Abstract
In many bacteria tenI is found clustered with genes involved in thiamin thiazole biosynthesis. However, while TenI shows high sequence similarity with thiamin phosphate synthase, the purified protein has no thiamin phosphate synthase activity and the role of this enzyme in thiamin biosynthesis remains unknown. In this paper, we identify the function of TenI as a thiazole tautomerase, describe the structure of the enzyme complexed with its reaction product, identify the substrate phosphate and histidine 122 as the acid/base residues involved in catalysis and propose a mechanism for the reaction. The identification of the function of TenI completes the identification of all of the enzymes needed for thiamin biosynthesis by the major bacterial pathway.
INTRODUCTION
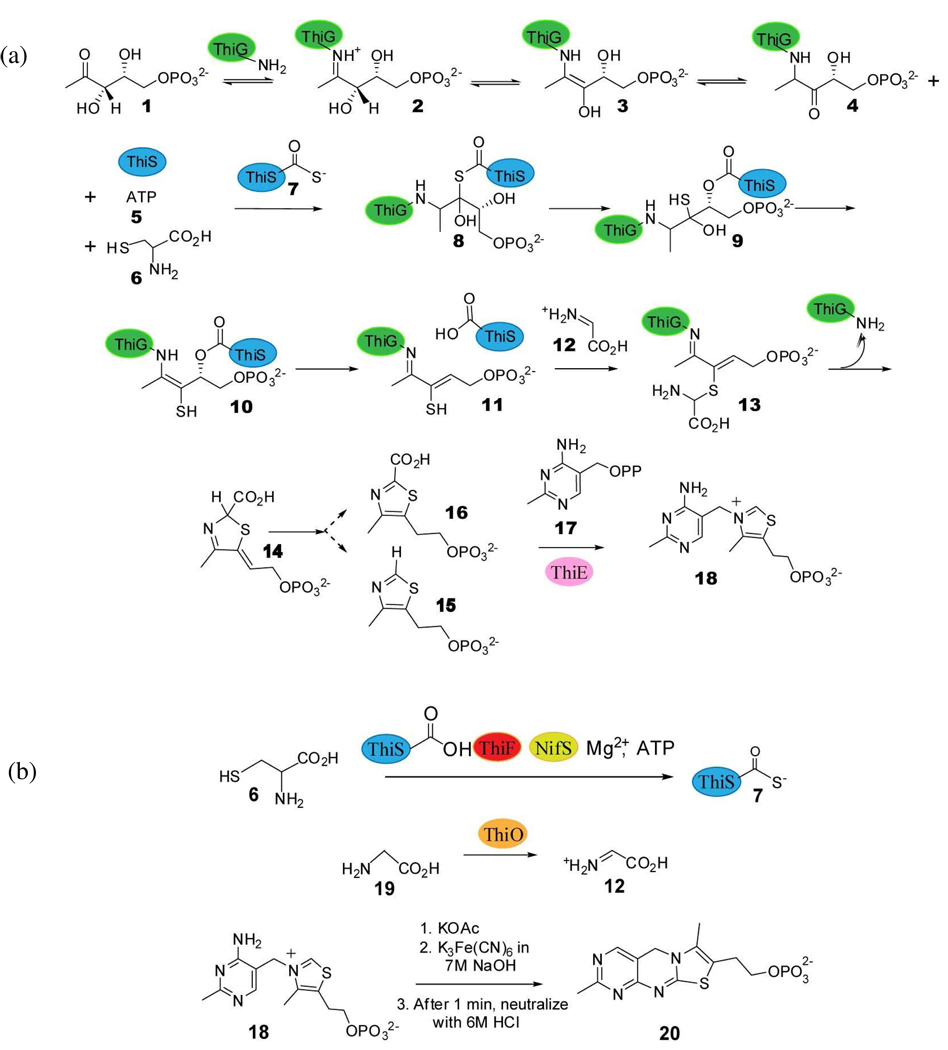
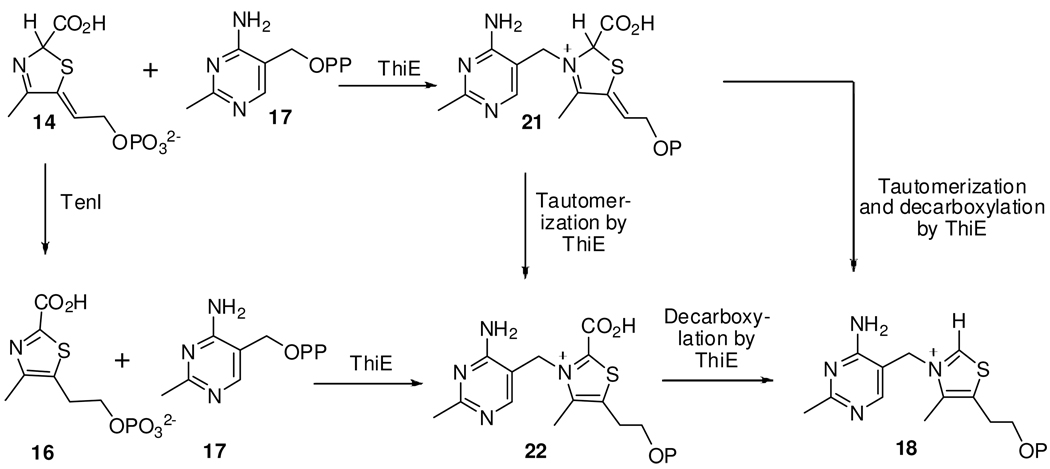
Thiamin is an important cofactor in prokaryotes, lower eukaryotes and plants and is biosynthesized by a complex enzymatic pathway. The biosynthesis of thiamin in microorganisms has been studied extensively and most of the genes involved have been characterized.1,2 The thiazole and the pyrimidine are biosynthesized by two separate unique mechanisms in bacteria and Saccharomyces cerevisiae. In Bacillus subtilis, the thiazole tautomer cThz*-P 14 is formed by an oxidative condensation of glycine, DXP 1 and cysteine3–6 and HMP-PP 17 is produced by rearrangement of aminoimidazole ribonucleotide followed by phosphorylation7–9. Thiamin phosphate 18 is then formed by the coupling of the pyrimidine and the thiazole heterocycles (Figure 1a and 1b).6,10 Thz-P 15 and cThz-P 16 are potential intermediates in this coupling reaction. A final phosphorylation gives thiamin pyrophosphate, the biologically active form of the cofactor.
Figure 1.
(a) Mechanistic proposal for the formation of the thiamin thiazole in B. subtilis. The thiazole synthase (ThiG) reaction product 14 is coupled to HMP-PP 17 to form thiamin phosphate 18. Thz-P 15 and cThz-P 16 are possible intermediates in this coupling reaction. (b) Reactions showing conversion of ThiS by ThiF and NifS into ThiS-COSH 7, glycine oxidase ThiO converting glycine 19 to glycine imine 12 and the oxidation of thiamin phosphate 18 to thiochrome 20.
In bacteria such as Bacillus subtilis, Bacillus licheniformis, Geobacillus kaustophilus and Geobacillus stearothermophilus, the tenA and tenI genes were found clustered with genes involved in thiamin thiazole biosynthesis8 (Figure 1a, Supplementary Information). Previous studies indicated the involvement of TenA and TenI in the regulation of the production of extracellular enzymes like neutral protease, alkaline protease, and levansucrase11. However, since both TenA and TenI are strongly repressed by thiamin12, these regulatory roles did not provide any insights into the actual biochemical function of these two proteins. Subsequently, TenA was characterized as a thiaminase II involved in pyrimidine salvage from base-degraded thiamin13,14. The two genes do not co-occur with high frequency and TenI is less widely distributed than TenA.14,15 B. subtilis TenI is a 22,783 Da protein14 and it shares high sequence similarity with thiamin phosphate synthase (ThiE) (Figure 2, Supplementary Information). Orthologs of TenI have been identified in bacilli, clostridiae, members of the CFB (cytophagea/flavobacter/bacteriodes) group, Campylobacter jejuni, Fusobacterium nucleatum, Chlorobium tepidum, and Aquifex aeolicus and various other classes of bacteria and a ThiD-TenI-ThiE fusion has been found in Porphyromonas gingivalis. (Figure 1b, Supplementary Information). We have previously reported a crystal structure of TenI, but the function of this protein in thiamin biosynthesis remained unknown.14 In this paper, we identify the function of TenI as a thiazole tautomerase, describe the structure of the enzyme complexed with its reaction product, and propose a mechanism for the reaction.
Figure 2.
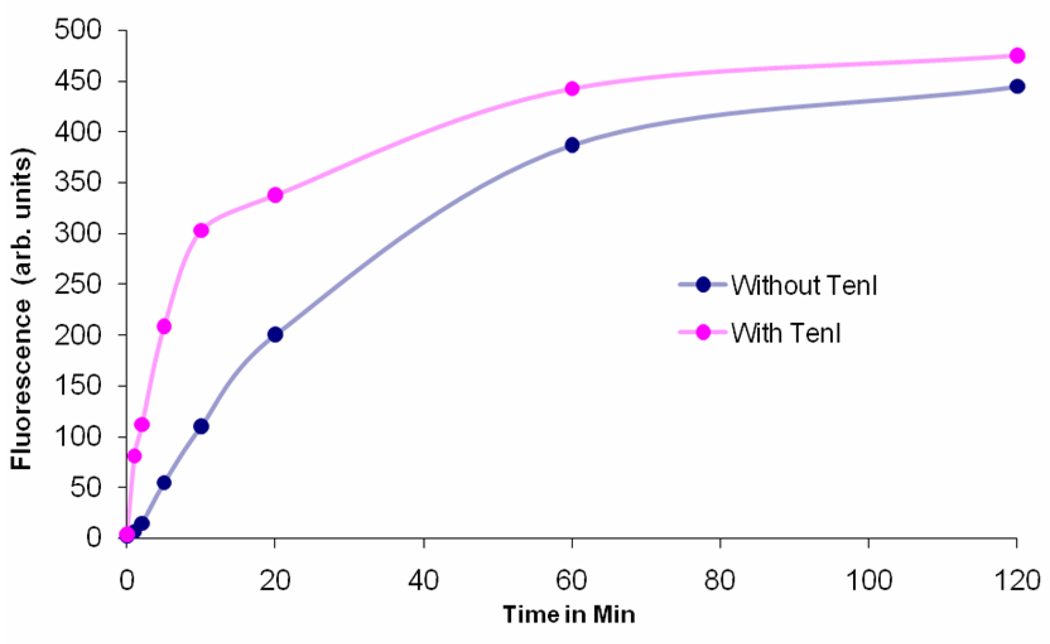
Time course for the thiazole reconstitution reaction run in the presence and absence of TenI.
RESULTS and DISCUSSION
TenI accelerates the rate of thiazole formation
To study the effect of TenI on the rate of in vitro thiazole formation, the reaction product was coupled with HMP-PP 17 and the rate of formation of thiamin phosphate 18 was measured. The thiazole reconstitution was accomplished by first incubating ThiF, NifS and ThiSG with cysteine and ATP for 1.5 h to form ThiS-COSH. Glycine, ThiO, HMP-PP and ThiE were added, and the reaction was then initiated by adding DXP 1. 50 µL aliquots were quenched after 0 min, 1 min, 2 min, 5 min, 10 min, 20 min, 60 min and 120 min. To quantitate the amount of thiamin phosphate 18 formed, each aliquot was oxidized to thiochrome phosphate, which was assayed by HPLC with fluorescence detection.3 An identical reconstitution was run in the presence of TenI.
At short reaction times, TenI has a significant enhancement effect on the rate of thiazole formation. (Figure 2 and Supplementary Information Figure 3). However, this effect is reduced at longer reaction times. This suggests that while TenI enhances the rate of thiazole formation, it is not essential for the process. The thiazole reconstitution is complex. Therefore, the next task was to identify which of the reaction components are enhanced by TenI.
TenI does not affect glycine oxidase, thiamin phosphate synthase, or ThiS-COSH formation
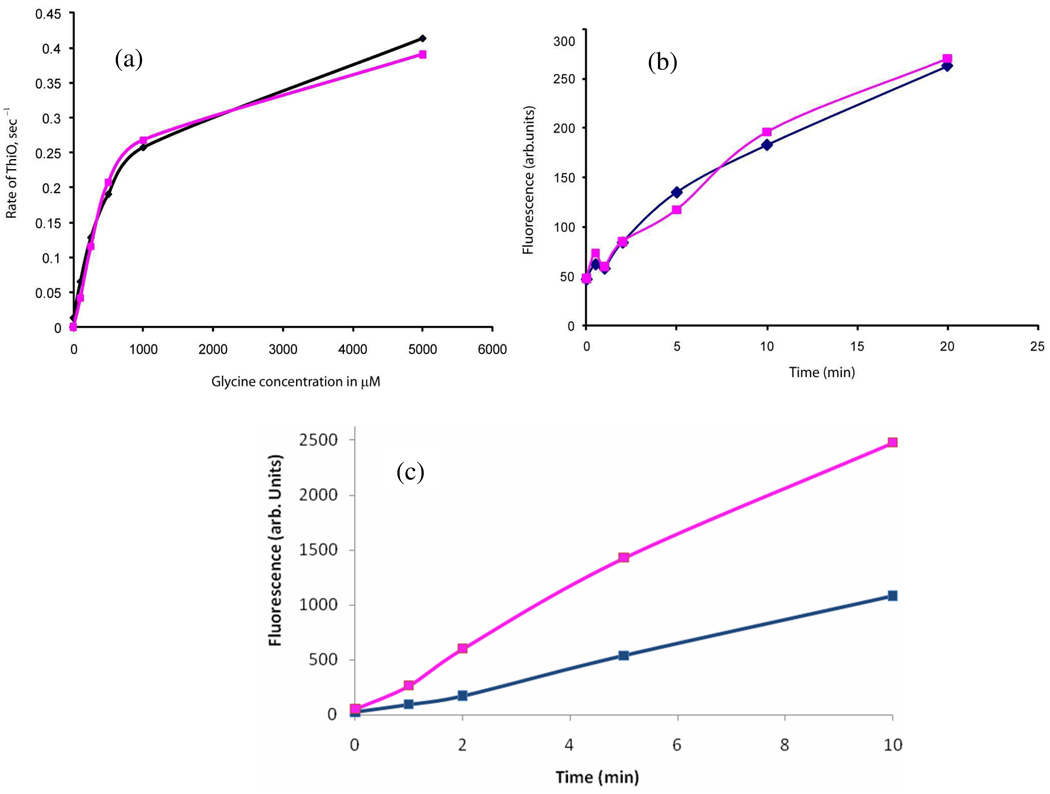
The glycine oxidase activity of ThiO was assayed in the presence and absence of TenI by monitoring hydrogen peroxide formation.16 This experiment demonstrated that the rate of hydrogen peroxide production with increasing concentrations of glycine was unaffected by TenI (Figure 3a).
Figure 3.
TenI does not affect glycine oxidase, thiamin phosphate synthase or ThiS-COSH formation. The blue trace is the reaction in the absence of TenI and the pink trace is the reaction in the presence of TenI. (a) The presence of TenI does not affect the rate of glycine oxidase (ThiO). (b) The presence of TenI does not affect the rate of the HMP-PP/Thz-P coupling reaction catalyzed by thiamin phosphate synthase. (c) The rate of the thiazole synthase catalyzed reaction, using preformed ThiS-COSH, is enhanced by TenI.
The involvement of TenI in the thiamin phosphate synthase (ThiE) catalyzed coupling of Thz-P and HMP-PP was similarly probed. In each case thiamin phosphate was assayed by HPLC after oxidation to thiochrome phosphate.17 TenI also has no effect on this reaction (Figure 3b).
To evaluate the effect of TenI on ThiS-COSH formation, the thiazole reconstitution reaction was run using preformed ThiS-COSH in the presence and absence of TenI. If TenI was acting on ThiS-COSH formation, no TenI effect should be observed in this reconstitution. In the event, a clear increase in the rate of production of thiochrome could again be seen at the early stages of the reaction in the presence of TenI (Figure 3c). This suggests that TenI is not influencing the rate of ThiS-COSH formation and leaves only two possible functions for TenI: enhancement of the ThiG-catalyzed formation of cThz*-P 14 or catalysis of the conversion of cThz*-P 14 to a better substrate for thiamin phosphate synthase.
Thz-P 15 is the characterized substrate for thiamin phosphate synthase and all of our mechanistic studies with this well-studied enzyme have been carried out using this compound.10,17,18 The substrate tolerance of this enzyme now becomes important in the context of TenI function. In the reconstitution of the thiazole biosynthesis described in Figure 2, cThz*-P 14, the product of the thiazole synthase6 is clearly a substrate for thiamin phosphate synthase. Therefore if TenI is catalyzing the conversion of cThz*-P to a better substrate, Thz-P 15 and cThz-P 16 are the only possibilities (Figure 1). We therefore next evaluated cThz*-P, cThz-P and Thz-P as substrates for thiamin phosphate synthase.
Thiamin phosphate synthase (ThiE) can use Thz-P (15), cThz-P (16) and cThz*-P (14) as substrates
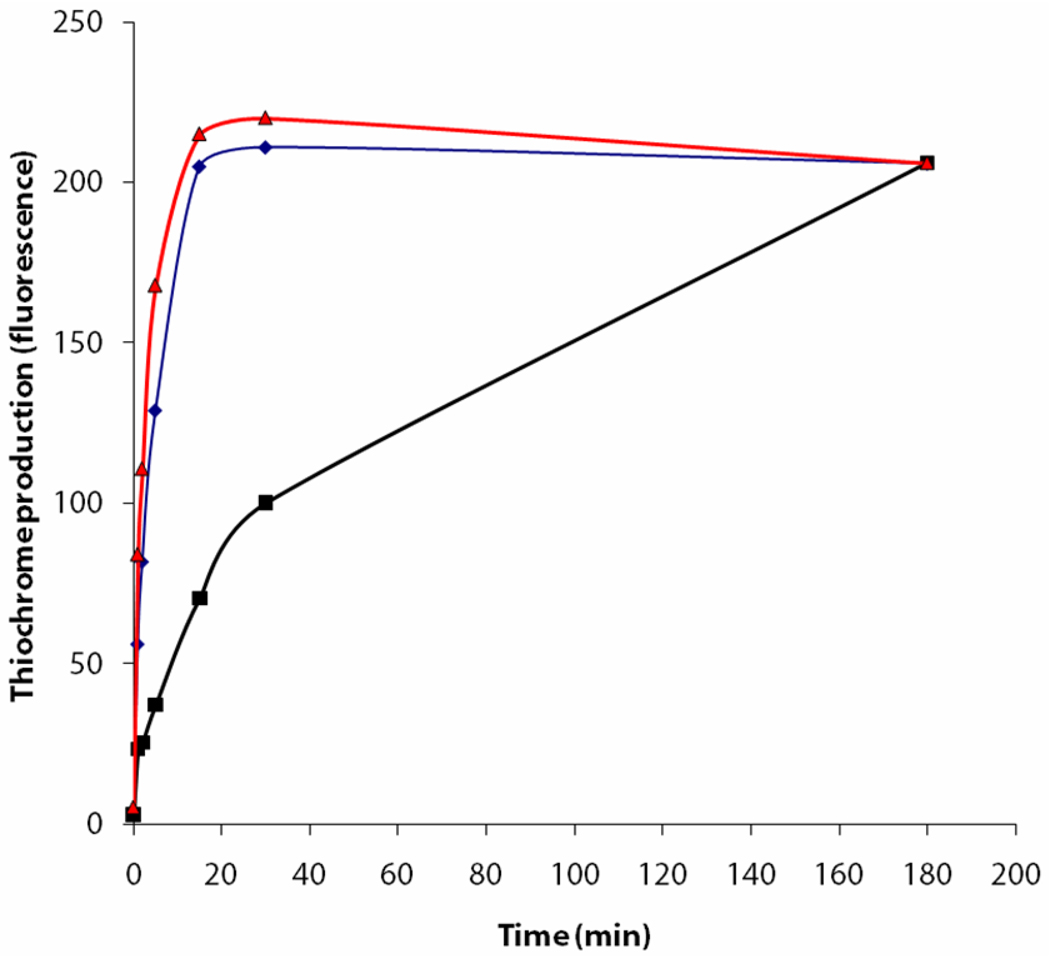
The relative rates of the coupling of Thz-P, cThz-P and cThz*-P with HMP-PP 17 were measured. The reactions were quenched at 0, 1, 2, 5, 15, 30 and 180 mins and the thiamin phosphate formed was converted to thiochrome phosphate for HPLC analysis with fluorescence detection. The results are shown in Figure 4. Early in the course of the reaction, cThz-P and Thz-P are significantly better substrates than cThz*-P. As expected from the data shown in Figure 2, the yield of thiamin phosphate is the same for all three substrates at longer reaction times. This experiment suggests that the enhanced formation of thiamin phosphate shown in Figures 2 and 4 may be a consequence of TenI catalyzed aromatization of 14 to generate 15 or 16 which are better substrates for thiamin phosphate synthase. Previous experiments have conclusively shown the formation of thiamin phosphate by coupling of Thz-P 15 with HMP-PP.17,19 Plausible routes by which cThz-P 16 and cThz*-P 14 may be converted to thiamin phosphate are shown in Figure 5.
Figure 4.
Substrate selectivity of thiamin phosphate synthase. The plot shows the time course for the alkylation of cThz*-P 14 (black trace), Thz-P 15 (blue trace) and cThz-P 16 (red trace) by HMP-PP 17.
Figure 5.
Routes for the conversion of cThz*-P 14 and cThz-P 16 to thiamin phosphate 18. In bacteria lacking TenI, ThiE can catalyze the coupling of the thiazole tautomer 14 and HMP-PP 17 to give 21, which is converted to 18 directly or via intermediate 22.
Stability of cThz*-P
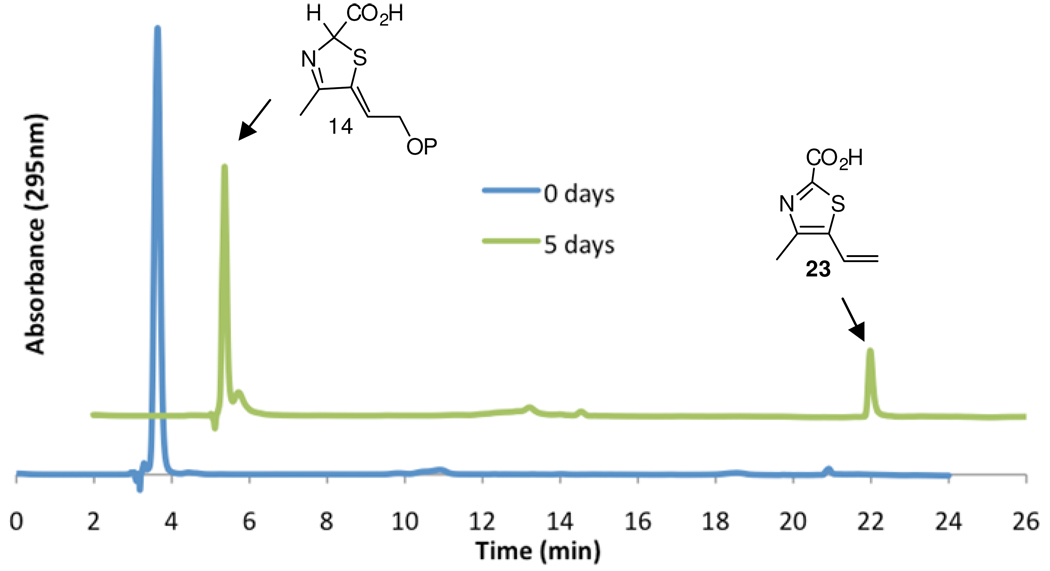
cThz*-P is surprisingly stable and survives purification and structural analysis without undergoing aromatization.6 On standing at room temperature, it slowly degrades to the carboxy vinylthiazole 23, with a half life of approximately 13.5 days (Figure 6 and Supplementary Information, Figure 6). As B. subtilis has a dividing time of about 30 minutes, in vivo aromatization of 14 clearly requires enzymatic catalysis.
Figure 6.
Reverse-phase HPLC analysis of the degradation of cThz*-P 14 to carboxy vinylthiazole 23. The blue trace shows pure cThz*-P, and the green trace shows the formation of the carboxy vinylthiazole as a degradation product after 5 days. Chromatograms are offset by 2 minutes.
TenI catalyzes the aromatization of cThz*-P to cThz-P
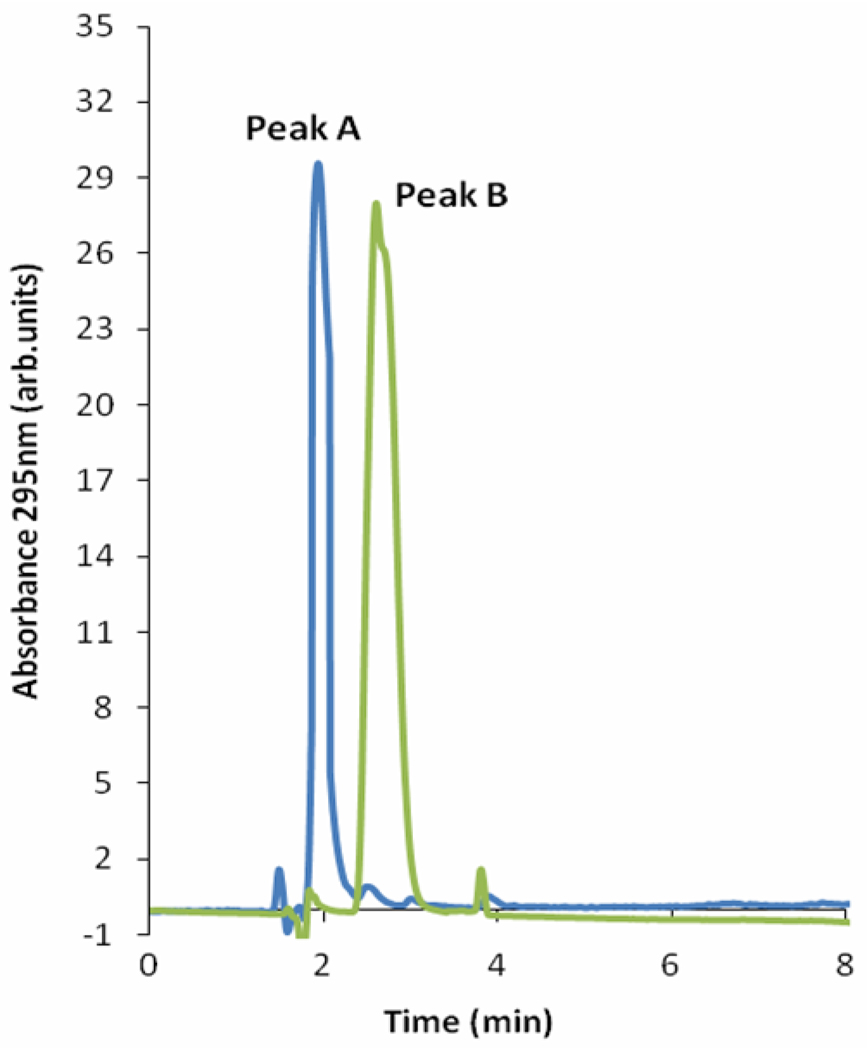
Incubation of cThz*-P 14 with TenI followed by HPLC analysis gave the chromatogram in Figure 7 showing the consumption of 14 (Peak A) and the production of a new compound (Peak B). Peak B was identified as cThz-P 16 and not Thz-P 15 by comigration with authentic reference compounds.6 It was not possible to carry out kinetics studies due to the scarcity of cThz*-P which was prepared from a metabolite bound to the S. cerevisiae thiazole synthase (cThz*-P preparation in Supplementary Information).20,21
Figure 7.
HPLC analysis demonstrating that TenI catalyzes the conversion of cThz*-P 14 (blue trace, Peak A) to a new compound (green trace) that comigrates with cThz-P 16 (Peak B).
This conversion of cThz*-P to cThz-P catalyzed by TenI was also verified by negative mode ESI-MS analysis of the reaction (both cThz*-P to cThz-P have distinct fragmentation patterns, Figure 7, Supplementary Information).
Addition of TenI to the thiazole reconstitution yields cThz-P
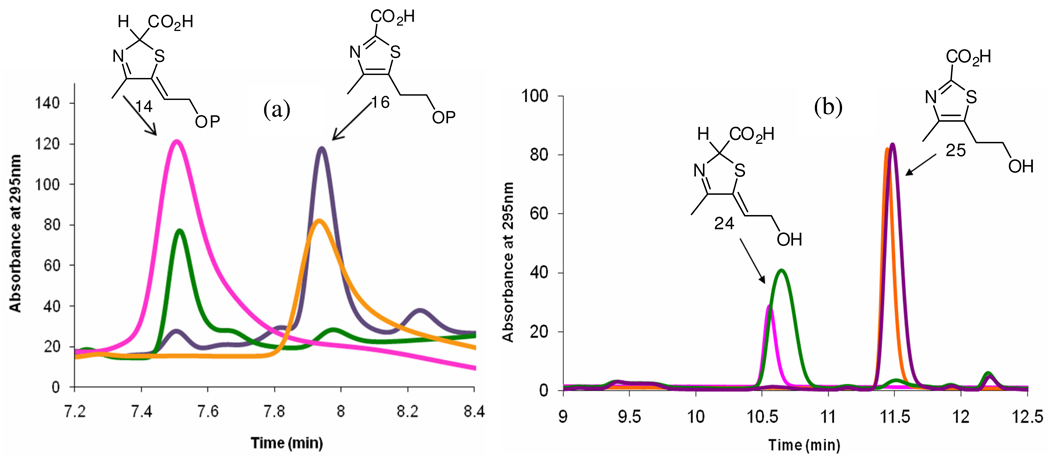
The thiazole reconstitution reaction was run in the presence and absence of TenI as described above and analyzed by HPLC using a strong anion exchange column (Figure 8a). In the absence of TenI, cThz*-P was the only thiazole product formed while in the presence of TenI, cThz-P was the major reaction product accompanied by a very small amount of cThz*-P.
Figure 8.
Reconstitution of thiazole biosynthesis in the presence and absence of TenI. (a) Comigration of the product of thiazole biosynthesis in the absence of TenI (green) with cThz*-P (pink) and comigration of the product of thiazole biosynthesis in the presence of TenI (purple) with cThz-P (orange). (b) Comigration of the alkaline phosphatase treated product of thiazole biosynthesis in the absence of TenI (green) with cThz* (pink) and comigration of the alkaline phosphatase treated product of thiazole biosynthesis in the presence of TenI (purple) with cThz (orange).
To further confirm the identity of the reaction product, cThz*-P and cThz-P and the products of the two reconstitution reactions were dephosphorylated by treatment with alkaline phosphatase and the resulting alcohols were reanalyzed by reverse phase HPLC. Again, the dephosphorylated product of the reconstitution reaction without TenI comigrated with cThz* 24, while the product of the reconstitution reaction run in the presence of TenI comigrated with cThz 25 (Figure 8b).
Reversibility of the aromatization reaction catalyzed by TenI
Incubation of TenI with cThz-P did not result in the formation of detectable quantities of cThz*-P by NMR analysis. In addition, if TenI catalyzes the de-aromatization of cThz-P, even at very low levels, it should be possible to observe TenI dependent H/D exchange at C2 of the phosphoethyl side chain of cThz-P. In the event, incubation of cThz-P and TenI (1:1 ratio) in 50% D2O:H2O did not result in any detectable H/D exchange after 12 hours. This is consistent with a large free energy difference between cThz-P and cThz*-P.
Structure of the TenI-cThz-P Complex
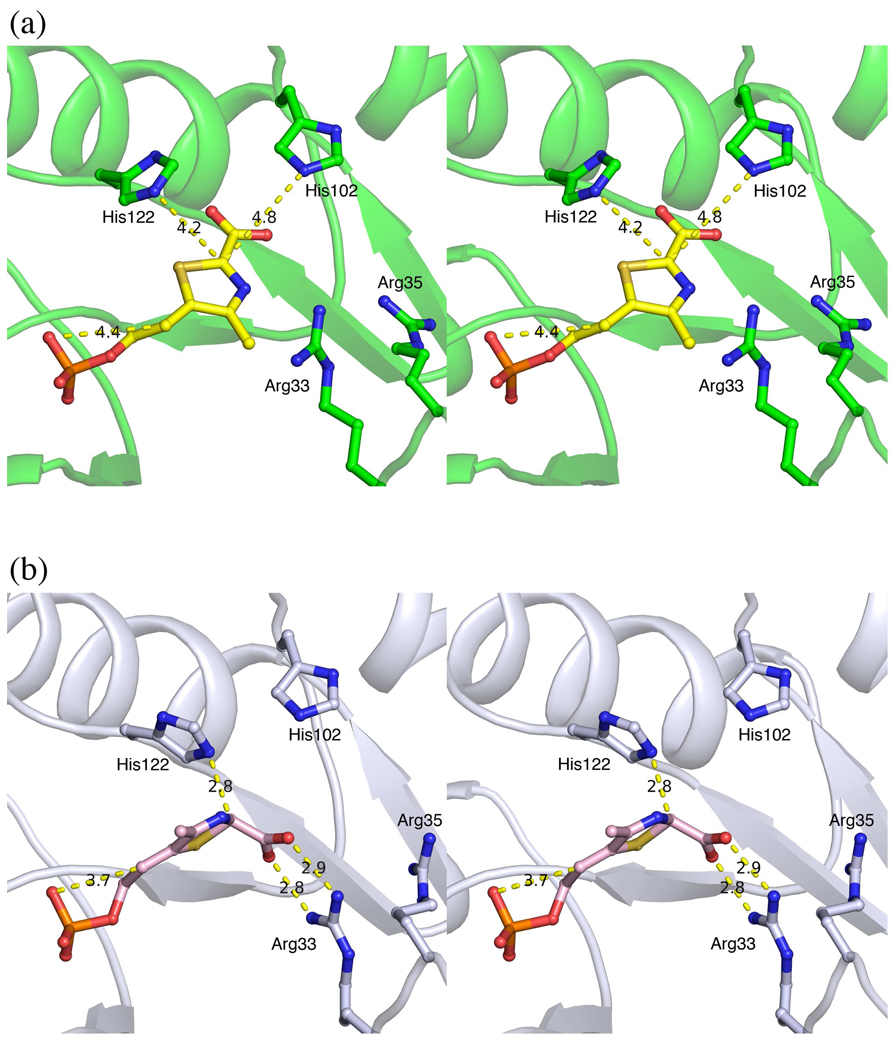
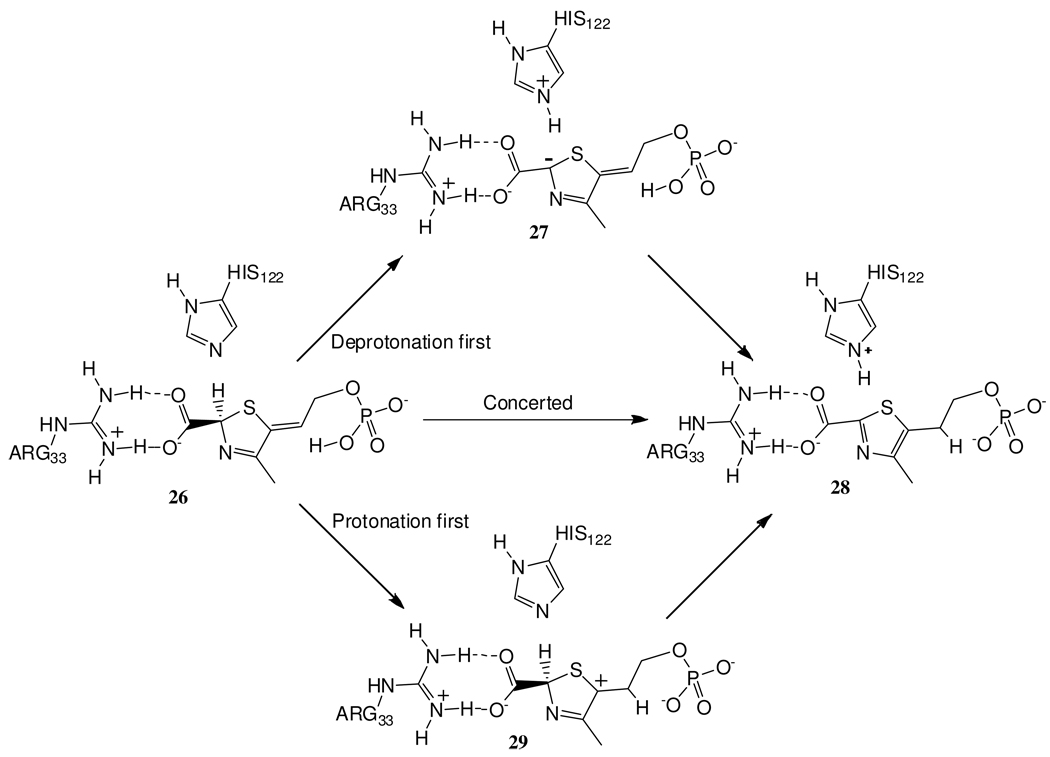
To clarify the mechanism of the thiazole tautomerization reaction, a structure of TenI with the reaction product cThz-P 16 bound at the active site was obtained. The active site structure of the enzyme is shown in Figure 9a. From this structure, it is not possible to unambiguously identify the acid and base required to catalyze the aromatization reaction. His102 and His122 are too far from the C2 of the thiazole and are improperly oriented for the deprotonation. In addition, an acidic residue to protonate the side chain is not apparent. The only possibility is the phosphate located 4.4 Å away. Assuming that this non-optimal arrangement of active site residues was due to the substantial change in the structure of the substrate following aromatization, we modeled cThz*-P into the active site. In this model (Figure 9b), His122 is reasonably positioned to deprotonate C2 of the thiazole tautomer and the substrate phosphate is suitably positioned for protonation of the side chain double bond. In addition, the substrate carboxylate forms hydrogen bonding/electrostatic interactions with Arg33. In support of this model, the H122Q and H122A mutants are inactive (detection limit < 0.5% wild type activity), and cThz* is not a substrate for TenI indicating that the phosphate group is essential.
Figure 9.
a) Stereoview of the active site of the TenI with bound cThz-P. b) Stereoview of a model of the active site of TenI with bound cThz*-P.
A mechanistic analysis of the thiazole aromatization reaction is outlined in Figure 10. Each reaction involves a phosphate-mediated protonation and a histidine mediated deprotonation sequence. At this point, it is not possible to determine the order of the protonation/deprotonation reactions.
Figure 10.
Mechanistic proposals for the TenI-catalyzed thiazole aromatization reaction.
Conclusions
cThz*P 14 is the product of the bacterial thiazole synthase. This thiazole tautomer is surprisingly stable and its aromatization requires enzymatic catalysis. In this paper we demonstrate that TenI is the missing thiazole tautomerase and catalyzes an irreversible aromatization reaction converting cThz*-P 14 to cThz-P 16. We also prove that while thiamin phosphate synthase can convert each of cThz*-P 14, Thz-P 15, and cThz-P 16 into thiamin phosphate, cThz-P 16 is likely the true substrate in vivo. A model for the structure of cThz*-P 14 bound at the active site of TenI, based on the structure of the TenI/cThz-P complex, identifies the substrate phosphate and His122 as the acid/base residues involved in catalysis. The identification of the function of TenI completes the identification of all of the enzymes needed for thiamin biosynthesis by the major bacterial pathway.
Experimental Methods
Source of Chemicals
All chemicals and snake venom nucleotide pyrophosphatase were purchased from Sigma-Aldrich Corporation (USA) unless otherwise mentioned. Calf intestinal phosphatase was obtained from New England Biolabs. LB medium was obtained from EMD Biosciences. Kanamycin, ampicillin and IPTG were purchased from LabScientific Inc. NTA resin was the NTA superflow by Qiagen. E. coli BL21(DE3) competent cells were obtained from Invitrogen. The microcon membrane filters were from Millipore. TALON metal affinity resin was purchased from BD Biosciences. Analytical HPLC (Agilent 1100 instrument) was carried out using a Phenomenex Gemini C18 110A (150×4.6 mm, 5 µm ID) reverse phase column for thiochrome analysis, a Supelco LC-18-T (150×4.6 mm, 3 µm ID) column for thiazole reconstitution analysis and a Phenosphere Strong Anion-Exchange 80A (250×4.6 mm, 5 µm ID) column for the anion exchange chromatography. HPLC purifications were carried out using a semi-prep Supelco LC-18-T (250×10 mm, 5 µm ID) column. HPLC grade solvents were obtained from Fisher Scientific. The Superdex 200 gel-filtration column was obtained from Pharmacia. A previously synthesized stock of [1-13C]-DXP22 was used as the substrate of the thiazole reconstitution reactions and previously synthesized cThz-P and cThz6 were used as reference compounds for the TenI catalyzed reaction.
Enzyme overexpression and purification
E. coli BL21(DE3) containing the ThiSG overexpression plasmid (ThiG is co-purified with ThiS for stability) in pET16b was grown in LB medium containing ampicillin (40 µg/mL) with shaking at 37 °C until the OD600 reached 0.6. At this point, protein overexpression was induced with IPTG (final concentration = 2 mM) and cell growth was continued at 15°C for 16 h. The cells were harvested by centrifugation and the resulting cell pellets were stored at −80°C. To purify the protein, the cell pellets from 1L of culture were resuspended in 25 mL lysis buffer (10 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, pH 8) and lysed by sonication (Heat Systems Ultrasonics model W-385 sonicator, 2 s cycle, 50% duty). The resulting cell lysate was clarified by centrifugation and the ThiSG protein was purified on Ni-NTA resin following the manufacturer’s instructions. After elution, the protein was desalted using a 10-DG column (BioRad) pre-equilibrated with 50 mM Tris-HCl buffer, pH 7.8. The remaining proteins ThiF (pET22), NifS (pET16), ThiO (pET22) ThiE (pQE32 and pREP4), TenI (pET28b), H122Q TenI (pET28b) and H122A TenI (pET28b) were overexpressed and purified in a similar manner. NifS, ThiO and ThiE were stored in aliquots at −80 °C in 20% glycerol. ThiSG, TenI and ThiF were purified immediately before use.
Reconstitution of the thiazole synthase catalyzed reaction on an analytical scale (in the presence and absence of TenI)
All solutions were made with 50 mM Tris buffer, pH 8. Final concentrations of the reactants are given in parentheses. Cysteine (0.35 mM), DTT (0.70 mM), ATP (0.60 mM) and MgCl2 (3.5 mM) were incubated with purified ThiSG (1.25 µM), ThiF (1.24 µM) and 70 µL NifS (1.38 µM) for 1.5 hours. Total volume of this solution was 425 µL. Glycine (6.50 mM), DXP (0.33 mM), MgCl2 (3.5 mM) and ThiO (6.8 µM) were then added to this reaction mixture and the final volume of the reconstitution mixture now was 610 µL. TenI was added in the reconstitution reaction to a final concentration of 10 µM to check for the acceleration of the rate of thiazole formation. In a control reaction set up in exactly the same way, the same volume of buffer was added into the reaction instead of TenI. This mixture was incubated for an additional 2 hours. The reaction mixture was then analyzed for product formation using the thiochrome assay (see below). In this reconstitution, 16% of the DXP was converted to product. This is a 3-fold improvement over our previously reported reconstitution, and corresponds to about 12 turnovers by the thiazole synthase.
Thiochrome Assay:3,4
The thiochrome assay involves conversion of the thiazole product of the reconstitution to thiamin phosphate 18 and further to thiochrome phosphate. The product of the thiazole reconstitution is reacted with HMP-PP 17 (0.5 mM) in the presence of thiamin phosphate synthase (ThiE) (1.00 µM). The reaction is allowed to stand at room temperature for 2 hours and then quenched with an equal volume of 10% TCA. Potassium acetate (50 µL of 4M) is added to 100 µL of the quenched reaction followed by oxidative cyclization to thiochrome phosphate using 50 µL of a saturated solution of K3Fe(CN)6 in 7M NaOH. The oxidation reaction is neutralized after 1 minute with 6M HCl and analyzed by reverse phase HPLC with fluorescence detection (excitation at 365 nm, emission at 450 nm). The following linear gradient, at a flow rate of 1 mL/min, was used. Solvent A is water, solvent B is 100 mM K2HPO4, pH 6.6, solvent C is methanol. 0 min: 100% B; 2 min: 10% A, 90%B; 10 min: 25% A, 15% B, 60% C; 12 min: 25% A, 15% B, 60%; 15 min: 100% B; 17 min: 100%B.
ThiO activity assay:16
25 mL of assay solution containing 4 mM phenol, 100 mM 4-amino-antipyrene and 2 units/mL HRP was made. To 500 uL of the assay solution, 10 mM, 5 mM, 1 mM, 500 µM, 250 µM and 100 µM glycine was added and the volume each time was diluted to 505 uL. In each case the reaction was initiated by the addition of a final concentration of 6.6 µM ThiO and 10 µM TenI. A parallel set of reactions was similarly run in the absence of TenI. The rate of glycine oxidation was measured, at various glycine concentrations, by monitoring the absorbance change at 500 nm for 600sec.
ThiE activity assay:17
437 µM of Thz-P and 485 µM HMP-PP were mixed with 10 µM ThiE and 10 µM TenI in a final volume of 700uL of 50 mM Tris-HCl buffer, 2 mM MgCl2, pH 7.8. An identical reaction lacking TenI was run under the same conditions. 100 µL aliquots of each reaction mixture were quenched after 0 min, 0.5 min, 1 min, 2 min, 5 min, 10 min and 20 min and oxidized to thiochrome phosphate. Reaction mixtures were analyzed by HPLC with fluorescence detection.
ThiG activity assay:3,4
To evaluate the effect of TenI on ThiG, the thiazole reconstitution reaction was run using preformed ThiS-COSH in the presence and absence of TenI. Cysteine (0.35 mM), DTT (0.70 mM), ATP (0.60 mM) and MgCl2 (3.5 mM) were incubated with purified ThiSG (1.25 µM), ThiF (1.24 µM) and NifS (1.38 µM) for 1.5 hours. The total volume of this solution was 425 µL. Desalting through a 10-DG column (BioRad) pre-equilibrated with 50 mM Tris-HCl, pH 7.8 yielded ThiS-COSH. Two sets of the thiazole reconstitution reactions were carried out as described above, one in the presence of TenI and the other in its absence. 100 µL aliquots of these reaction mixtures were quenched at time points of 0 min, 1 min, 2 min, 5 min and 10 min and oxidized to thiochrome phosphate which was detected by reverse phase HPLC with fluorescence detection.
Activity assay for TenI with cThz*-P
cThz*-P was obtained as described in Supplementary Information (Figure 4 and 5, Supplementary Information). 1 µM TenI was added to 100 µM cThz*-P in a final assay volume of 300 µL of 50mM Tris buffer, pH 7.6. The mixture was incubated for 120 min, filtered using a 10 kDa MW cut-off filter, and analyzed by HPLC. Mutants were similarly analyzed except the reaction time was increased to 12 hours.
HPLC conditions for separation of cThz*-P and cThz-P using analytical strong anion exchange chromatography
The following linear gradient, at a flow rate of 1 mL/min, on the Phenosphere Strong Anion-Exchange 80A (250×4.6 mm, 5 µm ID) column was used: solvent A is water, solvent B is 100 mM triethylammonium acetate, pH 7.8. 0 min: 100% A; 1 min: 100% A; 4 min: 100% B; 7 min: 100% A; 10 min: 100%A.
HPLC conditions for separation of cThz* and cThz using analytical reverse chromatography
The following linear gradient, at a flow rate of 1 mL/min, on the Supelcosil LC-18-T column (150×4.6 mm, 3 µm ID) was used: solvent A is water, solvent B is 100 mM KPi, pH 6.6, solvent C is methanol. 0 min: 100% B; 2 min: 100% B; 4min 10% A, 90%B; 9 min: 10% A, 25% B, 65% C; 14 min: 10% A, 25% B, 65% C; 16 min: 100%B; 20min 100%B.
Equilibration of cThz-P/CThz*-P in the presence of TenI
540 µM TenI in 50mM KPi pH 7.6 in 50% D2O: H2O was added to 540 µM cThz-P (TenI:cThz-P ratio approximately 1:1) in a total volume of 650 µL. The reaction mixture was analyzed by NMR after 12 hours.
Expression and Purification of Bacillis subtilis TenI for crystallography
The TenI overexpression strain was grown at 37 °C with vigorous agitation (200 rpm) LB medium containing 30 µg/mL kanamycin to an OD600 of 0.7, at which point the cells were induced with 500 µM IPTG and allowed to incubate overnight at 22 °C under conditions of mild mixing (180 rpm). The cells were harvested by centrifugation (6,000 g) for 15 min at 4 °C and stored at −80 °C for later use. All purification steps were carried out at 4 °C. The cell pellet was suspended in 50 mL binding buffer (50 mM sodium phosphate, pH 7.0 and 300 mM NaCl), and lysed by sonication. The crude extract was centrifuged at 4 °C for 30 min at 50,000 g and the resulting supernatant was augmented with 5 mM imidazole and loaded onto a column containing 2 mL of TALON metal affinity resin equilibrated with 50 mL binding buffer. The column was washed with 20 column volumes of binding buffer plus 5 mM imidazole, followed by 5 column volumes of binding buffer plus 10 mM imidazole. The His6-tagged TenI was eluted from the column with elution buffer (50 mM sodium phosphate, pH 7.0, 300 mM NaCl and 300 mM imidazole). The recombinant TenI was further purified on a Superdex 200 gel-filtration column and eluted in the storage buffer (25 mM Tris-HCl, pH 8.5, 150 mM NaCl, and 1 mM thiamin-phosphate). The fractions containing pure TenI were combined and concentrated to 12 mg/mL using a 10 kDa cutoff concentrator (Vivaspin) and stored at −80 °C. Protein concentration was determined by the Bradford method with bovine serum albumin as the standard. The purity of TenI was determined by SDS-PAGE analysis and found to be greater than 99%.
Crystallization of TenI with the cThz-P 16 bound in the active site
Crystals of ligand free TenI were grown from 1.65 – 1.75 M ammonium sulfate, 100 mM bicine, pH 8.7 – 9.6, 2% PEG400 (w/v), and 8 mM L-cysteine using the hanging drop vapor diffusion method.12 To obtain the product complex, the crystals were first dialyzed into 2.38 M sodium malonate (pH 7.0) (Hampton Research) to remove the sulfate ions, by gradually increasing the sodium malonate concentration and decreasing the ammonium sulfate concentration in 30 steps with 5 min incubation for each step. Subsequently the crystals were soaked overnight in 2.38 M sodium malonate, 21.5 mM bicine (pH 9.0), 0.01% PEG400 (w/v), 0.5 mM L-cysteine, 4% glycerol and 11.5 mM cThz-P, followed by flash freezing in liquid nitrogen.
X-ray Data Collection and Processing
The X-ray intensity data from the TenI-cThz-P complex were measured at the A1 beamline of the Cornell High Energy Synchrotron Source (CHESS) using a Quantum 210 CCD detector (Area Detector Systems Corp.). Data were collected over 180° using a 10 s exposure time and 1° oscillation per frame with a crystal to detector distance of 200 mm. The data were integrated and scaled using HKL2000.23
Structure Determination and Refinement
The structure of TenI-cThz-P complex was determined by Fourier synthesis using the previously reported TenI structure (PDB: 1YAD) as the starting model. The first round of rigid-body refinement with the starting model by REFMAC524 reduced the R-factor to 0.286 (Rfree 0.294). cThz-P was modeled in based on clear electron density. The model was refined through iterative cycles of restrained refinement by REFMAC5 and PHENIX25, and manual rebuilding in COOT.26 Refinement statistics are shown in Table 1, Supplementary Information.
Modeling of cThz*-P in the active site
The initial coordinates of proposed intermediate cThz*-P were 26generated using program SKETCHER of CCP4 suite.27 Using the crystal structure of TenI complexed with Thz-P as the template, the intermediate cThz*-P was docked into the active site and the key interacting residues were adjusted manually in COOT.26
Supplementary Material
Acknowledgement
We thank David Hilmey for the synthesis of cThz 25 and Kathryn McCulloch for helpful discussions. This research was supported by the Robert A Welch Foundation (A-0034) and NIH grants DK44083 to TPB and DK67081 to SEE. The Macromolecular Diffraction beamlines at CHESS (MacCHESS) facility are supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
Abbreviations
- Thz-P
2-(4-methylthiazol-5-yl)ethyl phosphate
- cThz-P
2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate
- cThz*-P
(R,Z)-2-(2-carboxy-4-methylthiazol-5(2H)-ylidene)ethyl phosphate
- cThz
5-(2-hydroxyethyl)-4-methylthiazole-2-carboxylic acid
- cThz*
(R,Z)-5-(2-hydroxyethylidene)-4-methyl-2,5-dihydrothiazole-2-carboxylic acid
- cThz-ADP
adenylated 2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate
- cThz*-ADP
adenylated (R,Z)-2-(2-carboxy-4-methylthiazol-5(2H)-ylidene)ethyl phosphate
- HRP
horse radish peroxidase
- DXP
1-deoxy-D-xylulose-5-phosphate
- HMP-P
4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate
- HMP-PP
4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate
- DTT
dithiothreitol
- TCEP
tris(2-carboxyethyl)phosphine
- IPTG
isopropyl-β-D-thiogalactopyranoside
- KPi
potassium phosphate buffer
- NTA
nitrilotriacetic acid
- Tris
tris(hydroxymethyl)aminomethane
- NMR
nuclear magnetic resonance
- ThiS, ThiF, NifS, ThiG, ThiO
thiazole biosynthetic proteins
- TenA
thiaminase II
- ThiE
thiamin phosphate synthase
- tenI
gene of unknown function
- TenI
corresponding protein of unknown function
Footnotes
The coordinates of TenI-cThz-P complex has been deposited in the Protein Data Bank under accession number 3QH2.
Supporting Information Available. Figures for the gene neighborhood for TenI and TenA, comparison of primary sequence of TenI with ThiE, time course for effect of TenI on thiazole reconstitution, procedure for preparing and assaying cThz*-P, NMR analysis of cThz*-P decomposition, mass spectrum of cThz-P and cThz*-P and HPLC analysis of TenI mutants with cThz*-P and cThz- P/TenI crystal structure data and refinement statistics. This information is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Steven E. Ealick, Email: see3@cornell.edu.
Tadhg P. Begley, Email: begley@chem.tamu.edu.
REFERENCES
- 1.Jurgenson CT, Begley TP, Ealick SE. Annual Review of Biochemistry. 2009;78:569. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley TP, Chatterjee A, Hanes JW, Hazra A, Ealick SE. Current Opinion in Chemical Biology. 2008;12:118. doi: 10.1016/j.cbpa.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J-H, Dorrestein PC, Zhai H, Kinsland C, McLafferty FW, Begley TP. Biochemistry. 2003;42:12430. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]
- 4.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Chem. Biol. 2004;11:1373. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Settembre EC, Dorrestein PC, Zhai H, Chatterjee A, McLafferty FW, Begley TP, Ealick SE. Biochemistry. 2004;43:11647. doi: 10.1021/bi0488911. [DOI] [PubMed] [Google Scholar]
- 6.Hazra A, Chatterjee A, Begley TP. J. Am. Chem. Soc. 2009;131:3225. doi: 10.1021/ja806752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Gomez NC, Downs DM. Biochemistry. 2008;47:9054. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat. Chem. Biol. 2008;4:758. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee A, Hazra AB, Abdelwahed S, Hilmey DG, Begley TP. Angewandte Chemie, International Edition. 2010;49:8653. doi: 10.1002/anie.201003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peapus DH, Chiu H-J, Campobasso N, Reddick JJ, Begley TP, Ealick SE. Biochemistry. 2001;40:10103. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]
- 11.Pang ASH, Nathoo S, Wong SL. J. Bacteriol. 1991;173:46. doi: 10.1128/jb.173.1.46-54.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J-M, Zhang S, Saha S, Santa Anna S, Jiang C, Perkins J. J. Bacteriol. 2001;183:7371. doi: 10.1128/JB.183.24.7371-7380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. Nat. Chem. Biol. 2007;3:492. doi: 10.1038/nchembio.2007.13. [DOI] [PubMed] [Google Scholar]
- 14.Toms AV, Haas AL, Park J-H, Begley TP, Ealick SE. Biochemistry. 2005;44:2319. doi: 10.1021/bi0478648. [DOI] [PubMed] [Google Scholar]
- 15.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. J. Biol. Chem. 2002;277:48949. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 16.Settembre EC, Dorrestein PC, Park J-H, Augustine AM, Begley TP, Ealick SE. Biochemistry. 203;42:2971. doi: 10.1021/bi026916v. [DOI] [PubMed] [Google Scholar]
- 17.Reddick JJ, Nicewonger R, Begley TP. Biochemistry. 2001;40:10095. doi: 10.1021/bi010267q. [DOI] [PubMed] [Google Scholar]
- 18.Hanes JW, Ealick SE, Begley TP. J. Am. Chem. Soc. 2007;129:4860. doi: 10.1021/ja0679634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu H-J, Reddick JJ, Begley TP, Ealick SE. Biochemistry. 1999;38:6460. doi: 10.1021/bi982903z. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. J. Am. Chem. Soc. 2006;128:7158. doi: 10.1021/ja061413o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. J. Am. Soc. 2007;129:2914. doi: 10.1021/ja067606t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SV, Vu LD, Begley TP, Schoerken U, Grolle S, Sprenger GA, Bringer-Meyer S, Sahm H. J. Org. Chem. 1998;63:2375. [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;D53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;D58:1948. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;D60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Bailey S. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;D50:760. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.