Abstract
Purpose
Mouse double minute 4 (MDM4), a homolog of MDM2, is a key negative regulator of p53, and its amplification or over-expression contributes to carcinogenesis by inhibiting the p53 tumor suppressor activity. We investigated the association between MDM4 polymorphisms and risk of squamous cell carcinoma of the head and neck (SCCHN).
Methods
We genotyped three MDM4 tagging polymorphisms, two in the 3′ untranslated region (3′ UTR: rs11801299G>A and rs10900598G>T) and one in intron 1 (rs1380576C>G), in a case-control study of 1,075 non-Hispanic white SCCHN patients and 1,084 cancer-free controls and evaluated their associations with SCCHN risk.
Results
Although none of these three polymorphisms individually had a statistically significant effect on risk of SCCHN, nor did their combined number of putative risk genotypes (i.e., rs11801299GG, rs1380576CG+GG, and rs10900598GG) (OR = 1.16 and 95% CI=0.93–1.45), we found that individuals with 1–3 risk genotypes had statistically significantly increased risk of oropharyngeal cancer (OR = 1.32 and 95% CI = 1.00–1.73), particularly for those with T1–2 stage (OR = 1.40; 95% CI = 1.02–1.94), those with regional lymph node metastases (N1–3) (OR = 1.44; 95% CI = 1.07–1.95), and those with late stages (III and IV) (OR = 1.34; 95% CI = 1.01–1.77).
Conclusion
Our results suggest that the joint effect of MDM4 variants may contribute to the risk of oropharyngeal cancer in non-Hispanic whites. Additional studies are warranted to unravel whether the particular stage distribution of oropharyngeal cancer with the strongest association (T1–2, N1–3, and III–IV) is a possible link with human papillomavirus-related oropharyngeal cancers.
Keywords: MDM4 polymorphism, case-control, genetic susceptibility, molecular epidemiology, head and neck neoplasms, oropharyngeal cancer
Introduction
Squamous cell carcinomas of the head and neck (SCCHN), comprising cancers of the oral cavity, pharynx, and larynx, is a common cancer worldwide, but the incidence is dropping in the developed world as smoking prevalence has declined, with the exception of the oropharyngeal subsite [1]. In the United States, there were estimated 49,260 new cases of SCCHN and an estimated 11,480 deaths from the disease in 2010 [2]. Although tobacco smoking and alcohol use are well-established etiologic factors for SCCHN, only a small fraction of smokers develop SCCHN. Additionally, the oropharyngeal subsite of SCCHN appears strongly associated with human papillomavirus (HPV) [3, 4], and similarly it would appear that while many are exposed to HPV few ever develop oropharyngeal cancer. These discrepancies between the numbers exposed and the numbers ultimately developing cancer suggest that genetic susceptibility contributes to the etiology of this disease.
The p53 tumor suppressor protein is a key regulator of the cellular response to genotoxic stresses that include DNA damage, hypoxia, UV light exposure, and oncogene activation. Under such stresses, p53 become activated and then induce the transcription of genes involved in DNA repair, cell-cycle arrest, senescence, and apoptosis [5]. The p53 stress-response pathway has been shown to be crucial in preventing tumor formation, and the p53 protein is dysfunctional in most human solid cancers [6]. Furthermore, HPV oncoproteins play an important role in head and neck carcinogenesis by disrupting the function of both the Rb and p53 pathways [7, 8]. The murine double minute protein MDM2 and its homolog MDM4 are the key regulators of the p53 activity. MDM2 not only binds to and inhibits the p53 transactivation domain, but also promotes ubiquitin-dependent proteasomal degradation of p53 by functioning as an E3 ubiquitin ligase [9]. Similarly, MDM4 also directly binds to the transactivation domain of p53 and inhibits p53 transcriptional activity [10, 11]. Recent data have shown that MDM4 stabilizes the p53-MDM2 complex and enhances the function of the E3 ubiquitin ligase of MDM2, accelerating degradation of p53 [12]. Furthermore, MDM4 can also directly bind to MDM2 and inhibit MDM2 degradation, regulating the role of MDM2 in inhibiting the p53 activity [13, 14]. The human MDM4 gene, comprising of 11 exons and encoding a protein of 490 amino acids [15] is located on chromosome region 1q32, a region that is frequently amplified in cancer [16, 17]. Indeed, the amplification or over-expression of the human MDM4 gene has been observed in numerous tumor cell lines retaining the wild-type p53 and a large subset of human tumors including SCCHN; Interestingly, many of these human tumors express the will-type p53, indicating that MDM4 may cause p53 inactivation during carcinogenesis [18–20]. For example, Valentin-vega et al. reported that over-expression of MDM4 was detected in 50% (28/56) of SCCHN, all of which retained the wild-type p53, suggesting that increased levels of MDM4 may substitute for p53 mutations in SCCHN and therefore contribute to tumorigenesis [20]. It was also reported that over-expression of MDM4 was associated with tumor progression and poor prognosis [21–23]. Taken together, these data suggest that altered MDM4 expression may play an important role in tumor development.
Although the functions of MDM4 have been well studied, the association between MDM4 polymorphisms and cancer risk has rarely been assessed. Considerable attention has been focused on the role of MDM2 single nucleotide polymorphisms (SNP), particularly for SNP 309T>G (rs2279744) within the promoter region of MDM2, in cancer risk. The well-known T to G change of SNP309 results in high levels of MDM2 mRNA and protein, thereby attenuating the p53 pathway and increasing tumor susceptibility. A number of studies showed that SNP309 was associated with the risk or the onset age of different types of cancers, including SCCHN [9, 24–32]. Unlike MDM2, only few studies have investigated the role of MDM4 SNPs in risk of some types of human cancers [33, 34]. To date, however, no published study has evaluated the association between MDM4 SNPs and SCCHN risk. Therefore, we hypothesized that genetic variants in MDM4 are associated with risk of SCCHN, and we tested this hypothesis in our ongoing hospital-based case-control study of SCCHN.
Methods
Patients and controls
The study population included 1,084 patients with newly diagnosed SCCHN between October 1999 and October 2007, identified at The University of Texas M. D. Anderson Cancer Center. Patients with second primary tumors, primary tumors of the skin, nasopharynx, sinonasal tract, or any histopathologic diagnoses other than squamous cell carcinoma were excluded. All patients were non-Hispanic whites and had not received any treatment at the time of recruitment. Approximately 93% of the eligible patients contacted chose to participate in this study. The stages of SCCHN were classified according to the tumor-node-metastasis (TNM) staging system and were categorized as I, II, III and IV [35]. The regional lymph node (N) involvement was defined as follows [35]: N0, no regional node metastasis; N1, metastasis in a single ipsilateral lymph node, 3 cm or less in greatest dimension; N2, metastasis in a single ipsilateral lymph node, more than 3 cm but not more than 6 cm in greatest dimension; or in multiple ipsilateral lymph nodes, none more than 6 cm in greatest dimension; or in bilateral or contralateral lymph nodes, none more than 6 cm in greatest dimension; N3, metastasis in a lymph node more than 6 cm in greatest dimension.
This study also included 1,090 cancer-free controls who were recruited in the same period as the cases and were genetically unrelated visitors or companions of patients seen at The M. D. Anderson Cancer Center clinics. The cases and controls were frequency matched by age (± 5 years), sex and ethnicity. The response rate of the eligible controls was approximately 85%.
Having signed a written informed consent during the interview, all subjects enrolled in the study were interviewed to gather demographic data and history of tobacco and alcohol use. Each subject donated 30-mL of blood, of which 1 ml was used for genomic DNA extraction with a DNA blood Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacture’s instructions. The research protocol was approved by the M.D. Anderson Institutional Review Board.
Tag SNPs selection and genotyping
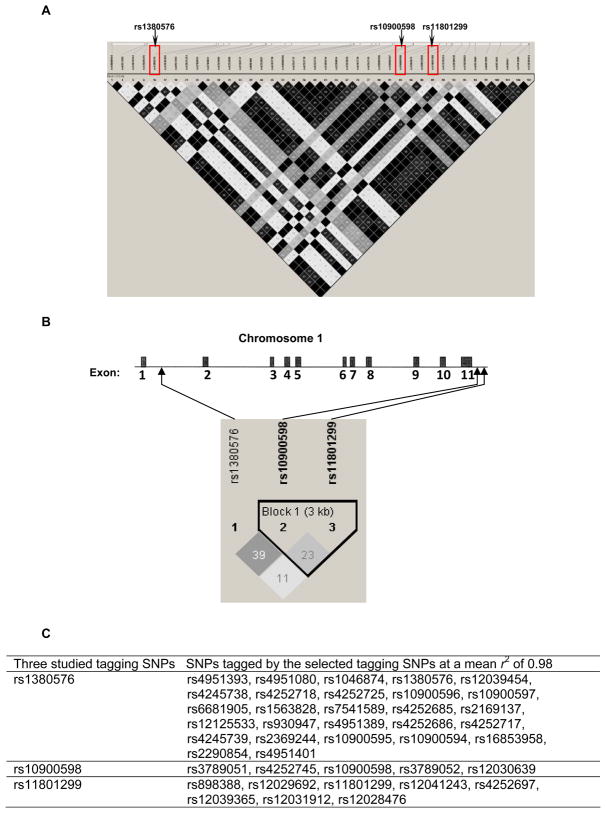
We used the public HapMap SNP database (http://www.hapmap.org/) to identify the MDM4 tagging SNPs by using a tagger with a greedy algorithm [36], for which all SNPs either were directly genotyped or exceeded a threshold level of linkage disequilibrium (LD) value (r2) with a genotyped SNP. We searched for SNPs of the MDM4 gene within an about 34-kb region on chromosome 1q32 (i.e., from 202,752,134 bp to 202,786,349 bp) among a European population (CEPH: Utah residents with ancestry from northern and western Europe). The tagging SNPs were selected based on their pairwise LD with the r2 threshold of 0.8 and minor allele frequency (MAF) ≥ 0.10. As a result, we identified three tagging SNPs (i.e., rs11801299G>A, rs1380576C>G, and rs10900598G>T). The selected tagging SNPs tagged 40 common SNPs with the mean r2 of 0.98 (Fig. 1A). Among these three selected SNPs, both rs11801299and rs10900598 are located in the 3′ untranslated region (3′ UTR) of the MDM4 gene, whereas rs1380576 is located in intron 1 of the MDM4 gene (Fig. 1B).
Fig. 1.
(A) The linkage disequilibrium structure of 40 common SNPs across the MDM4 region (chr1: 202,752,134 to 202,786,349) tagged by the 3 selected tagging SNPs among a European population (http://www.hapmap.org/). The matrix indicates the r2 value between each pair of SNPs. The darker, the stronger r2, with dark black representing r2 = 1 and pure white representing r2 = 0. (B) Gene structure of human MDM4, location of the three tagging SNPs in MDM4, and the linkage disequilibrium structure of the 3 selected tagging SNPs in 1,079 controls. The matrix indicates the r2 value between each pair of SNPs. (C) Selected tagging SNPs and SNPs tagged by the tagging SNPs.
The genotyping was performed using the Applied Biosystems TaqMan genotyping platform according to the manufacture’s recommendation. Briefly, the reactions were prepared by using TaqMan Universal Master Mix, 80×SNP Genotyping Assay Mix, Dnase-free water, and 10-ng genomic DNA in a final volume of 5 μL per reaction. Both negative and positive controls and three samples with known genotypes were included in each plate to ensure the accuracy of the genotyping. The PCR amplification was run, and the plate was read using a TaqMan 7900 HT sequence detection system (Applied Biosystems). The analyzed fluorescence results were then auto-called into the genotypes using the built-in SDS2.3 software of the system.
Statistical analysis
We used Chi-square test to evaluate the differences in the frequency distributions of selected demographic variables, smoking status, alcohol use, and each allele and genotype of the three MDM4 SNPs between the cases and the controls. Unconditional univariate and multivariate logistic regression analyses were performed to obtain the crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs). The multivariate adjustment included age, sex, smoking status, and alcohol use. Considering the potential gene-gene interaction of the MDM4 SNPs on SCCHN risk, the associations between the combined genotypes of the three SNPs and SCCHN risk were evaluated. The combined genotype data were further stratified by subgroups of age, sex, smoking status, alcohol use, and primary tumor sites. Finally, we calculated the false positive report probability (FPRP) for observed statistically significant associations in a range of prior probabilities, and used a threshold of noteworthiness of FPRP ≤0.5 in the present study [37]. Two-sided tests of statistical significance were conducted by using the SAS software (version 9.2; SAS Institute Inc., Cary, NC).
Results
In this study, 1,084 SCCHN cases and 1,090 controls of non-Hispanic whites were genotyped for the selected tagging SNPs, but 9 cases and 11 controls failed in the genotyping assays, which were excluded from the final analysis that included 1,075 SCCHN patients and 1,079 controls. Among the 1,075 SCCHN patients, 315 (29.3%) had cancer of the oral cavity, 546 (50.8%) had cancer of the oropharynx, 171 (15.9%) had cancer of the hypopharynx, and 43 (4%) had cancer of the larynx.
As shown in Table 1, the cases and controls were adequately frequency-matched by age and sex: the mean age was 57.1 years old for cases (±11.2 years; range, 18–90 years) and 56.7 years old for controls (±11.0 years; range, 20–87 years) (P = 0.341), and 75.2% of the cases and 76.6% of the controls were men (P = 0.451). However, there were more current smokers (37.8%) and current drinkers (51.0%) among the cases than among the controls (14.5 % and 40.4%, respectively), and these differences were statistically significant (P < 0.001). All of these variables were further adjusted for in the multivariate logistic regression analysis for any residual confounding effect.
Table 1.
Demographic characteristics of SCCHN cases and cancer-free controls
| Variables | Cases
|
Controls
|
P valuea |
|---|---|---|---|
| No. (%) | No. (%) | ||
| All subjects | 1,075 (100%) | 1,079 (100%) | |
| Age (years) | 0.559 | ||
| Mean | 57.1 ± 11.2 | 56.7 ± 11.0 | |
| ≤ 45 | 153 (14.2) | 170 (15.7) | |
| 46–60 | 525 (48.8) | 523 (48.5) | |
| >60 | 397 (36.9) | 386 (35.8) | |
| Sex | 0.451 | ||
| Male | 808 (75.2) | 826 (76.5) | |
| Female | 267 (24.8) | 253 (23.5) | |
| Smoking Status | <0.0001 | ||
| Never | 299 (27.8) | 526 (48.8) | |
| Former | 370 (34.4) | 396 (36.7) | |
| Current | 406 (37.8) | 157 (14.5) | |
| Alcohol use | <0.0001 | ||
| Never | 294 (27.3) | 468 (43.4) | |
| Former | 233 (21.7) | 175 (16.2) | |
| Current | 548 (51.0) | 436 (40.4) |
Two-sided Chi-square test.
Association between the MDM4 SNPs and SCCHN risk
The genotype frequencies of the three tagging SNPs among the controls were all in agreement with the Hardy–Weinberg equilibrium (Chi-square test: P = 0.084 for rs11801299, P = 0.712 for rs1380576, and P = 0.398 for rs10900598). Table 2 summarizes distributions of genotype and allele frequencies of the three MDM4 SNPs in case patients and control subjects and their associations with SCCHN risk. No significant difference in the genotype distributions was found between the cases and the controls (P = 0.557 for rs11801299, P = 0.453 for rs1380576, and P = 0.827 for rs10900598).
Table 2.
Genotype frequencies of MDM4 polymorphisms between cases and controls and their associations with risk of SCCHN
| Genotypes | Case (n = 1,075) | Control (n = 1,079)a | Pb | OR (95% CI)c | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| rs11801299 | ||||||
| GG | 684 | 63.6 | 665 | 61.6 | 0.557 | 1.00 |
| AG | 351 | 32.7 | 376 | 34.9 | 0.90 (0.74–1.08) | |
| AA | 40 | 3.7 | 38 | 3.5 | 0.95 (0.59–1.54) | |
| AG+AA | 391 | 36.4 | 414 | 38.4 | 0.338 | 0.90 (0.75–1.08) |
| A allele | 0.200 | 0.209 | 0.465 | |||
| rs1380576 | ||||||
| CC | 487 | 45.3 | 518 | 48.0 | 0.453 | 1.00 |
| CG | 477 | 44.4 | 455 | 42.2 | 1.15 (0.96–1.39) | |
| GG | 111 | 10.3 | 106 | 9.8 | 1.17 (0.87–1.60) | |
| CG+GG | 588 | 54.7 | 561 | 52.0 | 0.208 | 1.15 (0.97–1.37) |
| G allele | 0.325 | 0.309 | 0.258 | |||
| rs10900598 | ||||||
| GG | 307 | 28.6 | 296 | 27.4 | 0.827 | 1.00 |
| GT | 545 | 50.7 | 552 | 51.2 | 0.95 (0.77–1.17) | |
| TT | 223 | 20.7 | 231 | 21.4 | 0.91 (0.70–1.17) | |
| GT+TT | 768 | 71.4 | 783 | 72.6 | 0.561 | 0.94 (0.77–1.14) |
| T allele | 0.461 | 0.470 | 0.556 | |||
| No. of combined risk genotypesd | ||||||
| 0 | 206 | 19.2 | 228 | 21.1 | 0.255 | 1.00 |
| 1–3 | 869 | 80.8 | 851 | 78.9 | 1.16 (0.93–1.45) | |
The observed genotype frequency among control subjects was in agreement with the Hardy-Weinberg equilibrium (Chi-square = 2.98, P = 0.084 for rs11801299, Chi-square = 0.136, P = 0.712 for rs1380576, and Chi-square = 0.713, P = 0.398 for rs10900598).
Two-side Chi-square test for either genotype distribution or allele frequency between cases and controls.
Adjusted for age, sex, smoking status, and alcohol use in a logistic regression model.
MDM4 rs11801299GG, rs1380576CG+GG, and rs10900598GG were considered as risk genotypes.
As shown in Table 2, the frequencies of the A allele of rs11801299 and the T allele of rs10900598 among the cases (0.20 and 0.46, respectively) were slightly lower than those in the controls (0.21 and 0.47, respectively); in contrast, the frequency of the G allele of rs1380576 among the cases (0.33) was slightly higher than those among the controls (0.31), suggesting that the G alleles of rs11801299, rs10900598, and rs1380576 may be the putative risk alleles. Although none of the variant genotypes was alone associated with significantly altered risk, both the A allele of rs11801299 and the T allele of rs10900598 tended to be associated with non-significantly reduced SCCHN risk (OR = 0.90 and 95% CI = 0.75–1.08 for AG+AA of rs11801299, and OR= 0.94 and 95% CI = 0.77–1.14 for GT+TT of rs10900598), and the G allele of rs1380576 tended to be associated with non-significantly increased SCCHN risk (OR = 1.15 and 95% CI = 0.97–1.37 for CG+GG of rs1380576).
Since the three SNPs are not in LD to each other (r2 = 0.23 and D′ = 1.0 for rs11801299 and rs10900598, r2 = 0.11 and D′ = 1.0 for rs11801299 and rs1380576, r2 = 0.39 and D′ =0.99 for rs10900598 and rs1380576) in the controls (Fig. 1B), we combined their genotypes by the number of the putative risk genotypes (i.e., rs11801299 GG, rs1380576 CG+GG, and rs10900598 GG genotypes) to assess their possible combined effects on risk of SCCHN;, the combined genotypes with 1–3 risk genotypes was associated with non-significantly increased overall risk of SCCHN (OR = 1.16 and 95% CI=0.93–1.45) (Table 2).
To investigate the possible subgroup effect, we further stratified the combined genotypes of these three SNPs by subgroups of age, sex, smoking status, alcohol drinking status, and the primary tumor site. No association was observed in the subgroups of age, sex, smoking and drinking status (never vs. ever) (Table 3). However, we found that the combined 1–3 genotypes was associated with a statistically significantly increased risk of oropharyngeal cancer (OR = 1.32 and 95% CI = 1.00–1.73) but for not other tumor sites (Table 4).
Table 3.
Stratification analyses between the combined genotypes of MDM4 polymorphisms and SCCHN risk
| Variables | No. (Case / Control) | Combined genotypes (case / control)
|
OR (95% CI)b | |||
|---|---|---|---|---|---|---|
| 0 risk genotypea | 1–3 risk genotypesa | |||||
| n | % | n | % | |||
| Age (years) | ||||||
| ≤45 | 153 / 170 | 28 / 32 | 18.3 / 18.8 | 125 / 138 | 81.7 / 81.2 | 1.06 (0.59–1.87) |
| 46–60 | 525 / 523 | 108 / 109 | 20.6 / 20.8 | 417 / 414 | 79.4 / 79.2 | 1.06 (0.77–1.45) |
| >60 | 397 / 386 | 70 / 87 | 17.6 / 22.5 | 327 / 299 | 82.4 / 77.5 | 1.41 (0.97–2.05) |
| Sex | ||||||
| Female | 267 / 253 | 48 / 52 | 18.0 / 20.5 | 219 / 201 | 82.0 / 79.5 | 1.31 (0.81–2.09) |
| Male | 808 / 826 | 158 / 176 | 19.6 / 21.3 | 650 / 650 | 80.5 / 78.7 | 1.12 (0.87–1.43) |
| Smoking status | ||||||
| Never | 299 / 526 | 55 / 111 | 18.4 / 21.1 | 245 / 415 | 81.6 / 78.9 | 1.18 (0.82–1.69) |
| Ever | 776 / 553 | 151 / 117 | 19.46 / 21.2 | 625 / 436 | 80.54 / 78.8 | 1.11 (0.84–1.46) |
| Drinking status | ||||||
| Never | 294 / 468 | 52 / 102 | 17.7 / 21.8 | 242 / 366 | 82.31 / 78.2 | 1.29 (0.88–1.87) |
| Ever | 781 / 611 | 154 / 126 | 19.7 / 20.6 | 627 / 485 | 80.28 / 79.4 | 1.09 (0.83–1.44) |
This represents the number of risk genotypes: rs11801299GG, rs1380576CG+GG, and rs10900598GG.
Adjusted for age, sex, smoking status, and alcohol use in a logistic regression model.
Table 4.
Association between the combined genotypes of MDM4 polymorphisms and SCCHN risk stratified by tumor sites
| Genotypes | Oropharynx | Oral cavity | Larynx and Hypopharynx | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 1,079) | Case (n = 546) | P | OR (95% CI)a | Case (n = 315) | P | OR (95% CI)a | Case (n = 214) | P | OR (95% CI) a | |
| rs11801299 | ||||||||||
| GG | 665 | 305 | 0.110c | 1.00 | 191 | 0.600c | 1.00 | 138 | 0.690c | 1.00 |
| AG | 376 | 165 | 0.80 (0.63–1.01) | 116 | 1.07 (0.81–1.42) | 70 | 0.87 (0.61–1.24) | |||
| AA | 38 | 26 | 1.24 (0.73–2.11) | 8 | 0.65 (0.29–1.49) | 6 | 0.54 (0.20–1.43) | |||
| AG+AA | 414 | 191 | 0.182d | 0.84 (0.67–1.05) | 124 | 0.749d | 1.03 (0.79–1.36) | 76 | 0.432d | 0.83 (0.59–1.18) |
| P for trende | 0.342 | 0.876 | 0.199 | |||||||
| rs1380576 | ||||||||||
| CC | 518 | 250 | 0.677c | 1.00 | 141 | 0.586c | 1.00 | 96 | 0.539c | 1.00 |
| CG | 455 | 238 | 1.08 (0.87–1.35) | 140 | 1.20 (0.90–1.59) | 99 | 1.35 (0.95–1.92) | |||
| GG | 106 | 58 | 1.15 (0.80–1.65) | 34 | 1.29 (0.82–2.04) | 19 | 1.11 (0.61–2.02) | |||
| CG+GG | 561 | 296 | 0.397d | 1.09 (0.89–1.35) | 174 | 0.310d | 1.21 (0.93–1.59) | 118 | 0.400d | 1.31 (0.93–1.83) |
| P for trende | 0.373 | 0.154 | 0.260 | |||||||
| rs10900598 | ||||||||||
| GG | 296 | 166 | 0.279c | 1.00 | 87 | 0.230c | 1.00 | 54 | 0.789c | 1.00 |
| GT | 552 | 257 | 0.86 (0.67–1.10) | 174 | 1.02 (0.75–1.40) | 114 | 1.09 (0.73–1.62) | |||
| TT | 231 | 123 | 0.97 (0.72–1.31) | 54 | 0.76 (0.51–1.13) | 46 | 1.02 (0.62–1.65) | |||
| GT+TT | 783 | 380 | 0.210d | 0.89 (0.71–1.12) | 228 | 0.948d | 0.94 (0.70–1.27) | 160 | 0.508d | 1.07 (0.73–1.56) |
| P for trende | 0.738 | 0.218 | 0.909 | |||||||
| No. of combined | ||||||||||
| risk genotypesf | ||||||||||
| 0 | 228 | 93 | 0.050g | 1.00 | 71 | 0.592g | 1.00 | 42 | 0.621g | |
| 1–3 | 851 | 453 | 1.32 (1.00–1.73) | 244 | 0.98 (0.71–1.35) | 172 | 1.23 (0.81–1.87) | |||
Adjusted for age, sex, smoking status, and alcohol use in a logistic regression model.
Two-side Chi-square test for the distribution of three genotypes.
Two-side Chi-square test for the distribution of combined genotypes under a dominant model.
Adjusted by age, sex, smoking status, and alcohol use.
This represents the number of risk genotypes rs11801299GG, rs1380576CG+GG, and rs10900598GG.
Two-side Chi-square test for the distribution of combined genotypes.
Association between the MDM4 combined genotypes and stages at presentation of oropharyngeal cancer
Because the risk associated with the combined 1–3 risk genotypes was more pronounced in oropharyngeal cancer, we then evaluated whether these SNPs were associated with a particular stage at presentation of oropharyngeal cancer. As shown in Table 5, we found that the combined 1–3 risk genotypes were associated with a statistically significantly increased risk of late-stage (III and IV) oropharyngeal cancer (OR = 1.34; 95% CI = 1.01–1.77) but not with that of early-stage (I and II) oropharyngeal cancer (OR = 1.13; 95% CI = 0.54–2.39), compared with the combined genotype with 0 risk genotype; Furthermore, the combined 1–3 risk genotypes were associated with a statistically significantly increased risk of oropharyngeal cancer with T1–2 stage (OR = 1.40; 95% CI = 1.02–1.94) and regional lymph-node metastasis (N1–3) (OR = 1.44; 95% CI = 1.07–1.95) but not with that of oropharyngeal cancer with T3–4 stage (OR = 1.16; 95% CI = 0.78–1.73) and without regional lymph-node metastasis (OR = 0.86; 95% CI = 0.51–1.44), respectively. Similar results were not observed for cancers of the oral cavity and larynx / hypopharynx (data not shown). Furthermore, we did not observe any effect of the variant alleles or putative risk genotypes on the age of disease onset either in SCCHN or in oropharyngeal cancer (data not shown).
Table 5.
Association of the combined genotypes of MDM4 polymorphisms with stage at presentation of oropharyngeal cancer
| Combined genotypes (no. of risk genotypes)a | T1–2 | T3–4 | ||
|---|---|---|---|---|
| Case (%) / Control (%) | OR (95% CI)b | Case (%) / Control (%) | OR (95% CI)b | |
| 0 | 58 (16.1) / 228 (21.1) | 1.00 | 35 (18.8) / 228 (21.1) | 1.00 |
| 1–3 | 302 (83.9) / 851 (78.9) | 1.40 (1.02–1.94) | 151 (81.2) / 851 (78.9) | 1.16 (0.78–1.73) |
| Combined genotypes (no. of risk genotypes)a | No regional lymph nodes metastasis (N0) | Involvement of regional lymph nodes (N1–3) | ||
|
| ||||
| Case (%) / Control (%) | OR (95% CI)b | Case (%) / Control (%) | OR (95% CI)b | |
|
| ||||
| 0 | 21 (23.9) / 228 (21.1) | 1.00 | 72 (15.7) / 228 (21.1) | 1.00 |
| 1–3 | 67 (76.1) / 851 (78.9) | 0.86 (0.51–1.44) | 386 (84.3) / 851 (78.9) | 1.44 (1.07–1.95) |
| Combined genotypes (no. of risk genotypes)a | Stage I–II | Stage III–IV | ||
|
| ||||
| Case (%) / Control (%) | OR (95% CI)b | Case (%) / Control (%) | OR (95% CI)b | |
| 0 | 9 (19.2) / 228 (21.1) | 1.00 | 84 (16.8) / 228 (21.1) | 1.00 |
| 1–3 | 38 (80.8) / 851 (78.9) | 1.13 (0.54– 2.39) | 415 (83.2) / 851(78.9) | 1.34 (1.01–1.77) |
This represents the number of risk genotypes used in the calculation: rs11801299GG, rs1380576CG+CC, and rs10900598GG.
Adjusted for age, sex, smoking status, and alcohol use in a logistic regression model.
Finally, because multiple tests had been performed, we estimated the false positive reporting probability (FPRP) for all the statistically significant results of oropharyngeal cancer. As shown in Table 6, with the assumption of a moderate prior probability of 0.1 and the observed ORs or an expected OR of 1.5, the FPRPs for all of the above–mentioned significant results were below the threshold of 0.5.
Table 6.
False positive reporting probability values for associations between the combined genotypes of MDM4 polymorphisms and oropharyngeal cancer risk
| Prior probability | Oropharyngeal cancer | Oropharyngeal cancer with T1–2 | Oropharyngeal cancer with regional lymph nodes | Oropharyngeal cancer with stage III–IV | ||||
|---|---|---|---|---|---|---|---|---|
| Observed ORa | Expected ORb | Observed ORa | Expected ORb | Observed ORa | Expected ORb | Observed ORa | Expected ORb | |
| 1.32 | 1.5 | 1.40 | 1.5 | 1.44 | 1.5 | 1.34 | 1.5 | |
|
| ||||||||
| 0.25 | 0.220 | 0.152 | 0.079 | 0.148 | 0.186 | 0.066 | 0.207 | 0.148 |
| 0.1 | 0.458 | 0.350 | 0.204 | 0.343 | 0.407 | 0.175 | 0.439 | 0.342 |
| 0.01 | 0.903 | 0.856 | 0.738 | 0.852 | 0.883 | 0.700 | 0.896 | 0.851 |
| 0.001 | 0.990 | 0.984 | 0.966 | 0.983 | 0.987 | 0.959 | 0.989 | 0.983 |
| 0.0001 | 1.000 | 0.998 | 0.999 | 0.998 | 0.999 | 0.996 | 0.999 | 0.998 |
Calculation of FPRP using the observed OR with the common combined genotypes.
Calculation of FPRP using study subjects to detect an OR of 1.5 with the common combined genotypes.
Discussion
To the best of our knowledge, this is the first study aimed at evaluating whether tagging SNPs in MDM4 have any impact on the overall risk of SCCHN. Although we did not find any significant main effect of each MDM4 tagging SNP on risk of SCCHN, we found statistical evidence of a joint effect of three MDM4 tagging SNPs on risk of oropharyngeal cancer. Given the role of MDM4 as a key inhibitor of p53, it is biologically plausible that the MDM4 SNPs may collectively modulate risk of oropharyngeal cancer.
The p53 pathway can be functionally inactivated in human cancer through either amplification or over-expression of MDM4 that directly binds to the p53 transactivation domain and thus inhibits its activity. As a key negative regulator of p53, it is conceivable that MDM4 may play an important role in tumor formation and growth, because increased levels of MDM4 inhibit the protective functions of p53 and thus could lead to oncogenesis. Terizian et al. recently demonstrated that haplo-insufficiency of MDM4 loci leads to an increased p53 activity, also exhibiting an increased sensitivity to DNA damage, decreased transformation potential, and reduced tumorigenesis [38]. These data suggested that genetic changes in MDM4 could alter the levels of MDM4 and thus affect the p53 pathway and subsequent development of cancers [33].
To date, very few association studies have been conducted to evaluate the roles of MDM4 SNPs in cancer risk. Recently, Atwal et al. investigated the association between MDM4 SNPs and breast and ovarian cancer risk [33] in which they found that the minor alleles of MDM4 SNPs rs10900594, rs2369244, and rs12039454 were associated with a greater risk of developing breast cancer in three independent Ashkenazi Jewish (AJ) case/control populations. They also demonstrated that the minor allele of another SNP rs1563828 in MDM4 was associated with an earlier age of onset in both of familial and sporadic ovarian cancers in the Caucasian case/control populations. These results suggested that MDM4 variants may contribute to human breast and ovarian cancers. In another independent US study, the variant allele of rs1563828 was found to be associated with an earlier age of onset of estrogen receptor (ER)-negative breast cancer but not with that of ER positive breast cancer [34]. It is worth noting that all the above-mentioned SNPs in these two published studies are tagged by our selected tagging SNP rs1380576 with r2 = 1.0 and D′=1 in the European CEU samples used in the Haplotype Mapping Project (HapMap) (Fig. 1). In particular, with a mean r2 of 0.98, the tagging SNP rs1380576 tags 8 out of 10 SNPs used for characterizing the haplotype structure of MDM4 locus in three different populations in the Atwal’s study [33]. The pair-wise correlation estimates (r2) among these 10 SNPs in the CEU populations seem somewhat different between the Atwal’s report and ours, which may result from different sample sizes of the CEU study populations. In the study of Atwal et al., the pair-wise correction statistics D′ and r2 among the ten SNPs were determined in 38 Coriell Caucasians, whereas in our study, the statistics D′ and r2 amongthoseSNPs were obtained from 60 Europeans (CEPH: Utah residents with ancestry from northern and western Europe) used in the HapMap SNP database available publicly (http://www.hapmap.org/).
In the present study, we found that the frequencies of the minor A allele of rs11801299 and the minor T allele of rs10900598 among the cases (0.20 and 0.46, respectively) were slightly lower than those in the controls (0.21 and 0.47, respectively), and the ORs had the same trends at less than 1 for both heterozygous and homozygous variant genotypes for these two SNPs compared with the corresponding wild genotypes; in contrast, the frequency of the minor G allele of rs1380576 among the cases (0.33) was slightly higher than those among the controls (0.31), and the ORs had the same trend at greater than 1 for both heterozygous and homozygous variant genotypes compared with the wild genotype, suggesting that the major G alleles of rs11801299 and rs10900598, and the minor G allele of rs1380576 may be the putative risk alleles in ourthe study population. Although we did not find any main effect on the overall risk of SCCHN, nor on the age of disease onset, we did find that those who carried the 1–3 combined risk genotypes (i.e., rs11801299 GG, rs10900598 GG, and rs1380576 CG+GG under a dominant genetic model) appeared to have an increased risk of oropharyngeal cancer, particularly in the late stages (III and IV) and for those with early primaries (T1–2 stages) but with advanced nodal staging (N1–3 stages). Since the apparent discrepancy of early primary but advanced nodal stage is the classic stage at the presentation for HPV-related oropharyngeal cancer [39–41], our findings suggest a possible association between these tagging SNPs in MDM4 and risk of HPV-related oropharyngeal cancer.
It has been recognized that SNPs in the 3′ UTR or introns of a gene can influence alternative splicing events, mRNA stability or protein translation, thereby resulting in altered functions of the gene [42–45]. In addition to the full-length mRNA, several transcript variants of MDM4 have been identified, most of which were derived from a gene splicing process [21, 46]. MDM4-S is one of the shorter isoforms, which is functionally more efficient than full-length MDM4 in binding and inhibiting p53-induced transcription [47]. Human MDM4 gene amplification or over-expression of human MDM4-S mRNA splice variant was observed in malignant gliomas, papillary thyroid tumors, and soft tissue sarcomas with the expression of the splice variant being associated with an unfavorable prognosis [21, 22, 48, 49]. MDM4-G and MDM4-211 are other two isoforms that attribute to the stabilization of MDM2 levels [46, 50]. Two (i.e., rs11801299 and rs10900598) of the three SNPs we studied are located in the 3′-UTR, whereas the third, rs1380576, is in the first intron. Therefore, it is possible that these three SNPs in MDM4 or other untyped functional ones in linkage with them may promote expression of one of the shorter isoforms of MDM4 with more efficient binding to p53 or MDM2, thereby attenuating the p53 tumor suppressor pathway and leading to tumorigenesis in humans. In addition, HPV16, which encodes E6 and E7 oncoproteins, is the most common type in HPV-positive oropharyngeal cancer [3, 4, 41]. It has been shown that HPV16 E6 binds to p53 and targets it for degradation by proteasome [7, 8]. Hence, we postulate that HPV16 E6 and MDM4 oncoproteins may act synergistically in development of HPV-positive oropharyngeal cancer through the common pathways that cause p53 degradation. However, these hypotheses need to be tested by further functional studies.
The FPRP analysis showed that with the assumption of a prior probability of 0.1, the FPRP values for the statistically significant findings in risk of oropharyngeal cancer were below 0.5, providing some measures of robustness for our observations. The FPRP is used to separate noteworthy discoveries from spurious reports [37]. However, it should be noted that the FPRP approach has limitations. One potential limitation of the FPRP approach is that it assigns a prior probability and yields the posterior probability of the null hypothesis given a significant report [37]. Furthermore, it can yield inferential errors and generate false-positive results [51]. Hence, our findings in the risk of oropharyngeal cancer should be interpreted with caution and replicated in large studies in other study populations.
As in all other case-control studies, our study has some limitations. Firstly, we had included only non-Hispanic white subjects in the study, and the result might not be generalizable to other ethnic populations. Secondly, because our study was hospital-based case-control study, an uncontrolled selection bias may exist. Thus, it is possible that our findings could be due to chance. However, in our study, cases were well matched to controls on age and sex, and the inadequacy in matching on smoking and alcohol exposure was further controlled by additional adjustment in multivariate analysis, which might minimize the influence of selection bias. Finally, HPV is another etiologic factor, in addition to tobacco and alcohol, for SCCHN, especially for oropharyngeal cancer [52, 53]. However, when we evaluated the modification effects of MDM4 variants on the risk of SCCHN / oropharyngeal cancer, HPV status was not taken into account because HPV16 serological status was available only for a few samples (procured between October 1999 and October 2007) overlapping with that of a previously published study using samples procured between May 1996 and May 2002 [54], which did not allow us to formally analyze the effect of interaction between the MDM4 genetic variants and HPV exposure on the risk of SCCHN. This analysis will be performed in our future expanded studies.
In summary, we have genotyped three tagging SNPs (i.e., rs11801299, rs10900598, and rs1380576) of the MDM4 gene in SCCHN cases and controls, none of which had a main effect on SCCHN cancer risk. However, given only an expected modest effect size of a single SNP, evaluating their joint effects may help understand comprehensively the role that MDM4 SNPs may play in cancer etiology. Indeed, we found that the combined genotypes of these three tagging SNPs (i.e., rs11801299 GG, rs1380576 CG+GG, and rs10900598 GG) were significantly associated with risk of oropharyngeal cancer, suggesting that rs11801299, rs1380576, and rs10900598 SNPs may jointly contribute to the development of oropharyngeal cancer. However, large studies are required to validate these findings. Further functional studies on these and/or related variants in MDM4 are warranted to elucidate the underlying molecular mechanisms of the observed association in our study. Because our significant findings were limited to oropharyngeal cancer, a disease that has a more complex etiology than cancers of the oral cavity and larynx / hypopharynx, it is possible that MDM4 SNPs are particularly important for the risk of HPV associated oropharyngeal cancer. Additional follow-up studies are underway in our laboratory to explore these questions.
Acknowledgments
Sponsorship: This study was supported in part by the National Institute of Health grants R01 CA131274 and R01 ES011740 (Q. Wei), ES015587 (D.G. Johnson), P50 CA097007 (S. Lippman), and P30 CA016672 (The University of Texas M. D. Anderson Cancer Center).
We thank Min Zhao, Jianzhong He, Kejing Xu, and Yawei Qiao for their laboratory assistance, and Dakai Zhu for his technical support. This study was supported in part by the National Institute of Health grants R01 CA131274 and R01 ES011740 (Q. Wei), ES015587 (D.G. Johnson), P50 CA097007 (S. Lippman), and P30 CA016672 (The University of Texas M. D. Anderson Cancer Center). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110:1429–35. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–75. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 4.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 5.Jin S, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139–40. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 8.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 9.Bond GL, Hu W, Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 10.Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun. 2005;331:750–60. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 11.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 12.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–34. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 13.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–96. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 14.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 15.Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43:34–42. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 16.Ried T, Just KE, Holtgreve-Grez H, du Manoir S, Speicher MR, Schröck E, et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415–23. [PubMed] [Google Scholar]
- 17.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–84. [PubMed] [Google Scholar]
- 18.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–43. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61:1839–42. [PubMed] [Google Scholar]
- 20.Valentin-Vega YA, Barboza JA, Chau GP, El-Naggar AK, Lozano G. High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum Pathol. 2007;38:1553–62. doi: 10.1016/j.humpath.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel F, Schulz J, Blumke K, Kappler M, Bache M, Schmidt H, et al. HDMX amplification and high levels of HDMX-S splice variant are correlated with a poor prognosis in soft tissue sarcomas. Verh Dtsch Ges Pathol. 2004;88:199–206. [PubMed] [Google Scholar]
- 22.Bartel F, Schulz J, Bohnke A, Blümke K, Kappler M, Bache M, et al. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer. 2005;117:469–75. doi: 10.1002/ijc.21206. [DOI] [PubMed] [Google Scholar]
- 23.Sampieri K, Mencarelli MA, Epistolato MC, Toti P, Lazzi S, Bruttini M, et al. Genomic differences between retinoma and retinoblastoma. Acta Oncol. 2008;47:1483–92. doi: 10.1080/02841860802342382. [DOI] [PubMed] [Google Scholar]
- 24.Bai J, Dai J, Yu H, Shen H, Chen F. Cigarette smoking, MDM2 SNP309, gene-environment interactions, and lung cancer risk: a meta-analysis. J Toxicol Environ Health A. 2009;72:677–82. doi: 10.1080/15287390902840930. [DOI] [PubMed] [Google Scholar]
- 25.Economopoulos KP, Sergentanis TN. Differential effects of MDM2 SNP309 polymorphism on breast cancer risk along with race: a meta-analysis. Breast Cancer Res Treat. 120:211–6. doi: 10.1007/s10549-009-0467-1. [DOI] [PubMed] [Google Scholar]
- 26.Fang F, Yu XJ, Yu L, Yao L. MDM2 309 T/G polymorphism is associated with colorectal cancer risk especially in Asians: a meta-analysis. Med Oncol. 2010 doi: 10.1007/s12032-010-9577-1. [DOI] [PubMed] [Google Scholar]
- 27.Gui XH, Qiu LX, Zhang HF, Zhang DP, Zhong WZ, Li J, et al. MDM2 309 T/G polymorphism is associated with lung cancer risk among Asians. Eur J Cancer. 2009;45:2023–6. doi: 10.1016/j.ejca.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z, Jin G, Wang L, Chen F, Wang X, Shen H. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: evidence from 21 case-control studies. Cancer Epidemiol Biomarkers Prev. 2007;16:2717–23. doi: 10.1158/1055-9965.EPI-07-0634. [DOI] [PubMed] [Google Scholar]
- 29.Huang SF, Chen IH, Liao CT, Wang HM, Liou SH, Hsieh LL. Combined effects of MDM2 SNP 309 and p53 mutation on oral squamous cell carcinomas associated with areca quid chewing. Oral Oncol. 2009;45:16–22. doi: 10.1016/j.oraloncology.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima M, Kondo S, Shimizu Y, Wakisaka N, Murono S, Furukawa M, et al. Impact of MDM2 single nucleotide polymorphism on tumor onset in head and neck squamous cell carcinoma. Acta Otolaryngol. 2008;128:808–13. doi: 10.1080/00016480701724904. [DOI] [PubMed] [Google Scholar]
- 31.Wilkening S, Bermejo JL, Hemminki K. MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis. 2007;28:2262–7. doi: 10.1093/carcin/bgm191. [DOI] [PubMed] [Google Scholar]
- 32.Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamira, Jackson JG, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–30. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci U S A. 2009;106:10236–41. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni DA, Vazquez A, Haffty BG, Bandera EV, Hu W, Sun YY, et al. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer. Carcinogenesis. 2009;30:1910–5. doi: 10.1093/carcin/bgp224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Joint Committee on Cancer. Manual for Staging of Cancer. 4. J.B. Lippincott; Philadelphia: 1988. [Google Scholar]
- 36.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–85. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27:6213–21. doi: 10.1200/JCO.2009.23.1670. [DOI] [PubMed] [Google Scholar]
- 40.Smith EM, Ritchie JM, Summersgill KF, Klussmann JP, Lee JH, Wang D, et al. Age, sexual behavior and human papillomavirus infection in oral cavity and oropharyngeal cancers. Int J Cancer. 2004;108:766–72. doi: 10.1002/ijc.11633. [DOI] [PubMed] [Google Scholar]
- 41.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15:6758–62. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 42.Duan J, Wainwright MS, Comeron JM, Saiton N, Sanders AR, Gelernter J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 43.Jupe ER, Badgett AA, Neas BR, Craft MA, Mitchell DS, Resta, et al. Single nucleotide polymorphism in prohibitin 39 untranslated region and breast-cancer susceptibility. Lancet. 2001;357:1588–9. doi: 10.1016/s0140-6736(00)04747-4. [DOI] [PubMed] [Google Scholar]
- 44.Knight JC. Functional implications of genetic variation in non-coding DNA for disease susceptibility and gene regulation. Clin Sci (Lond) 2003;104:493–501. doi: 10.1042/CS20020304. [DOI] [PubMed] [Google Scholar]
- 45.Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 46.Mancini F, Conza GD, Moretti F. MDM4 (MDMX) and its Transcript Variants. Curr Genomics. 2009;10:42–50. doi: 10.2174/138920209787581280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rallapalli R, Strachan G, Cho B, Mercer WE, Hall DJ. A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J Biol Chem. 1999;274:8299–308. doi: 10.1074/jbc.274.12.8299. [DOI] [PubMed] [Google Scholar]
- 48.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–6. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 49.Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6. [PubMed] [Google Scholar]
- 50.Giglio S, Mancini F, Gentiletti F, Spareco G, Felicioni L, Barassi F, et al. Identification of an aberrantly spliced form of HDMX in human tumors: a new mechanism for HDM2 stabilization. Cancer Res. 2005;65:9687–94. doi: 10.1158/0008-5472.CAN-05-0450. [DOI] [PubMed] [Google Scholar]
- 51.Lucke JF. A critique of the false-positive report probability. Genet Epidemiol. 2009;33:145–50. doi: 10.1002/gepi.20363. [DOI] [PubMed] [Google Scholar]
- 52.Marklund L, Hammarstedt L. Impact of HPV in Oropharyngeal Cancer. J Oncol. 2011:509036. doi: 10.1155/2011/509036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ringstrom E, Peters E, Hasegawa M, Posner M, Liu M, Kelsey KT. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clin Cancer Res. 2002;8:3187–92. [PubMed] [Google Scholar]
- 54.Chen X, Sturgis EM, Lei D, Dahlstrom K, Wei Q, Li G. Human papillomavirus seropositivity synergizes with MDM2 variants to increase the risk of oral squamous cell carcinoma. Cancer Res. 2010;70:7199–208. doi: 10.1158/0008-5472.CAN-09-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]