Abstract
The method of Directional Milestoning is revisited. We start from an exact and more general expression and state the conditions and validity of the memory-loss approximation. An algorithm to compute a reaction coordinate from Directional Milestoning data is presented. The reaction coordinate is calculated as a set of discrete jumps between Milestones that maximizes the flux between two stable states. As an application we consider a conformational transition in solvated Adenosine. We compare a long molecular dynamic trajectory with Directional Milestoning and discuss the differences between the maximum flux path and minimum energy coordinates.
I. Introduction
Computational chemists are charged with predicting thermodynamics and kinetics of molecular processes. The charge implies the determination of the stable states of the system, the statistical weights of the states, and the rate of transitions between states. In the present manuscript we focus on computational studies of transitions and rates (or kinetics).
Predicting kinetics and mechanisms of molecular processes by computer simulations is complex. Indeed, significant advances in computer simulations of kinetics in the condensed phases have been made only recently. In contrast to equilibrium calculations in which the basic objects we sample and use to compute statistical mechanics averages are structures, studies of kinetics use trajectories as the basic objects. To describe a reaction we consider trajectories that start at the reactant state and end at the product state. Clearly trajectories are more complex entities than a single configuration, making the sampling and computational averages of kinetics more expensive compared to equilibrium calculations.
Trajectory sampling can be done by direct integration. That is, sampling initial conditions (velocities and coordinates) at the reactant state, integrating the equations of motion, and hoping that in a reasonable number of integration steps trajectories will reach the product state. Trajectories can also be computed by shooting, starting somewhere between reactants and products and integrating the trajectory backward (to reactants) and forward (to product) in time, like is done in TPS 1. Yet another direction is the sampling of boundary value trajectories that are guaranteed to be productive (by construction every computed trajectory reaches the products from the reactants), Boundary value trajectories are, however, significantly more expensive to compute 2 compared to initial value calculations. Boundary value trajectories can be computed approximately with large time steps 3,4 but in this case their statistical weights are not accurate enough to proceed with calculations of ensemble averages and kinetic.
One of the kinetic observables of significant interest is the mean first passage time <t> (or MFPT). It is the time averaged over an ensemble of trajectories initiated at the reactant state that are terminated when they reach the product state for the first time. For reactions that obey first order kinetic the inverse of the MFPT is the rate constant. The initial distribution of the trajectories is determined by the preparation of the reactant state and in many cases is simply the canonical distribution restricted to the domain of the reactant.
The need to compute an ensemble of trajectories from reactants to products explains the significant difficulties in simulating kinetics on the computers. The time scale of a single MD trajectory is restricted in routine calculations to the sub-microsecond time scale, far shorter than many processes of interest. For example, channel gating and some folding events are in a millisecond time scale. Molecular machines operate and conformational changes occur at a time range from microseconds to seconds. While massive parallelization and special purpose machines can push trajectories to milliseconds5, individual trajectories provide a poor sample for kinetics at that timescale. For numerous molecular processes with typical time scales of microseconds or longer, straightforward sampling of trajectories is an exceptionally challenging task. Theories and algorithms were therefore developed to address the problem and extend the time scales of atomically detailed simulations. Atomic simulations in biophysics provide the most comprehensive picture of the process under investigation.
The oldest theory of molecular reaction kinetic (Transition State Theory6) still provides a powerful and accurate computational tool for a set of reactions in which the reactants and products are separated by a large barrier. A large barrier crossing is energetically costly and occurs rarely. If crossing is assumed to happen only once during the reaction the calculations of trajectories can be avoided and only equilibrium sampling at the transition state and reactant state remain. Dynamical effects (re-crossing) can be addressed with the transmission coefficient7. Trajectories are computed from the top of the barrier until they are trapped either in the reactant or in product states. The probability of making it to the product states is computed conditioned so the initial configuration is on the transition state and the velocity points to the product. In the Transition State Theory it is assumed to be one, since no-crossings are allowed. The transmission coefficient is a useful computational tool if the barrier is sufficiently high such that re-crossing events, if they do occur, are diminished rapidly as a function of time.
Large barriers are exploited also in Transition Path Sampling (TPS)1. It is observed that while very rare, actual transitional trajectories pass over large barriers rapidly and quickly re-settle in a new or old free energy minimum. This observation is in the spirit of the convergence condition of the transmission coefficient. The rate is slow (even with rapid transitional trajectories) because these trajectories are rare. Individually each of these trajectories is actually very fast. It is not uncommon to find individual reactive trajectories of picoseconds for systems with an overall rate of seconds. The transitional trajectories do not include the typically very long waiting time at the minimum until a rare fluctuation pushes the system over the barrier. It is therefore possible to compute with moderate computer resources the transitional trajectories if a sampling algorithm for these rare trajectories with proper weights is available. Such an algorithm is offered by TPS.
A large number of molecular processes occur however without one clear and dominating barrier; for example protein folding8, RNA folding9, conformational transitions10, and ion permeation through membranes11. In fact, diffusive multi-barrier kinetics is the rule rather than the exception in biophysics. In these processes, the individual transitional trajectories (i.e. a trajectory that left the basin of the reactant and did not yet reach the basin of the product) are very long and are conducting diffusive motion on a rough energy landscape in many dimensions. Long individual trajectories (e.g. milliseconds) pose a significant challenge to theories and algorithms that require the calculations of an ensemble of complete trajectories, or of trajectories that “decide” quickly their final state. Therefore a number of algorithms and theories have emerged in the last ten years to address the problem of multi-barrier diffusive systems and Exceptionally Long Dynamics (ELD). These approaches include the PPTIS method 12, the Markov State Model 13-15, and Milestoning16-17.
All current technologies for ELD are using the same concept of breaking the long trajectory to fragments that are assumed independent. The fragments can be run in trivial parallelism. The split to trajectory fragments and their sampling must be done with care to ensure correctness, and is of course approximate. It is the central point of all ELD approaches and is discussed extensively in the context of the Milestoning theory. If done properly the results are accurate and a tremendous saving in computational resources can be obtained. Historically, the three major approaches to ELD, which are PPTIS, Milestoning, and Markov State Model, started from very different viewpoints. PPTIS developed from TPS in which the statistics of complete trajectories from reactants to products in TPS is replaced by the statistics of trajectory fragments along a pre-determined order parameter. The Markov State Model started from an analysis of long trajectories to identify metastable states and evolved to include additional sampling of trajectories between metastable states. Milestoning the method we focus on in the present article, was initiated as a technique to find time scales in systems for which the reaction coordinate is known. Recent developments removed the need for a reaction coordinate and introduced the Directional Milestoning DiM16. In the present manuscript we extend the theory of DiM to a more general form and consider the limits in which the approximations are satisfied. We further illustrate how Directional Milestoning can be used to determine a reaction coordinate.
II. Algorithm
Consider two molecular states A and B. We wish to determine the rate in which A is converted into B and their relative thermodynamic weight. A phase space (velocity and coordinate) vector X determines uniquely the state of the molecule. A phase point in A is denoted by XA and similarly we have XB. A trajectory is somewhere between A and B at time t and is denoted by X(t).
The first step of Directional Milestoning (DiM) is the identification of anchors. Here we follow the ideas of Markovian Milestoning with Voronoi Tesselation18 though our space representation is not tessellation. Anchors, Aa, are configurations in the full coordinate space of the system (for N atoms A is a vector of length 3N). They are spread over conformational space to provide representation of every important portion. An example for the use of anchors in the present study is given in figure 5 in which the anchors are clustered from a long molecular dynamics trajectory. We have used a similar approach in the past20. “Long molecular dynamics trajectories” means trajectories that are long enough to sample relevant conformations at least once, but not long enough to provide accurate thermodynamics or kinetics. This is similar to the approach taken in the Markov State Model13,14,21,22. Replica exchange simulation (that provides adequate sampling for thermodynamics but not kinetics) is yet another way to generate candidates for anchors. We derive anchors also from reaction path calculations23,24. The anchors are important since they mark the initial space in which we compute the kinetics. However, optimal determination is not required. The selection of the anchors can be re-visited in the calculations that follow.
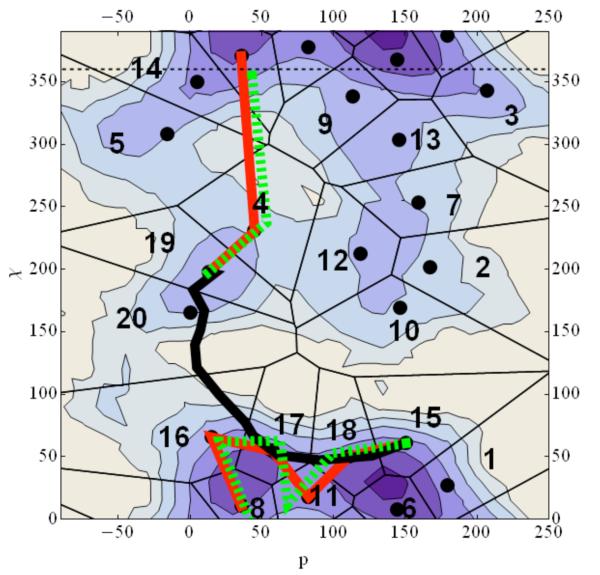
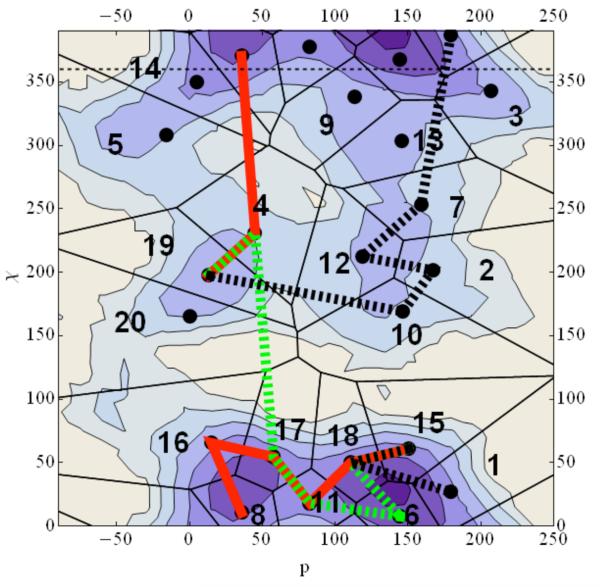
Figure 5.
A contour plot of the two-dimensional free energy profile for adenosine. Each contour corresponds to one kcal/mol change in free energy with dark blue being the lowest energy while white is the highest. Black dots are the cluster centers used for the Directional Milestoning. The black solid line is a steepest descent path from C3a to C2s computed with the functional “scalar force”37 while the red line is the Maximum flux reaction coordinate calculated from Directional Milestoning. The green dashed line is the SDP path if the initial guess for the SDP algorithm is the Maximum Flux Path. The black and red lines clearly illustrate the existence of multiple channels. Note that six coarse variables were used in the Milestoning calculation and the picture shown is a projection onto two dimensions. The projection is not always successful. For example, in the transition from χ≅220 to χ≅360 it seems (incorrectly) that several Milestones are crossed in one transition.
We define a distance between any pair of phase space points Xi an Xj as the distance in lower dimension using coarse variables.
| (1) |
The Q-s are coarse variables that we use to follow the progress of the reaction. For example, in describing the folding of a peptide20, the set of backbone soft torsions (all the φ,ψ dihedrals) makes the set of Q. In general, Q-s are defined in the space of coordinates and momentum. However, in all the applications we pursued so far20,23,24 and in the present study of conformational transition of a sugar the coarse variables Q include coordinates only. The set of coarse variables identifies every anchor uniquely. If we cannot identify all the anchors uniquely it suggests that either some anchors are redundant or that the set of the coarse variables is insufficient and needs to be expanded; for example, by adding one more torsion. In the next step we identify Directional Milestones (DiMs). A DiM is an interface separating two anchors. It is directional since its location may change if the trajectory is arriving from the neighborhood of anchor a or the neighborhood of anchor b. The Milestone is placed in such a way that it is closer to the target anchor compared to the anchor it started from (Figure 1).
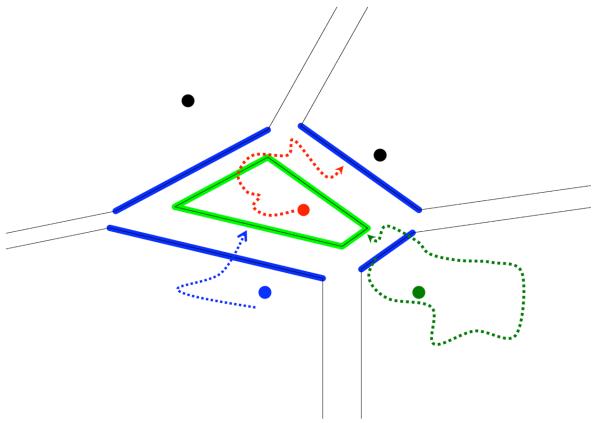
Figure 1.
A schematic representation of Directional Milestoning. The circles denote anchors, the straight lines denote interfaces or Milestones, and the curves are trajectory fragments. Consider the red anchor. The Directional Milestones that mark entry to the domain of the red anchor are the green lines. The red trajectory, which starts from the red anchor, illustrates an exit from the red anchor domain. The blue curve denotes a trajectory that enters from the blue domain to the red domain. Note the asymmetry in the position of the entry and exit Milestones. The green curve illustrates the type of trajectories that we use in Milestoning calculations (a trajectory from an interface or a Milestone to another Milestone). The green trajectory starts at an entry to the domain of the green anchor and continues until it enters the red domain. It is then terminated and its time and terminating Milestone are recorded. See text and reference 16 for more details.
The Directional Milestones are denoted by M(a → b). They are used to record transitions from the domain of anchor a to the domain of anchor b and are defined as the set of all phase points X such that
| (2) |
Where the distance was defined in Eq. (1). The term Δa causes the asymmetry between Milestones M(a → b) and M(b → a), and is Δa = minb≠a d(Xb, Xa). The asymmetry is introduced to better satisfy the condition of memory loss (see III. Theory). In brief, without the shift Δ the Milestone representation is that of Voronoi cells18 and they may cross. Trajectories between crossing Milestones can terminate very quickly and therefore “live” shorter times than required relaxation period25.
In Milestoning we are focusing on transitions between the interfaces (e.g. the green curve in figure 2) that are analyzed with statistical mechanics tools to obtain complete description of kinetics and thermodynamics. Consider a long reactive trajectory X(t). We follow this trajectory as a function of time and label it by the last Milestone that it passed (e.g. Xα(t)). The additional labeling provides a coarser description of the trajectory. Passing Milestone α implies that the system is in a certain part of phase space and this is the only label we use in Milestoning. The Milestone resolution is clearly less precise than X(t) but may be sufficient for our purpose. A sketch of a trajectory and its Milestones is given in Figure 2 where the different colors correspond to different Milestone transitions.
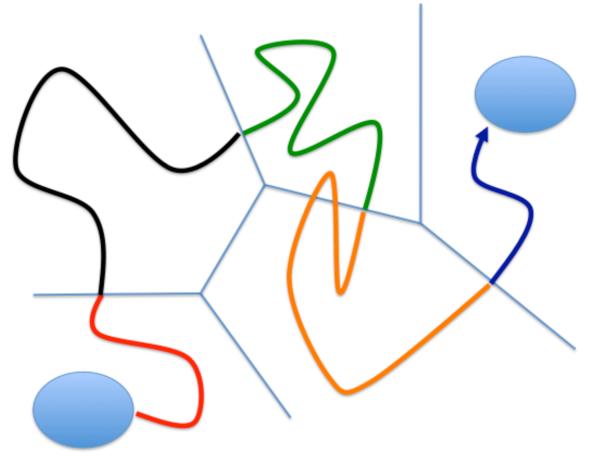
Figure 2.
A schematic representation of an exact trajectory partitioned to fragments between Milestones (or interfaces). Every color denotes a different trajectory fragment. We do not initiate a new fragment when the trajectory re-crosses the same Milestone it started from (see orange trajectory). For clarity we do not differentiate between incoming and outgoing Milestones in this figure.
It is obvious that every long trajectory can be broken to fragments of trajectories between Milestones. The reverse (combining fragments to complete trajectories) is less obvious. To combine the fragments we need to know how to initiate the trajectories at the Milestone, how to terminate them, and their proper weight (without running the complete ensemble of trajectories, which can be prohibitively expensive). In Milestoning we terminate a trajectory fragment the first time it hits a Milestone different from the one it started from. Therefore the distribution generated at the next Milestone by incoming trajectories is a first hitting distribution (FHD). This distribution should be used to generate continuations of trajectory fragments to yet other Milestones.
Algorithmically, the termination step is clear. More complex is the start at the Milestone in which we sample initial conditions from a FHD. A FHD is generated by trajectories hitting for the first time the Milestone we want to start from. Looking backward and forward at the history of generating FHDs it seems that in order to generate a FHD we need an ensemble of complete trajectories. Such complete generation of trajectories is possible and is essentially the approach taken by the Transition Interface Sampling (TIS)26 and the Forward Flux27 (FF) methods. However, since complete trajectories are computed, these exact procedures are limited to rapid transitional events. The overall rate can still be slow if reactive trajectories are rare. TIS and FF are designed to sample such rare trajectories efficiently. However, diffusive barriers and individual long trajectories cannot be computed this way. For diffusive processes there is much to be gained by the additional assumption of fragment-independence, an assumption we exploit in Milestoning. In Milestoning we do not continue a trajectory from a termination point of a previous trajectory. Instead we sample independent phase space points from the FHD. The independently constructed configurations ensure proper sampling to start new trajectory fragments even if only a few fragments reach the current Milestone.
The following procedure is used in DiM to initiate trajectory fragments. We generate configurations at a Milestone α by restrained or constrained canonical sampling to the neighborhood of the Milestone25,28. The constrained sampling is made with the help of harmonic potentials as described in28, or with constraints that are implemented using Lagrange's multipliers23,25. In the past25 we used the distribution generated by constrained dynamics “as is” to approximate the FHD. However, in DiM we offered a correction which is very much in the spirit of the PPTIS calculations12. Every point sampled in the restrained calculations is freely integrated (without the restraint) back in time until it hits a Milestone. If the trajectory hits the Milestone it started from, we eliminate this point from the set since it is not a first hitting point. If however, another Milestone is hit then we keep the sampled point and use it for integration forward in time.
The integration forward in time is terminated when a new Milestone is hit. The trajectory is not stopped when re-crossing of the original Milestone is detected (in contrast to the integration backward in time). See for example the orange trajectory fragment in Figure 2. The orange trajectory starts at the end of the green trajectory. If we integrate it backward in time (follow the green trajectory) we find that the beginning of the orange trajectory is indeed a first hitting point on the desired Milestone. The orange trajectory fragment crosses the starting Milestone, but this is allowed at this phase, and we continue the integration until it hits a new Milestone (and terminates).
At the end of a successful calculation of a trajectory fragment in DiM we have the following information: “Touching” of three Milestones and two termination times. We know the Milestone (say α) on which we sample phase space points to start trajectory fragments. We know Milestone β that was reached after backward integration in time from α, and Milestone γ that was reached after forward integration. We also know the times forward and backward. Actually we know more than the list just discussed. We know the complete trajectory fragments. This is however more than we want to know. Our goal is to construct a coarse grained model for the system dynamics which is based on lower spatial resolution (using Milestone crossings) and not the full phase space. We prefer to state that “the system was last seen in Milestone α” as the spatial descriptor rather than X(t). Finally, in the current version of Milestoning we do not use the termination time obtained from backward integration nor the identity of the Milestone β. Extensions are in principle possible at additional computational costs (more statistics are needed to use additional labels). The backward integration is used only to verify that the initial phase point is sampled from FHD. We use the trajectory data to estimate the transitional probabilities, Kαβ (t), which are the probabilities that the system initiated at Milestone α at time 0 will pass Milestone β for the first time exactly at time t. If the total number of trajectories initiated at α is nα and if the number of trajectories that hit Milestone β at time t is nαβ(t) then we estimate Kαβ(t)≃nαβ(t)/nα . We describe how this information is used to compute overall kinetics and thermodynamics in the next section.
III. Theory
With a better picture of how trajectories are initiated and computed we now introduce the theory that uses the fragment statistics to investigate kinetics and thermodynamics. We define the following entities:
p(X̄α,t)-- the probability that at time t the last Milestone that was crossed was α at an interface point X̄α.
qα (X̄α,t)-- the probability density that a trajectory hit a Milestone coordinate X̄α exactly at time t.
Kαγ(X̄α,t; X̄γ,t′)-- transition probability from a specific phase space point X̄α at time t to another phase space point X̄γ at time t.
Below we write an expression for the probability distribution generated by Milestoning trajectories. Note that while the expression resembles Eq. 4 in the original Milestoning paper17 the expressions are different. The present expression is exact since the explicit coordinates within the interface are taken into account.
| (3) |
The notation ᾱ means the set of milestones that are directly accessible (without passing another milestone) from and to milestone α using trajectory fragments. We also call this set “neighbors to α“. The integration over V̄ is over the hypersurface that makes the Milestone. Verbally the expressions above are simple probability balance. Therefore they are exact and are independent of the specific dynamics that was used to calculate trajectories (can be Newtonian, Langevin, etc.). The first equation states that the probability that the last milestone passed is α (pα (X̄α,t)) is the probability that the α milestone was crossed at an earlier time t′ (qα(X̄α,t′)), no other milestone was crossed since then, and we sum over all earlier times. The second equation suggests that milestone α is crossed exactly at time t by one of the two mechanisms: it is exactly at α at the beginning (t=0) or it passes at an earlier time t′ a milestone neighbor to α, and then exactly at time t transitions to α.
The above expressions are exact but also extremely complex to compute. The matrix Kαγ(X̄α,t; X̄γ,t′) is of very high dimension. Assume that we use 100 bins to determine the position of every coordinate x. Assume further that the system under consideration has about 10 coarse degrees of freedom. The transition matrix (which is not symmetric) will have ~10020(!) elements to compute. Some simplifications are therefore necessary. We first assume a stationary process. As a result the transition probability depends on the time difference and not the absolute time. We have
| (4) |
The second simplification is an approximation and is the core of the theory. We assume that when a trajectory leaves milestone α on its way to terminate on milestone γ it no longer “remembers” the exact phase space point that came from milestone α; i.e., we can write
| (5) |
The trajectory still remembers the milestone it came from but no longer the exact phase space point it started in that milestone. This approximation allows for dramatic simplification and coarse-graining in space as we illustrate below.
When is this approximation valid? A limit in which this approximation is satisfied is when the trajectory has been running for a while before terminating on a neighboring Milestone, statistical mechanics takes its course and the precise location of the initial conditions is no longer important. This limit is clearly correct for a sufficiently long time in which initial conditions are forgotten. However, in practice memory-loss is evident for times much shorter than this particular limit. Note that the phase space point includes velocity. In reference 25 we illustrate that if the average termination time between milestones is longer than the time for velocity de-correlation, then the rate computed for solvated alanine dipeptide is accurate and insensitive to the number of Milestones. We have a reasonable control on this condition. The spatial separation between Milestones determines the de-correlation or memory-loss time and is determined by the number of Milestones we put between the cell of reactants and products. It is therefore a good practice to repeat rate calculations with more than one distribution of Milestones and to verify that the results remained unchanged as a function of reasonable separation of Milestones. A clear illustration of the stability of the Milestoning results as a function of the separation size is given in reference 25.
Another limit of Milestoning that leads to exact mean first passage time (MFPT) (but not higher moments of the first passage time) is discussed in 29. If the milestones happened to be iso-committer surfaces (i.e. hypersurfaces for which the probability to reach the product before the reactant is a constant), the transition probability does not depend on exact location in the Milestone (the probability is always the same) and the approximation above becomes exact. In the language of the kernel above we have
| (6) |
This limit is mathematically elegant and attractive. A concern is that exact determination of iso-committer surfaces is difficult and in most cases we can only approximate it. It is not clear how good the approximation of a committer surface is compared to the condition of equation (3). The second concern is that the iso-committer hypersurface is defined for a one-dimensional reaction coordinate and extension to several coarse-grained variables or that the DiM picture we advocate above is not obvious.
Majek and Elber16 formulated yet another condition that provides exact mean first passage time. Generation of a FHD at a Milestone is obtained from trajectories originating from at least two different Milestones. If the generated FHD is independent of the initial Milestone, then the overall mean first passage time is exact.
The Milestoning approximation makes it possible to remove the coordinate representation and replace it with the reduced representation of Milestone labels.
The time remains continuous. With the last adjustment and approximation (5) at hand we are ready for the next step; we have
| (7) |
We define the following functions
| (8) |
Integrating equation (7) over X̄γ and adding yet another integration over X̄α we obtain
| (9) |
Equation (9) is a central result. These are the Milestoning equations of motion that were written ad-hoc in reference 17.
The entities we are interested in at present are pα (∞), which is the equilibrium or the stationary distribution, and the stationary flux qα (∞). We assume that these distributions exist, in the sense that the probability and the flux become time independent and constants at the long time limit.
We will need a few mathematical tricks in order to derive the stationary distributions. Below we follow the derivation of25,30, however, more details are added to make some of the derivations easier to follow. In particular we give some Laplace transform results in the Appendix.
The Laplace transform of equations (9) are (see Appendix):
| (10) |
We can formally solve for the vector q̃(u) using the second equation in a matrix form.
| (11) |
where q and p are row vectors, and K is matrix such that (K̃(u))αβ = K̃αβ (u). Multiplying by the Laplace variable u and taking the limit to zero we have
| (12) |
We note that . We used the above formula to define a new matrix K, which is the probability matrix of transitions between neighboring Milestones integrated over all times. Because K is a probability we also have . The vector qstat is an eigenvector of the operator (I-K) with the eigenvalue of zero. In the calculation of qstat we need only K and not K(t) which is easier. In other words, only the zero moment of the time distribution, K(t), is needed.
The stationary flux is not influenced by the initial conditions, consistent with a statistical mechanics longtime limit (also for stationary flux).
An example of a transition matrix for three Milestones that leads to a stationary flux is below.
| (13) |
Let the total number of trajectories that we start at interface 1 be n1 and the number of trajectories that were terminated at interface 2 be n12; a numerical estimate of K12 is n12/n1. The absorbing boundary is the third state. The matrix is set in such a way that a trajectory that makes it to the absorbing state returns to the initial state which is the first Milestone. To create a stationary flux we must have a cycle in which particles that are lost are returned to the system. Otherwise loss of probability (and no stationary solution) is unavoidable.
It is important to note that the matrix used to compute the mean first passage time (see below) is different. If we do not terminate the trajectory at the absorbing boundary condition and just return it to the beginning, the trajectory time will be of infinite length in time (!). For the determination of the mean first passage time we will use a matrix that terminates every trajectory that touches the final state.
| (14) |
Armed with an expression for the stationary flux we are ready to proceed to compute the stationary distribution. We write equation (10) multiplied by the Laplace variable u. Note that this is not a matrix equation.
| (15) |
We have to be a little careful when we take the limit of u → 0 in the above expression since and 1-1 will give us zero. We therefore write
| (16) |
We are ready to write the stationary distribution of the probability that the last milestone passed is α. The result below is remarkably simple. It is given by the stationary flux through that interface multiplied by the average life-time of trajectories initiated at interface α - 〈tα〉
| (17) |
.
We proceed to consider kinetics. One of the most useful measures of kinetic is the mean first passage time and its moments. For simple processes that decay exponentially in time the inverse of the mean first passage time is the rate constant. The overall mean first passage time is defined as the average time it takes the trajectories to reach for the first time the final absorbing state. Formally we write
| (18) |
Where qf is the probability of entering the final absorbing state. Similar to the calculations of stationary state, we use Laplace transform to evaluate this entity. Instead of equation (18) we write
| (19) |
The plan is to compute q̃(u) (and q̃f(u)), take a derivative with respect to the Laplace variable, and then take the zero limit of the Laplace variable. We already have an expression for q̃(u) in equation (11). We write (with some of the manipulations given explicitly)
| (20) |
Remember that p is the vector of initial conditions. Now we should take the zero limit of the Laplace variable. We have
| (21) |
The probability of each transition between nearby milestones is the corresponding element of the matrix K, and the local mean-first-passage-time is an element of the matrix <t>. If we have M Milestones we have a maximum of M(M-1) transition probabilities and local first passage times. However, most of the time, the number of Milestones that are directly accessible from one interface is much smaller than the maximal number.
We now project the overall first passage time on the final absorbing state f by taking the limit of u → 0 and by multiplying the last vector equation of Eq. (19) by ef/- a vector, which is 1 at the absorbing state(s), and zero elsewhere. A closed expression for the overall first passage time follows
| (22) |
The above expression can be simplified further. We note that a summation over the rows of K (with the exception of the absorbing boundary) is giving exactly 1. The sum over a row is the sum of probabilities that the trajectory will end at different neighboring Milestones. Since the trajectory ends “somewhere” the sum must be 1. For a trajectory that reaches the absorbing boundary, it cannot go anywhere else and is bound to disappear. The summation of the row with the Milestone at the absorbing boundary is therefore zero (see Eq. (14)).
An interesting observation is the result of the multiplication by 1 vector. It is easier to consider an example (say the transition matrix introduced in Eq. (14))
| (23) |
We can write for a general number of Milestones
| (24) |
Now use (24) to simplify somewhat equation (22)
| (25) |
There is an important difference between <t> and <t>. The first is a matrix that contains the local first passage times between all connected Milestones (directly accessible by trajectory fragments). The second is a vector and each element is the lifetime of a Milestone. It is a sum over the corresponding row of the matrix <t> and it defines the time a trajectory is assigned to a particular Milestone before terminating on any other Milestone. For example the average lifetime of Milestone a is an element of the vector <t> and is defined by
| (26) |
Note that the vector of ones in Eq. (24) miraculously eliminates the dependence of the result on the terminating Milestone. Only the lifetimes of the initiating Milestones remain.
The critical equations of the present section are formulas 12,17 and 25. The derivation of the stationary flux, stationary probability, and overall mean first passage time are not new and followed the derivations in reference 25,30; they are provided here for completeness and additional clarification. The new elements that were introduced to the Theory section are the exact equations, and the derivation of the Milestoning approximation. Below we consider another new element of the present article the use of the stationary flux to compute reaction coordinates from a multi-dimensional Directional Milestones (DiM)16 study.
IV. Computing reaction coordinate from DiM results
Computations of reaction coordinates (RC) serve different goals. One purpose is to use a RC to conduct quantitative calculation of the free energy profile along the reaction coordinate and of the kinetics using theories like Transition State Theory31. Another use of a reaction coordinate is for better understanding of molecular mechanisms. This idea was proposed in the context of Transition Path Sampling (TPS)1 in which transitional trajectories are sampled first and the rate is computed directly from their statistics. Hence, the quantitative answers are already at hand, and what is sometimes missing is qualitative interpretation of mechanisms. Qualitative interpretation can be difficult due to the vast trajectory data and the diversity of their behavior in the general case. If a reaction coordinate can be computed from the ensemble of trajectories then better insight to the mechanism of the reaction is likely.
The definition of a reaction coordinate poses a significant challenge. It can be particularly vague if the goal is of qualitative understanding of mechanisms rather than quantitative calculation of thermodynamics and kinetics. Iso-committer surfaces1,32,33 were advocated as the true choice of the reaction coordinate. An iso-committer surface is a set of points with the same reaction probability (RP). The RP is the probability that trajectories initiated from these points will reach the product state before the reactant for the first time. This choice is appealing; however, the determination of the iso-committer surfaces is far from trivial and must be done approximately and numerically at significant computational cost. Trajectories are launched from different positions on the energy landscape and their fate (ending in the reactant or product state) is determined 34,35. In some implementations the iso-committer surface is given a functional form with parameters that are fitted to the trajectory data35,36.
Reaction coordinates have been computed as minimum energy paths for a longtime now. Optimization of functionals 37,40 is a particularly attractive technique to determine RC for large molecular systems and has been used extensively. One of the functionals that defines a reaction coordinate is based on the notion of flux. Rather than surfaces we seek a line connecting the reactants and products such that the flux leading from reactant to product along this line is a maximum. This is a theoretically appealing idea that is based on dynamics (instead of only the static energy surface). Use of a curvilinear line instead of a hypersurface leads to dramatic simplification of the calculation and in our ability to understand the underlying mechanism. The idea was put forward for overdamped systems by Berkowitz and McCammon 41 and made into a useful algorithm for molecular systems (Maxflux) by Huo and Straub39. The last algorithm was revisited recently by Zhao and co-wrokers 42. The calculations are based on a path integral analysis of overdamped Langevin trajectories with two fixed end points (reactants and products). The flux in an infinitesimal tube around the RC is optimized as a function of the reaction (curvilinear) coordinate.
A disadvantage of the Maxflux functional is that the procedure was limited so far to overdamped Langevin dynamics. Moreover, the restriction to an infinitesimal volume around the path makes it harder to estimate fluctuations that are likely to occur if the free energy landscape is not an infinitesimally narrow tube. The Maxflux principle should be valid for analysis of trajectories of arbitrary dynamics. The required operation is straightforward: collecting trajectories from reactant to products and estimating the reactive flux of every phase space point and determining a not-so-narrow tube of maximal flux from reactant to product. We propose to do exactly that in the context of the Milestoning algorithm.
Interestingly, a Directional Milestoning calculation provides all the fluxes between the cells or at the interfaces separating the cells (equation 12). The fluxes, qa are a discrete representation of the fluxes in the continuous system. A discrete path between the cells that maximizes the reactive flux from reactant to product solves the Maxflux problem (see Figure 5 for an example of such a graph). The advantage of doing it through Milestoning is that the dynamics in Milestoning are arbitrary and the results are no longer restricted to overdamped Langevin dynamics. The volumes in phase space that are attached to anchors in Milestoning cannot be made infinitesimal since cell volumes that are too small violate the principle of memory loss. However, infinitesimal volumes are not desired since entropic effects are ignored. We loosely use the term “cell” to denote phase space allocated to anchors. It is a loose term since it is possible for phase space points along trajectory fragments to have exactly the same coordinates and momenta and originate from different Milestones.
Consider a graph G with N nodes (which are the cells) and E edges which are Milestones connecting the cells. For each edge connecting (say) cells k and l we assign a weight Wkl=qkl-qlk which is the net forward flux of this edge. We seek in the graph a path that connects the reactant and product and that carries maximum flux. The procedure we described below is similar in spirit to an algorithm based on the Master equation43 and on a calculation based on the Transition Path Theory 44 for the analysis of folding trajectories. The Milestoning calculation provides the flux directly, therefore, the current analysis is simple and straightforward.
The following algorithm finds a path of maximum flux on the graph. In case of degeneracy (more than one path with a maximum value for the flux) we pick one of the degenerate reaction coordinates.
Algorithm:
Find the link emin with the smallest weight wmin=mini(wi) on the graph Gcurrent (the edge with the smallest flux) and mark it for elimination.
Check if after removing emin it is still possible to find a sequence of edges leading from reactants to products. If yes, remove emin from the graph and go to 1. If not, store emin as an edge in the desired path, store the resulting disconnected graphs, Gi and Gj, remove emin from the graph, and continue to 3.
After the removal of emin in 2 we have at least two disconnected graphs. Pick one of these graphs (say Gi). If the graph does not have edges, pick another graph from the set and repeat the check. If none of the graphs have edges, join all the emin edges discovered so far to create the maximum flux path and stop. If a graph with edges is discovered rename it Gcurrent. Re-assign reactant or product according the nodes of the edge that was removed and go to 1.
The above procedure is guaranteed to provide a maximum flux path in the discrete space generated in the Milestoning calculations. It may not be necessarily the most efficient; however, this is not an expensive calculation compared to the estimates of the transition kernel by trajectories and is sufficient for the present task.
It is also possible to seek the second (and higher order) best path. One approach is to remove all the edges of the max flux path and to analyze the resulting graph with the same algorithm described above. A disadvantage is that we can have different paths that share parts of the edges. How many edges should be different in the pair of the paths to make them distinct? Chemical intuition suggests a solution that we use: The paths are different if the transition states are different. We choose a “transition state” to be the edge along the maximum flux path that has the smallest flux. We remove that edge and re-analyze the path for a second best pathway. There are two possible outcomes. The first is that a completely new path is obtained. In this case we store the second path, and perhaps continue to search for a third path. The second outcome is that all the remaining edges of the original path are still picked and a short detour near the old transition state is found. This may suggest that an alternative path does not exist. We can proceed by removing another edge from the MFP. We remove an edge that is carrying the lowest flux among the remaining edges along the path and repeat the analysis until a new path is found.
V. Example for a Directional Milestoning calculation and determination of a reaction coordinate
To illustrate Milestoning on a realistic system of chemical interest we consider a conformational transition in the molecule Adenosine. It is a highly flexible molecule with multiple reaction coordinates and reactive tubes that further cross each other. This makes reaction coordinate predictions difficult. Adenosine is also an important model system for conformational transitions in polynucleotides such as DNA and RNA molecules 45. Experiments estimate the relaxation time as nanoseconds 46. This time scale is accessible to straightforward Molecular Dynamics simulations making the present system a useful benchmark in which we compare our approximate result with exact calculation of rate from MD simulation. The two states of interest (“reactant” and “products”) are shown in Figure 3. A similar system was studied by the Transition Path Sampling method 47.
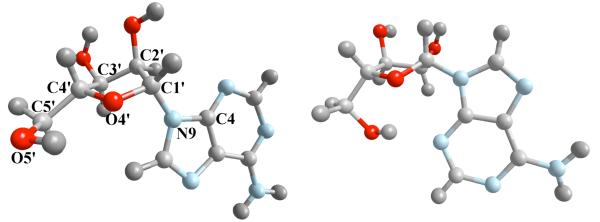
Figure 3.
Adenosine molecule in C3a (left) and C2s states (right). Labeled atoms are used to define the coarse variables.
The overall conformational transitions of the molecule can be described approximately by glycosyl torsion χ (O4`-C1`-N9-C4), which assumes anti or syn conformations at room temperature, and the pseudo rotation of the furanose sugar ring denoted here as p′. The pseudo rotation angle is commonly defined as 48
where the set {νi}i=1,5 are torsions corresponding to C1`-C2`-C3`-C4`, C2`-C3`-C4`-O4`, C3`-C4`-O4`-C1`, C4`-O4`-C1`-C2`,O4`-C1`-C2`-C3` respectively. To have always positive angles we replaced p′ with p = p′+180° when ν1 = 0° and p = mod(p′+360°) otherwise. Given the definition above, the sugar of adenosine is C3`endo at p ~ 30° and C2`endo at p ~ 150°.
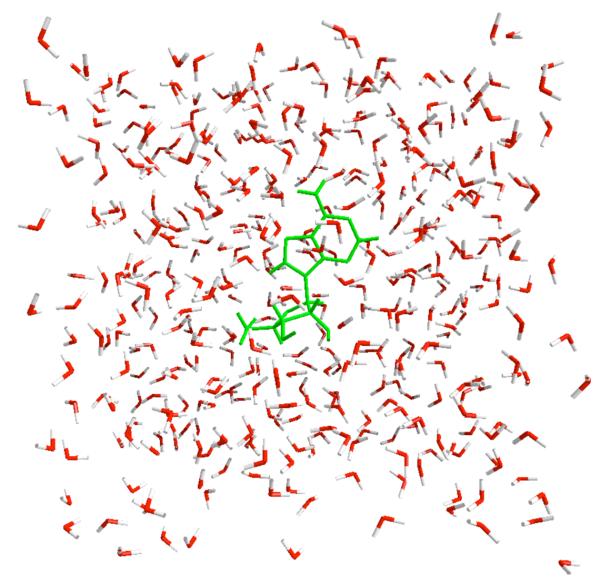
The molecule was solvated in a periodic box of water as shown in Figure 4.
Figure 4.
A periodic box of water that was used in the simulation of a conformational transition in adenosine. The molecule is solvated with 420 TIP3P water molecules 49 with a periodic cubic box of 23.8×23.8×23.8Å3. The box size is adjusted to give a pressure of ~1atm at 300K. We used a matrix variant50 of the SHAKE algorithm 51 to fix water bond lengths and angles while the solute is not constrained. 8Å cutoff is used for Lennard Jones and 8.5 Å for real space cutoff of electrostatics. Particle-mesh Ewald summation was used for long range electrostatics. The time step was 2fs. All simulations are performed with AMBER/OPLSAA force field where bonding terms are taken from Amber99 52 and the nonbonding terms are from OPLSAA 53. The parameters are available from MOIL 54 http://clsb.ices.utexas.edu/prebuilt/
To obtain an exact comparison to the Milestoning calculations we conducted first 1.1 μsec MD simulation at room temperature, which is sufficient to adequately estimate thermodynamic behavior (free energy surface) and the overall mean first passage time of the transition between the two states. The free energy surface is constructed for the two degrees of freedom p and χ by binning their sampled values along the trajectory. We choose cells 19 and 15 with cluster centers in the two dimensional variables (p,χ): C3a (p,χ=35°, 221°) and C2s (p,χ=154°,30°). The choice of C2s is rather arbitrary as we could also choose cells 6 or 18 as they have lower free energy on the contour map. Cell 15 is larger and it has larger volume and entropy. As a result it is sampled more frequently in the long Molecular Dynamics run we used for comparison.
The Directional Milestoning approach requires a set of anchors. As argued above this set of structures can come from numerous sources: replica exchange simulations 55, configurations along a pre-computed minimum energy or free energy path 24 and straightforward and long Molecular Dynamics trajectory (the present study). Interestingly, the present molecule is a poor candidate for Milestoning calculations based on minimum energy or minimum free energy pathways since multiple connected channels make such calculations difficult (Kirmizialtin and Elber, unpublished). Especially problematic is the use of the popular hyperplane approximation to determine the minimum energy (or free energy) pathways. We therefore use the configurations from the long time MD trajectory, and cluster them according to the six torsional angles described above.
The Euclidian distance between the six torsions of the selected atoms defined earlier (Eq. (1)) is used for clustering. We first reduced 275000 structures to 24560 by removing structures that are different by no more than 2 degrees per torsion. The remaining data is clustered by k-means algorithm56 into N=20 clusters that are used to define the anchors and the milestones. We conduct the Milestoning calculation in three steps: (1) search, (2) sample and (3) run to ensure efficiency16. The maximum number of possible interfaces is 20×19=380 which is large (reminder: the interfaces are directed and a transition from i->j is different from a transition from j->i). Rather than exhaustive enumeration of Milestones, and with an eye to more complex systems with a much larger number of plausible interfaces, we sample the most important milestones. The connectivity between anchors is estimated by first launching 500 trajectories of maximum length of 10ps from each anchor until they hit one of the nearby Milestones. We call this step the “search”. Interfaces that are “touched” are recorded and the touching configuration is used to initiate a sampling trajectory at the interface in a step of “sampling”. The sampling at the interface is performed with umbrella potential as described in reference 16. Each of the sampling points was tested by computing trajectories backward in time. Trajectories that return to the original interface during the test are not first hitting points, and are removed from the set as discussed in II Algorithms.
In the final step (called “run”) 400 sampled points were integrated forward in time until they hit a new interface. The identity of the interface and the termination time are recorded. Interfaces that were not detected in the “search” step can still be discovered in “run” since termination is tested for all possible interfaces. Newly discovered Milestones are added to the set and are “sampled” and “run”. After a few cycles of runs, in which additional interfaces are found and are added, the number of interfaces converges to 207. This is a high number (more than a half of the total possible), suggesting a very significant connectivity between the anchors. All simulations in the “run” step are in the NVT ensemble at 300K with a weak Anderson thermostat57. A time step of 0.5 fs is used during the DiM calculations. The rest of the computational details are the same as in the long MD simulation and described in the caption of Figure 4.
The states C3a and C2s correspond to cell numbers 19 and 15 respectively. We calculated the average first passage time in two directions from the long MD trajectory and from Milestoning.
The error in DiM calculations is estimated by dividing the data into two sets. The results are in reasonable agreement and are within the range of the statistical errors.
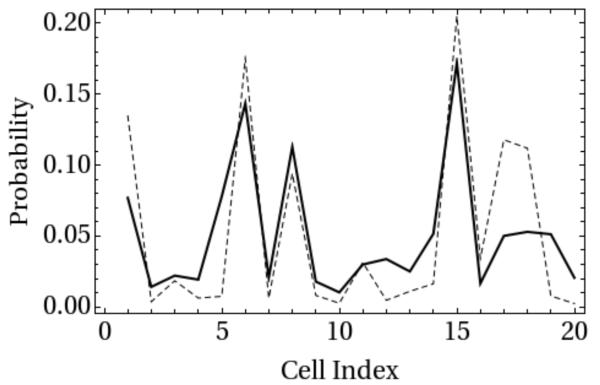
In Figure 6 we illustrate that the thermal probability of the individual sites is reasonably well reproduced in the Milestoning calculation.
Figure 6.
Equilibrium probability of being in a cell of an acnhor calculated from long MD simulation (dashed) is compared with stationary distribution of DiM. In DiM the stationary solution is a sum of all the probabilities of crossing a Milestone into a cell and remaining there. The contribution of one interface is given by Eq. (17).
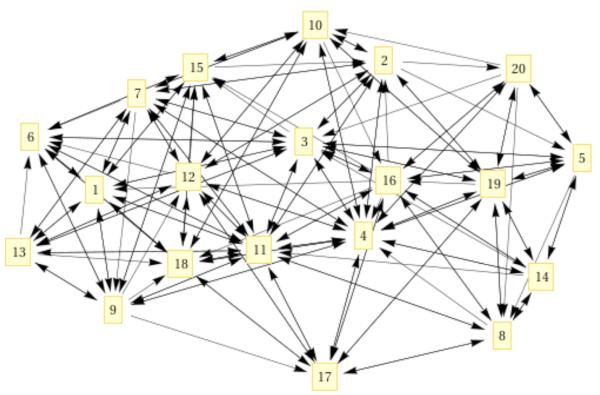
The next item of interest is the calculation of the reaction pathway. In Figure 7 we show the complete network between the cells.
Figure 7.
A connectivity graph of each cell with its neighbors. Nodes are centers of clusters and edges represent local transition pathways. The arrows indicate the direction of the transition. From detailed balance we expect every transition to be reversal. However, some transitions between nearby milestones are very rare and are not sampled. These transitions do not impact the overall rate.
With the network at hand we followed the algorithm of section IV and derive the maximum flux path (MFP) shown in Figure 5 in red. The MFP is plotted as linear red segments between the centers of the cells it connects. The MFP is computed in a space of six coarse variables and not in the two reduced variables that we use to represent the free energy landscape. Hence, what looks like “skipping” some interfaces at the top left of free energy surface is a result of a non-ideal projection of six dimensions into two.
The MFP is different from the Steepest Descent Path (SDP) computed with functional optimization as described in reference 37 and is shown in with the black thick line. The Steepest Descent Path is sometimes called the Intrinsic Reaction Coordinate. The SDP calculation starts with a linear interpolation between the two end structures. The algorithm finds a local minimum for the path that is not too far from the initial guess. When we start the SDP optimization from the MFP the algorithm settles in a path near the MFP (see Figure 5). It was probably not detected by the functional optimization since the initial guess of a straight line has a preference to nearby paths. Minimum energy path calculations provide a local minimum near the initial guess. The MFP graph analysis provides the global optimum over all path space, which is clearly an advantage. Of course in the graph analysis we rely on a discrete coarse-grained representation from Milestoning and the results will depend on the quality of the DiM sampling.
One can also search for alternative pathways to the MFP that is already found. The second and third best paths are calculated as outlined in IV and are shown in Figure 8. Note that the second best path (in green) is similar to the Steepest Descent Path (SDP) computed with functional optimization from straight-line interpolation.
Figure 8.
Two alternative paths when the graph is blocked at the two lowest bottleneck regions of the Maximum Path (solid red): green dashed when region 18-11 is blocked and black dashed when 4-19 is blocked.
Our free energy calculation and the thermal equilibrium probability shows that C2`endo is slightly more stable than C3`endo while χ has a lower energy at syn.
The reaction coordinates allow for better understanding of the conformational change. The molecule transitions from syn to anti in multiple pathways as is shown in Figure 8. The maximum Flux path suggests that the transition starts with a change of the puckering angle and proceeds with the glycosyl angle modification. Transition of the puckering angle from C2`endo (South) to C3`endo (North) is through the East barrier that is from O1′ endo and C4`exo (see Altona for details of the nomenculature48) and for the glycosyl angle the route is from 60° → 360° → 204°. The second path follows a route for χ from 60° → 180° → 204° after the puckering transition done in a similar way as in the first path. The third best path on the other hand starts with glycosyl angle change from 60° → 360° → 204° and followed by sugar puckering the same way before. Comparing the flux of the free paths we focus on the transition states. Path 1 (red line) the transition state is between cell 18 and 11, and the flux is 0.010. Path 2 (green dots) the transition state is between cells 6 and 11 and the flux is 0.007. Finally, path 3 (black dots) is between cells 10 and 19 and the flux is 0.002. The results suggest that while the third path can probably be ignored, the second path makes a significant contribution to the overall reactive flux.
Concluding remarks
We describe in detail the theory and application of Directional Milestoning (DiM). It makes it possible to study the kinetic and thermodynamic of complex systems using testable and physically sensible approximations. We also introduce a novel application of DiM in which a reaction coordinate, defined as the path of maximum flux, is computed with Milestoning.
Table 1.
Mean first passage time computed from exact (and long) Molecular Dynamics Trajectory (MD) and the method discussed in the present article Directional Milestoning (DiM). The times quoted are in picoseconds.
| <τ> | DIM | MD |
|---|---|---|
| 19→15 | 173±79 | 202±11 |
| 15→19 | 6461±710 | 5511±377 |
Acknowledgements
The research was supported by NIH grant GM59796. The help of Peter Majek in setting up the Directional Milestoning calculations is gratefully acknowledged. We acknowledge many useful discussions about Milestoning with Eric vanden Eijnden, Giovanni Ciccotti, and Maddalena Venturoli.
Appendix
We start by defining a Laplace transform
| (A.1) |
In the limit in which the Laplace variable u is approaching zero the integral is extended to infinite (or to very large values of t); i.e. most of the contribution will come from large values of t. In that limit f(t) is assumed to approach a stationary value (per our assumption above for the probability density and flux). In this case we can write
| (A.2) |
This result is useful since it correlates the Laplace transform with the asymptotic equilibrium distribution we are after. In short if we know the Laplace transform at the limit u → 0 we also know the equilibrium distribution. Another properly of Laplace transform that we need (and is quite famous) is that the transform of a convolution is a product of the transforms of the functions being convoluted. The Laplace transform of an integral is , which is a two-dimensional integral over t and t′.
Schematically we can draw the integration as
Figure A.1.
Laplace transform of an integral up to time t: case A
The vertical lines mean that for a fix value of t, we integrate t′ from 0 to t. This integration covers exactly half of the plane (case A). However there is another way of integrating over half of the plane, which is more convenient for the problem at hand, and we schematically draw as case B
Figure A.2.
The same as Figure 3 for case B. See text for more details
Here the integration is done by fixing the beginning of the integral over t to a particular value t′ and then integrating from t′ to infinite. Hence the above integral is translated to . The integral in the square brackets can be done explicitly to have . Hence the Laplace transform of an integral is the Laplace transform of the integrand divided by the Laplace variable. Another “useful-to-know” result of Laplace transform is that of convolution. Define the convolution . We write
| (A.3) |
References
- 1.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Annual Review of Physical Chemistry. 2002;53:291. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 2.Bai D, Elber R. Journal of Chemical Theory and Computation. 2006;2:484. doi: 10.1021/ct060028m. [DOI] [PubMed] [Google Scholar]
- 3.Elber R, Meller J, Olender R. Journal of Physical Chemistry B. 1999;103:899. [Google Scholar]
- 4.Olender R, Elber R. Journal of Chemical Physics. 1996;105:9299. [Google Scholar]
- 5.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan YB, Wriggers W. Science. 2010;330:341. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 6.Eyring H. Journal of Chemical Physics. 1935;3:107. [Google Scholar]
- 7.Chandler D. Journal of Chemical Physics. 1978;68:2959. [Google Scholar]
- 8.Cardenas AE, Elber R. Proteins-Structure Function and Genetics. 2003;51:245. doi: 10.1002/prot.10349. [DOI] [PubMed] [Google Scholar]
- 9.Ma HR, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. Journal of the American Chemical Society. 2006;128:1523. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 10.Bourgeois D, Vallone B, Arcovito A, Sciara G, Schotte F, Anfinrud PA, Brunori M. Proceedings of the National Academy of Sciences of the United States of America; 2006. p. 4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux B. Annual Review of Biophysics and Biomolecular Structure. 2005;34:153. doi: 10.1146/annurev.biophys.34.040204.144655. [DOI] [PubMed] [Google Scholar]
- 12.Moroni D, Bolhuis PG, van Erp TS. Journal of Chemical Physics. 2004;120:4055. doi: 10.1063/1.1644537. [DOI] [PubMed] [Google Scholar]
- 13.Sarich M, Noe F, Schutte C. Multiscale Modeling & Simulation. 2010;8:1154. [Google Scholar]
- 14.Hinrichs NS, Pande VS. Journal of Chemical Physics. 2007;126 doi: 10.1063/1.2740261. [DOI] [PubMed] [Google Scholar]
- 15.Buchete NV, Hummer G. Journal of Physical Chemistry B. 2008;112:6057. doi: 10.1021/jp0761665. [DOI] [PubMed] [Google Scholar]
- 16.Majek P, Elber R. Journal of Chemical Theory and Computation. 2010;6:1805. doi: 10.1021/ct100114j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faradjian AK, Elber R. Journal of Chemical Physics. 2004;120:10880. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 18.Vanden-Eijnden E, Venturoli M. Journal of Chemical Physics. 2009;130:13. doi: 10.1063/1.3130083. [DOI] [PubMed] [Google Scholar]
- 19.Muller K. Angewandte Chemie-International Edition in English. 1980;19:1. [Google Scholar]
- 20.Kuczera K, Jas GS, Elber R. J. Phys. Chem. A. 2009;113:7461. doi: 10.1021/jp900407w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noe F, Horenko I, Schutte C, Smith JC. J Chem Phys. 2007;126:155102. doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- 22.Hummer G, Kevrekidis IG. Journal of Chemical Physics. 2003;118:10762. [Google Scholar]
- 23.Elber R, West A. Proceedings of the National Academy of Sciences USA; 2010. p. 5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elber R. Biophysical Journal. 2007;92:L85. doi: 10.1529/biophysj.106.101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AMA, Elber R, Shalloway D. Journal of Chemical Physics. 2007;126 doi: 10.1063/1.2716389. [DOI] [PubMed] [Google Scholar]
- 26.van Erp TS, Moroni D, Bolhuis PG. Journal of Chemical Physics. 2003;118:7762. doi: 10.1063/1.1644537. [DOI] [PubMed] [Google Scholar]
- 27.Allen RJ, Frenkel D, ten Wolde PR. Journal of Chemical Physics. 2006;124:17. doi: 10.1063/1.2140273. [DOI] [PubMed] [Google Scholar]
- 28.Majek P, Elber R. Journal of Chemical Theory and Computation. 2010 doi: 10.1021/ct100114j. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanden Eijnden E, Venturoli M, Ciccotti G, Elber R. Journal of Chemical Physics. 2008;129:174102. doi: 10.1063/1.2996509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalloway D, Faradjian AK. Journal of Chemical Physics. 2006;124 doi: 10.1063/1.2161211. [DOI] [PubMed] [Google Scholar]
- 31.Truhlar DG, Garrett BC, Klippenstein SJ. Journal of Physical Chemistry. 1996;100:12771. [Google Scholar]
- 32.Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G. Journal of Chemical Physics. 2006;125 doi: 10.1063/1.2212942. [DOI] [PubMed] [Google Scholar]
- 33.E WN, Vanden-Eijnden E. Annual Review of Physical Chemistry, Vol 61. 2010;61:391. doi: 10.1146/annurev.physchem.040808.090412. [DOI] [PubMed] [Google Scholar]
- 34.Peters B, Beckham GT, Trout BL. Journal of Chemical Physics. 2007;127 doi: 10.1063/1.2748396. [DOI] [PubMed] [Google Scholar]
- 35.Ma A, Dinner AR. Journal of Physical Chemistry B. 2005;109:6769. doi: 10.1021/jp045546c. [DOI] [PubMed] [Google Scholar]
- 36.Peters B, Trout BL. Journal of Chemical Physics. 2006;125 doi: 10.1063/1.2234477. [DOI] [PubMed] [Google Scholar]
- 37.Olender R, Elber R. Theochem-Journal of Molecular Structure. 1997;398:63. [Google Scholar]
- 38.Jonsson H, Mills G, Jacobsen KW. In: Classical and quantum dynamics in condensed phase simulations. Berne BJ, Ciccotti G, Coker DF, editors. World Scientific; Singapore: 1998. p. 385. [Google Scholar]
- 39.Huo SH, Straub JE. Journal of Chemical Physics. 1997;107:5000. [Google Scholar]
- 40.Elber R, Karplus M. Chemical Physics Letters. 1987;139:375. [Google Scholar]
- 41.Berkowitz M, Morgan JD, McCammon JA, Northrup SH. Journal of Chemical Physics. 1983;79:5563. [Google Scholar]
- 42.Zhao RJ, Shen JF, Skeel RD. Journal of Chemical Theory and Computation. 2010;6:2411. doi: 10.1021/ct900689m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berezhkovskii A, Hummer G, Szabo A. Journal of Chemical Physics. 2009;130 doi: 10.1063/1.3139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Proceedings of the National Academy of Sciences of the United States of America; 2009. p. 19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rich A. Nature Structural Biology. 2003;10:247. doi: 10.1038/nsb0403-247. [DOI] [PubMed] [Google Scholar]
- 46.Rhodes LM, Schimmel PR. Biochemistry. 1971;10:4426. doi: 10.1021/bi00800a012. [DOI] [PubMed] [Google Scholar]
- 47.Radhakrishnan R, Schlick T. Journal of Chemical Physics. 2004;121:2436. doi: 10.1063/1.1766014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altona C, Sundaralingam M. JACS. 1972;94:8205. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- 50.Weinbach Y, Elber R. Journal of Computational Physics. 2005;209:193. [Google Scholar]
- 51.Ryckaert JP, Ciccotti G, Berendsen HJC. Journal of Computational Physics. 1977;23:327. [Google Scholar]
- 52.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. Journal of the American Chemical Society. 1995;117:5179. [Google Scholar]
- 53.Pranata J, Wierschke SG, Jorgensen WL. Journal of the American Chemical Society. 1991;113:2810. [Google Scholar]
- 54.Elber R, Roitberg A, Simmerling C, Goldstein R, Li HY, Verkhivker G, Keasar C, Zhang J, Ulitsky A. Computer Physics Communications. 1995;91:159. [Google Scholar]
- 55.Sugita Y, Okamoto Y. Chemical Physics Letters. 1999;314:141. [Google Scholar]
- 56.Duda R, Hart P, Stork D. Pattern Classification. John Wiley and Sons; New York: 2001. [Google Scholar]
- 57.Juraszek J, Bolhuis PG. Biophysical Journal. 2008;95:4246. doi: 10.1529/biophysj.108.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]