with anxiety disorder and resting EEG activity in an independent sample of AD individuals (Enoch et al., 2008). These findings along with our results on depressive symptoms imply genetic variation in CRHBP may play a role in affective disturbance in AD individuals.
All CRHBP markers tested were tagSNPs. Thus the significant SNP does not have any putative function. HapMap data show LD decays rapidly both proximally and distally from CRHBP, suggesting the observed associations cannot be explained by effects in surrounding genes. CRHBP encodes a 322 amino acid protein with 5 disulphide bonds essential for CRH binding. Single amino acid changes or intronic variants affecting exon content of CRHBP isoforms identified in brain might directly affect peptide sequence and alter folding, stability and/or CRH binding affinity. CRHBP is highly conserved across vertebrates including regions of high conservation in the 3′-UTR (possibly affecting mRNA turnover) and 3′-region introns (possibly influencing alternative exon usage).
A significant association was also observed for OPRM1. The mu-opioid receptor, which binds β-endorphin and the enkephalins, is typically activated in response to chronic or unpredictable stressful or noxious stimuli (Oswald and Wand, 2004). The opioidergic system is extensively studied in addiction and to a lesser extent in depression. Naltrexone, an opioid antagonist, is used in treatment of AD and reduces relapse. Depressed patients demonstrate antidepressant effects of opioid peptides and enkephalinase inhibitors (Broom et al., 2002; Jutkiewicz et al., 2006). Differences in mu-opioid receptor availability are reported among depressed patients compared to controls (Kennedy et al., 2006). One mechanism of action consistent with both the neuroendocrine hypothesis of depression and Koob's model of negative affect in the cycle of addiction may be the inhibitory role of opioid neurons in facilitating the termination of the biological stress response (Drolet et al., 2001; Kreek, 1996). Notably, a well-studied OPRM1 missense mutation with known consequences for receptor structure (rs1799971) has been associated with HPA axis activity (Chong et al., 2006; Wand et al., 2002), suicide (Hishimoto et al., 2008), alcohol sensitivity (Ray and Hutchison, 2004), and alcohol use disorders in adolescents and adults (Bart et al., 2005; Miranda et al., 2010) although a meta-analyses revealed several null associations with AD diagnosis (Arias et al., 2006). Genetic variation in OPRM1 may have functional consequences at the interface of disturbed HPA axis activity, depression, and addiction.
In our study, rs1799971 and three other OPRM1 markers reached nominal significance. One marker, rs650245, was strongly associated with depressive symptoms and survived a stringent test of multiple test correction. Early reports of the genetic architecture of OPRM1 identified this SNP as intronic. Recent work identifies this marker as exonic or in the 3′-UTR of some OPRM1 isoforms (Shabalina et al., 2009). Moreover, rs650245 is in high LD with and lies in the same haplotype block as known functional SNPs and a newly identified, potentially functional SNP (Shabalina et al., 2009). Markers in this haplotype block are associated with highly related phenotypes, including treatment response in MD individuals, pain threshold, negative mood, and substance dependence (Garriok et al., 2010; Shabalina et al., 2009). To our knowledge this is the first report of an OPRM1 marker associated with depressive symptoms. Taken together, these findings strongly suggest variation within the OPRM1 gene may contribute to risk for depressive and addictive disorders.
GABRB1 codes for the β1 subunit of gamma-aminobutyric acid A (GABAA) receptors. The GABAA receptor is a heteropentameric chloride channel that mediates inhibitory synaptic transmission via binding of GABA. There are 19 mammalian subunits; most GABA receptors contain α, β, and γ subunits. The majority of genes encoding human GABAA receptor subunits are organized in clusters, which may have functional significance. There is some evidence that altering the structure of one GABA subunit gene affects expression of the other genes in the same cluster (Uusi-Oukari et al., 2000).
Extensive research in rodents indicates variations in the GABA receptor genes contribute to differences in risk for alcoholism. Family based studies show modest association with alcohol dependence for GABRB1 (Reck et al., 2005; Song et al., 2003). A microsatellite polymorphism in GABRB1 has also been linked with AD (Parsian and Zhang, 1999). Association of GABRA2, which is clustered with GABRB1 but was only nominally associated with depressive symptoms in our study, has in other reports been associated with AD and resting EEG activity (e.g., Edenberg et al., 2004). The significant GABRB1 SNP in our study is intronic and in low LD with surrounding SNPs; thus the relation of our findings to previous reports or in terms of potential functional consequences is difficult to determine. To our knowledge there are no published reports examining association of GABRB1 with MD, although this gene has been investigated in relation to bipolar disorder (Coon et al., 1994; Crowe et al., 1997). Centrally involved in the neurobiology of addiction, GABA mediates the behavioral effects of ethanol. It is perhaps noteworthy that the β1 subunit is preferentially expressed in the hippocampus (Michels and Moss, 2007), which provides the major GABAergic inhibitory input to CRH neurons initiating the HPA axis and for which reduced volumes have been reported in depressed patients (Herman et al., 2005; Sheline et al., 1996). This finding is also consistent with Koob's model of the role of limbic system feedback of the HPA axis in the negative affect component of the addiction cycle.
Limitations and considerations
There are several limitations to the present results. The depressive symptom scale was created from a structured interview designed to assess clinical diagnoses. We opted to utilize the full range of scores for the number of symptom areas endorsed, rather than dividing individuals only above/below clinical threshold because of evidence suggesting the continuum of depressive symptoms has greater predictive power than standard diagnostic conventions (Kendler and Gardner, 1998). Our treatment of the data, which was non-normally distributed, as ordinal groups with one group representing all individuals below the clinical threshold for MD, and other groups reflecting increasing numbers of endorsed symptom areas, maximized comparability to diagnostic studies while maintaining the validity of the statistical results. In addition, lifetime histories of depression and alcohol use problems were assessed retrospectively, which may have resulted in unmeasured recall error or bias. Replication of key findings in a prospectively studied sample is warranted.
Participants were ascertained from hospitals and treatment facilities for alcohol use disorders. It is possible that observed associations were simply due to severe alcoholism. However, none of the markers associated with depressive symptom scores in this study were significantly associated with a comparable index of alcohol dependence symptoms. Although the analysis of alcohol dependence symptoms is of somewhat limited utility because of range restriction, it provides some confidence that observed associations were unique to the depressive symptoms. Moreover, the correlation among the number of depressive and alcohol dependence symptoms was modest.
Our sample was severely affected with AD, with the majority of participants endorsing most or all of the AD criteria. The rate of MD comorbidity was also high; however, our comorbidity rate was similar to another study of mostly male, severely affected (hospitalized) AD patients whose symptoms were assessed via structured clinical interview (Grant et al., 1989). Thus, our results must be interpreted in light of the severely affected nature of the sample. The rate of alcohol-induced MD (Furgusson et al., 2009) or something reminiscient of depressive-spectrum disorder (Winoker et al., 1971; see also Nurnberger et al., 2001) might be enhanced in samples of severe AD. Whether the associations observed here would be detected in AD individuals from a community-based sample or one in which the incidence of severe AD was lower is worthy of investigation.
Depressive symptoms may be causally involved in or result from alcohol dependence in different individuals. We were unable to distinguish with confidence the temporal or etiologic relationship of alcohol and depression problems in this sample. Thus we cannot determine whether genes associated with depressive symptom scores may be unique to depressive symptoms in AD individuals, susceptibility genes to both AD and MD, or modifier genes, that is, genes that modify susceptibility to one disorder in the presence of or genetic risk for the other. Reciprocal effects of the HPA, opioidergic and mesolimbic reward pathways implicated in alcohol use disorders suggest that altered activity in one system, owing to either genetic or other factors could simultaneously impact HPA axis function, mesolimbic dopamine production, and ethanol reinforcement (Oswald and Wand, 2004). Future research examining genetic contributions to depressive symptoms in the context of various developmental pathways to alcohol dependence is warranted. In addition, examining these genes in subjects ascertained initially for MD will be important to establish whether associations identified with depressive symptoms are common to or distinct from primary MD. It is perhaps noteworthy that some genes previously published for association with MD such as FKBP5 were not associated with depressive symptoms in this sample (Lekman et al., 2008). Similarly, GABRA2 and the 1799971 SNP in OPRM1, both of which have been associated with AD in other samples, were only nominally associated with depressive symptoms among AD individuals in this study. There is some evidence to suggest that AD/MD comorbid individuals may manifest a different course of illness than noncomorbid individuals (Grant et al., 2004) and that comorbid AD/MD tends to aggregate in relatives of individuals with this comorbidity (Nurnberger et al., 2001). This raises the possibility of genetic vulnerability to the comorbid phenotype. Careful attention to a history of comorbid symptoms is clearly warranted in future studies.
Conclusion
In sum, this study investigated the genetic contribution to depression symptoms in alcohol dependent individuals. Nominal associations were noted for markers in 20 genes. Following experimentwise permutation, markers in three genes were at or below the threshold for significance. The strongest signals were in the CRH system, which is potentially involved in both the negative affect component of the addiction cycle and the pathophysiology of major depression, and the opioidergic and GABAergic systems, both of which are implicated in AD and provide the major inhibitory input to the CRH system. These findings suggest further investigation may be warranted of genes involved in regulation of stress biology as they contribute to depressive symptoms comorbid with and potentially contributing to the cycle of addiction.
Supplementary Material
Acknowledgments
This work was supported by a National Institute on Alcohol Abuse and Alcoholism grant (AA011408) to Dr. Kendler and a National Research Service Award (MH020030) from the National Institute of Mental Health to Dr. Kertes through the Virginia Institute for Psychiatric and Behavioral Genetics. The authors thank C. Hodgkinson and D. Goldman for genotyping the Illumina array and Charles Gardner for statistical consultation.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. fourth. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83(3):262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, Heilig M. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacol. 2005;30(2):417–422. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- Behan DP, De Souza EB, Lowry PJ, Potter E, Sawchenko P, Vale WW. Corticotropin releasing factor (CRF) binding protein: a novel regulator of CRF and related peptides. Front Neuroendocrinol. 1995;16(4):362–382. doi: 10.1006/frne.1995.1013. [DOI] [PubMed] [Google Scholar]
- Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358(1):55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- Broom DC, Jutkiewicz EM, Rice KC, Traynor JR, Woods JH. Behavioral effects of delta-opioid receptor agonists: potential antidepressants? Jpn J Pharmacol. 2002;90(1):1–6. doi: 10.1254/jjp.90.1. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Chong RY, Oswald L, Yang X, Uhart M, Lin PI, Wand GS. The mu-opioid receptor polymorphism A118G predicts cortisol responses to naloxone and stress. Neuropsychopharmacology. 2006;31(1):204–211. doi: 10.1038/sj.npp.1300856. [DOI] [PubMed] [Google Scholar]
- Claes SJ. Corticotropin-releasing hormone (CRH) in psychiatry: from stress to psychopathology. Ann Med. 2004;36(1):50–61. doi: 10.1080/07853890310017044. [DOI] [PubMed] [Google Scholar]
- Coon H, Hicks AA, Bailey ME, Hoff M, Holik J, Harvey RJ, Johnson KJ, Darlison MG, Reimherr F, Wender P, Byerley W. Analysis of GABAA receptor subunit genes in multiplex pedigrees with manic depression. Psychiatr Genet. 1994;4(3):185–191. doi: 10.1097/00041444-199400430-00009. [DOI] [PubMed] [Google Scholar]
- Crowe RR, Wang Z, Noyes R, Jr, Albrecht BE, Darlison MG, Bailey ME, Johnson KJ, Zoëga T. Candidate gene study of eight GABAA receptor subunits in panic disorder. Am J Psychiatry. 1997;154(8):1096–1100. doi: 10.1176/ajp.154.8.1096. [DOI] [PubMed] [Google Scholar]
- DeRijk R, de Kloet ER. Corticosteroid receptor genetic polymorphisms and stress responsivity. Endocrine. 2005;28(3):263–270. doi: 10.1385/ENDO:28:3:263. [DOI] [PubMed] [Google Scholar]
- Drolet G, Dumont EC, Gosselin I, Kinkead R, Laforest S, Trottier JF. Role of endogenous opioid system in the regulation of the stress response. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25(4):729–741. doi: 10.1016/s0278-5846(01)00161-0. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Numberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: Identifying specific genes through family studies. Addict Biol. 2006;11(3-4):386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Ducci F, Yuan Q, Liu J, White KV, Albaugh B, Hodgkinson CA, Goldman D. Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PLoS ONE. 2008;3(10):e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20(4 Suppl):19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Fergusson DM, Boden JM, Horwood LJ. Tests of causal links between alcohol abuse or dependence and major depression. Arch Gen Psychiatry. 2009;66(3):260–266. doi: 10.1001/archgenpsychiatry.2008.543. [DOI] [PubMed] [Google Scholar]
- Garriock HA, Tanowitz M, Kraft JB, Dang VC, Peters EJ, Jenkins GD, Reinalda MS, McGrath PJ, von Zastrow M, Slager SL, Hamilton SP. Association of mu-opioid receptor variants and response to Citalopram treatment in major depressive disorder. Am J Psychiatry. 2010;167(5):565–573. doi: 10.1176/appi.ajp.2009.08081167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Grant BF, Hasin DS, Harford TC. Screening for major depression among alcoholics: an application of receiver operating characteristic analysis. Drug Alcohol Depend. 1989;23(2):123–131. doi: 10.1016/0376-8716(89)90017-3. [DOI] [PubMed] [Google Scholar]
- Grant BF, Stinson FS, Dawson DA, Chou SP, Dufour MC, Compton W, Pickering RP, Kaplan K. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2004;61(8):807–816. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- Greenfield SF, Weiss RD, Muenz LR, Vagge LM, Kelly JF, Bello LR, Michael J. The effect of depression on return to drinking: a prospective study. Arch Gen Psychiatry. 1998;55(3):259–265. doi: 10.1001/archpsyc.55.3.259. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Grant BF. Major depression in 6050 former drinkers: association with past alcohol dependence. Arch Gen Psychiatry. 2002;59(9):749–800. doi: 10.1001/archpsyc.59.9.794. [DOI] [PubMed] [Google Scholar]
- Herman JP, Ostrander MM, Mueller NK, Figueiredo H. Limbic system mechanisms of stress regulation: hypothalamo-pituitary-adrenocortical axis. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29(8):1201–1213. doi: 10.1016/j.pnpbp.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Hishimoto A, Cui H, Mouri K, Nushida H, Ueno Y, Maeda K, Shirakawa O. A functional polymorphism of the micro-opioid receptor gene is associated with completed suicides. J Neural Transm. 2008;115(3):531–536. doi: 10.1007/s00702-007-0853-y. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutkiewicz EM, Torregrossa MM, Sobczyk-Kojiro K, Mosberg HI, Folk JE, Rice KC, Watson SJ, Woods JH. Behavioral and neurobiological effects of the enkephalinase inhibitor RB101 relative to its antidepressant effects. Eur J Pharmacol. 2006;531(1-3):151–159. doi: 10.1016/j.ejphar.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Gardner CO., Jr Boundaries of major depression: an evaluation of DSM-IV criteria. Am J Psychiatry. 1998;155(2):172–177. doi: 10.1176/ajp.155.2.172. [DOI] [PubMed] [Google Scholar]
- Kennedy SE, Koeppe RA, Young EA, Zubieta JK. Dysregulation of endogenous opioid emotion regulation circuitry in major depression in women. Arch Gen Psychiatry. 2006;63(11):1199–1208. doi: 10.1001/archpsyc.63.11.1199. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Review. Neurobiological mechanisms for opponent motivational processes in addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363(1507):3113–3123. doi: 10.1098/rstb.2008.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ. Opioid receptors: some perspectives from early studies of their role in normal physiology, stress responsivity, and in specific addictive diseases. Neurochem Res. 1996;21(11):1469–1488. doi: 10.1007/BF02532387. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lekman M, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, Lipsky R, Wisniewski SR, Manji H, McMahon FJ, Paddock S. The FKBP5-gene in depression and treatment response--an association study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol Psychiatry. 2008;63(12):1103–1110. doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovallo WR. Cortisol secretion patterns in addiction and addiction risk. Int J Psychophysiol. 2006;59(3):195–202. doi: 10.1016/j.ijpsycho.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey MT. The comorbidity of alcohol dependence and affective disorders: treatment implications. Drug Alcohol Depend. 1998;52(3):201–209. doi: 10.1016/s0376-8716(98)00095-7. [DOI] [PubMed] [Google Scholar]
- McGue M. The behavioral genetics of alcoholism. Curr Directions Psychol Sci. 1999;8:109–115. [Google Scholar]
- Michels G, Moss SJ. GABAA receptors: properties and trafficking. Crit Rev Biochem Mol Biol. 2007;42(1):3–14. doi: 10.1080/10409230601146219. [DOI] [PubMed] [Google Scholar]
- Miranda R, Ray L, Justus A, Meyerson LA, Knopik VS, McGeary J, Monti PM. Initial evidence of an association between OPRM1 and adolescent alcohol misuse. Alcohol Clin Exp Res. 2010;34(1):112–122. doi: 10.1111/j.1530-0277.2009.01073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow AL, Porcu P, Boyd KN, Grant KA. Hypothalamic-pituitary-adrenal axis modulation of GABAergic neuroactive steroids influences ethanol sensitivity and drinking behavior. Dialogues Clin Neurosci. 2006;8(4):463–477. doi: 10.31887/DCNS.2006.8.4/amorrow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158(5):718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Oswald LM, Wand GS. Opioids and alcoholism. Physiol Behav. 2004;81(2):339–358. doi: 10.1016/j.physbeh.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Pacher P, Kecskemeti V. Trends in the development of new antidepressants. Is there a light at the end of the tunnel? Curr Med Chem. 2004;11(7):925–943. doi: 10.2174/0929867043455594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A, Zhang ZH. Human chromosomes 11p15 and 4p12 and alcohol dependence: possible association with the GABRB1 gene. Am J Med Genet. 1999;88(5):533–538. doi: 10.1002/(sici)1096-8628(19991015)88:5<533::aid-ajmg18>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Prendergast MA, Little HJ. Adolescence, glucocorticoids and alcohol. Pharmacol Biochem Behav. 2007;86(2):234–245. doi: 10.1016/j.pbb.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol Clin Exp Res. 1999;23(7):1136–1144. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11(6):603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28(12):1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- Reck BH, Mukhopadhyay N, Tsai HJ, Weeks DE. Analysis of alcohol dependence phenotype in the COGA families using covariates to detect linkage. BMC Genet. 2005;6 1:S143. doi: 10.1186/1471-2156-6-S1-S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Gold E, Risch C. Plasma cortisol levels following ethanol in sons of alcoholics and controls. Arch Gen Psychiatry. 1987;44(11):942–945. doi: 10.1001/archpsyc.1987.01800230022005. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, Tchivileva IE, Belfer I, Mishra B, Kiselycznyk C, Wallace MR, Staud R, Spiridonov NA, Max MB, Goldman D, Fillingim RB, Maixner W, Diatchenko L. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet. 2009;18(6):1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci USA. 1996;93(9):3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Koller DL, Foroud T, Carr K, Zhao J, Rice J, Numberger JI, Jr, Begleiter H, Porjesz B, Smith TL, Schuckit MA, Edenberg HJ. Association of GABA(A) receptors and alcohol dependence and the effects of genetic imprinting. Am J Med Genet B Neuropsychiatr Genet. 2003;117B(1):39–45. doi: 10.1002/ajmg.b.10022. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JBW. Structured clinical interview for DSM-III-R (SCID) New York Psychiatric Institute, Biometrics Research Institute; New York: 1985. [Google Scholar]
- Uusi-Oukari M, Heikkilä J, Sinkkonen ST, Mäkelä R, Hauer B, Homanics GE, Sieghart W, Wisden W, Korpi ER. Long-range interactions in neuronal gene expression: evidence from gene targeting in the GABA(A) receptor beta2-alpha6-alpha1-gamma2 subunit gene cluster. Mol Cell Neurosci. 2000;16(1):34–41. doi: 10.1006/mcne.2000.0856. [DOI] [PubMed] [Google Scholar]
- Valdez GR, Koob GF. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: implications for the development of alcoholism. Pharmacol Biochem Behav. 2004;79(4):671–689. doi: 10.1016/j.pbb.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Van Den Eede F, Van Broeckhoven C, Claes SJ. Corticotropin-releasing factor-binding protein, stress and major depression. Ageing Res Rev. 2005;4(2):213–239. doi: 10.1016/j.arr.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Van de Velde S, Bracke P, Levecque K. Gender differences in depression in 23 European countries. Cross-national variation in the gender gap in depression. Soc Sci Med. 2010;71(2):305–313. doi: 10.1016/j.socscimed.2010.03.035. [DOI] [PubMed] [Google Scholar]
- Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154(2):299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, Ali A. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology. 2002;26(1):106–114. doi: 10.1016/S0893-133X(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Winoker G, Cadoret R, Dorzab J, Baker M. Depressive disease: a genetic study. Arch Gen Psychiatry. 1971;24(2):135–144. doi: 10.1001/archpsyc.1971.01750080039006. [DOI] [PubMed] [Google Scholar]
- Zimmerman U, Spring K, Kunz-Ebrecht SR, Uhr M, Wittchen HU, Holsboer F. Effect of ethanol on hypothalamic-pituitary-adrenal system response to psychosocial stress in sons of alcohol-dependent fathers. Neuropsychopharmacology. 2004;29(6):1156–1165. doi: 10.1038/sj.npp.1300395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.