Abstract
Five sublacustrine thermal spring locations from 1 to 109 m water depth in Yellowstone Lake were surveyed by 16S ribosomal RNA gene sequencing in relation to their chemical composition and dark CO2 fixation rates. They harbor distinct chemosynthetic bacterial communities, depending on temperature (16–110°C) and electron donor supply (H2S <1 to >100 μM; NH3 <0.5 to >10 μM). Members of the Aquificales, most closely affiliated with the genus Sulfurihydrogenibium, are the most frequently recovered bacterial 16S rRNA gene phylotypes in the hottest samples; the detection of these thermophilic sulfur-oxidizing autotrophs coincided with maximal dark CO2 fixation rates reaching near 9 μM C h−1 at temperatures of 50–60°C. Vents at lower temperatures yielded mostly phylotypes related to the mesophilic gammaproteobacterial sulfur oxidizer Thiovirga. In contrast, cool vent water with low chemosynthetic activity yielded predominantly phylotypes related to freshwater Actinobacterial clusters with a cosmopolitan distribution.
Keywords: Yellowstone, hydrothermal vents, chemosynthesis, Sulfurihydrogenibium, Thiovirga, sublacustrine springs
Introduction
In-depth geophysical and geochemical exploration and lake floor mapping of Yellowstone Lake, the largest Alpine Lake in the United States, has revealed numerous sublacustrine hot vents and hydrothermal features in geothermally active areas on the lake bottom (Klump et al., 1988, 1995; Remsen et al., 1990; Morgan et al., 2003a, b). Direct observations by SCUBA and ROV have revealed a wide range of hydrothermal features, including large hydrothermal chimneys; gas fumaroles; seepage of hot, shimmering water; and sulfur-oxidizing microbial mats growing around hot water seeps and vents (Remsen et al., 2002). Examination of an ancient vent chimney revealed internal conduit structures with metal sulfide precipitates, indicating long-term hydrothermal activity (Cuhel et al., 2004).
In contrast to the well-studied photosynthetic and chemosynthetic aerial hot spring communities of Yellowstone Park terrestrial habitats (Ward et al., 1998; Spear et al., 2005), systematic analyses of these Yellowstone Lake hydrothermal microbial communities are in their early stages, but have already demonstrated the potential for autotrophic, thermophilic chemosynthetic microbial communities. For example, the vents at Steamboat Point and Mary Bay at the northern edge of the lake, and deep-water vents off Stevenson island in the center of the lake, harbor chemosynthetic bacteria that assimilate dissolved inorganic carbon (DIC) in the dark at rates typically 0.08–0.5 μM C h−1, with maxima at approximately 0.75 μM C h−1 (Cuhel et al., 2002). Thus, chemosynthetic rates often exceed the photosynthetic rate at the lake surface, approximately 0.25 μM C h−1 (Cuhel et al., 2002). Microbial biodiversity in Yellowstone Lake thermal springs is largely unexplored. The isolation of the thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, a deeply branching bacterial lineage from Yellowstone Lake thermal vents, implies untapped potential for further discoveries (Henry et al., 1994).
Here, we present results of a preliminary 16S rRNA gene clone library survey of the bacterial communities in five different thermal vent locations in Yellowstone Lake with distinct chemical signatures and distinct temperature-dependent chemosynthetic rates. The 16S rRNA gene clone libraries indicate the existence of distinct chemosynthetic bacterial communities, dominated either by Gammaproteobacteria affiliated with the mesophilic sulfur-oxidizing genus Thiovirga, or by phylotypes most closely related to cultured species and strains of the extremely thermophilic Aquificales.
Materials and Methods
Sampling and DNA extraction
Hydrothermal vent waters were collected in July 2003 from five locations (Table 1) in Yellowstone Lake for microbial community analysis by 16S rRNA gene sequencing (Table 1). The first two samples come from the Mary Bay area near the northeastern shore of the lake, one of the hydrothermally most active areas of Yellowstone Lake, with high heat flux and numerous hydrothermal vents (Morgan et al., 2003b): Mary Bay West 12 is a surface water sample taken above a nearshore bubbling warm fumarole in shallow water (1 m), and Mary Bay Canyon 28 represents warm deep water (52 m) below the sill of an underwater canyon in Mary Bay. The third and hottest water sample, Stevenson Island 72, comes from a deep trench east of Stevenson Island in the central portion of the lake, where small, well-developed hydrothermal vents coalesce along northwest-trending deep fissures that reach maximally 133 m depth, the deepest point in the lake (Morgan et al., 2003b). The fourth sample, West Thumb Canyon 129, represents hot vent water from a sublacustrine explosion crater in the western part of West Thumb basin, in the westernmost part of the lake (Morgan et al., 2003b). The fifth sample from the West Thumb Basin, West Thumb 98, represents a cooler, low activity water sample from the West Thumb area. In situ temperatures measured by ROV, and chemistry of the syringe-sampled waters are shown in Table 1.
Table 1.
Yellowstone Lake vent samples: summary of chemistry, dark C fixation, and 16S rRNA gene sequence data.
| Sampling location and sample code | Mary Bay West 12 MBW | Mary Bay Canyon 28 V03-13P | Stevenson Island 72 V03-28P | West Thumb Canyon 129 V03-46P | West Thumb 98 V03-36P | Yellowstone Lake station LW-02 |
|---|---|---|---|---|---|---|
| Sample volume and date | 310 mL 7-16-03 | 190 mL 7-18-03 | 303 mL 7-22-03 | 150 mL 7-29-03 | 303 mL 7-25-03 | N/A 7-1-03 |
| Water depth (m) | 1 | 47.9 | 109.3 | 52.4 | 21.7 | 0 |
| Temperature (°C) | 19 | 49 | 110 | 77 | 16 | Ambient |
| DOC (mg/l) | 1.77 | 1.89 | n.d. | n.d. | 1.45 | n.d. |
| H2S (μM) | 2.2 | 119.3 | 59.5 | 17.2 | 0.7 | 0 |
| (μM) | 1.3 | 4.5 | 5.3 | 8.4 | 3.7 | 0 |
| (μM) | 0.1 | 1.4 | 19.9 | 1.52 | 0.5 | 0 |
| (μM) | 104 | 122 | 93 | 187 | 86 | 83 |
| (μM) | 1.5 | 12.7 | 11.0 | 8.8 | 0.0 | 0.7 |
| (μM) | 0.1 | 0.2 | 0.9 | 1.0 | 0.0 | 0.0 |
| SRP (μM) | 0.21 | 1.84 | 0.68 | 2.51 | 0.59 | 0.32 |
| SiO2 (μM) | 192 | 325 | 197 | 1865 | 159 | 178 |
| Cl− (μM) | 146 | 210 | 132 | 1971 | 156 | 125 |
| Summary of dark CO2 fixation rates (μM C h−1) | 1.08, 15°C 6.46, 15°C with S | 1.7 at 15°C 8.6 at 60°C no. S stimulation | <0.01, 15°C 0.4 at 50°C 0.67 at 60°C | Near 0.1 at 50 and 60°C 0.83–1.66, with S, 60°C | Near 0.003 with S, 50°C < 0.003 otherwise | 0.003 |
| No. of bacterial 16S rRNA gene sequences | 44 | 23 | 37 | 54 | 38 | |
| Aquificales | – | 2 | 16 | 13 | – | |
| Gammaproteobacteria | 21 | 5 | 1 | – | 1 | |
| Betaproteobacteria | 8 | 3 | 6 | 10 | 13 | |
| Alphaproteobacteria | 1 | 1 | – | – | – | |
| Deltaproteobacteria | – | 7 | – | – | – | |
| Epsilonproteobacteria | – | 1 | – | – | – | |
| Actinobacteria | 6 | – | 7 | 10 | 16 | |
| Planctomycetales | – | – | 4 | – | 2 | |
| CFB phylum | 2 | 1 | – | 3 | – | |
| Chloroflexi | 2 | – | – | 6 | 1 | |
| Acidobacteria | – | 3 | – | – | – | |
| Cyanobacteria | 2 | – | 1 | – | 3 | |
| Thermodesulfovibrio | – | – | – | 7 | – | |
| Verrucomicrobium | 1 | – | 2 | – | 2 | |
| OP10 | 1 | – | – | – | – | |
| Chlorobia | – | – | – | 2 | – |
Water was collected into 2-L polycarbonate piston-style syringe samplers mounted on a tethered ROV (Eastern Oceanics, Inc.) using an articulated arm outfitted with a thermistor probe at the end to measure the temperature of the water as it was collected (Aguilar et al., 2002). Using a checkvalve system, each piston sample was rinsed with 300–500 mL of vent sample prior to filling. The ROV is fully operator-controlled with thrusters that allow for lake floor reconnaissance and positioning for sampling, and included live video for vent identification and guided sampling (Lovalvo et al., 2010); it is deployed from RV Cutthroat, the Yellowstone Lake research vessel owned and operated by the National Park Service. Vent water samples were retrieved from the ROV collection syringes and put into smaller, all-plastic syringes through a three-way valve without exposure to air. Subsamples of 150–310 mL volume (Table 1) were used for filtration and cell capture on 0.22 μm polycarbonate filters. The filters were frozen at −80°C until DNA isolation in the laboratory in Chapel Hill, by phenol/chloroform extraction (Teske et al., 1996). Near-complete 16S rRNA genes were PCR-amplified with bacterial primers 8F (5′-AGRGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CGGCTACCTTGTTACGACTT-3′); these primers were successful in recovering extremely diverse bacterial communities, including members of novel phyla (Teske et al., 2002). PCR products were cloned using the TOPO XL PCR cloning kit (Invitrogen Corporation, Carlsbad, CA, USA) following the manufacturers instructions, and sequenced at the sequencing center of the University of North Carolina with primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′).
Chemical analysis and dark C fixation rate measurements
Analytical equipment was transported to Yellowstone National Park and set up as a field laboratory at the National Park Service Lake Station. Freshly collected samples for stable analytes were filtered through 0.2-μm filters (Supor-200, Pall Corp.) and aliquoted for the different analyses. Dissolved compounds were measured by flow injection analysis (silicate, nitrate, nitrite), ion chromatography (chloride, sulfate), and spectrophotometry (ammonium, phosphate) according to standard methods (APHA, 1992).
All labile species were analyzed on site within 1 day of collection and analytical preparation. Rubber-free all polypropylene (PP) syringes with three-way valves were used to collect water from the 2-L ROV-mounted piston syringe samplers without introducing headspace. The ROV’s screw drive was used to squeeze the water into receiving syringes to maximally retain dissolved gases. Total DIC was analyzed by the Teflon-membrane flow injection method of Hall and Aller (1992), in which the sipper tube was inserted through the three-way valve and the syringe plunger used to prevent formation of headspace during injection. For deep samples from Stevenson Island, unavoidable degassing effects were reduced by shaking the syringe thoroughly just before sipping, as the method measures all forms of CO2 including dissolved gas. Reduced sulfur compounds (hydrogen sulfide, thiosulfate, sulfite) were quantified by a scaled-up modification of the microbore high-performance liquid chromatographic (HPLC) method of Vairavamurthy and Mopper (1990), using dithio-bis-nitropyridine (DTNP) derivatization. Samples were collected in PP syringes and after rinsing, exactly 10 mL were squeezed through 0.2 μm nylon syringe filters (Whatman Acrodisc) into acid-washed 20 mL liquid scintillation vials (LSV). Using a positive displacement repeating pipette, the DTNP reagent was added, the vial capped with a cone-seal cap, and mixed vigorously before storing in a cooler with ice. After return to the laboratory, the precipitated unreacted DTNP was removed with another syringe and nylon filter during injection into the 100-μL HPLC sample loop. Each analysis required 45 min in a dual mobile phase gradient separation on a 250-mm C18 5 μm HS reversed phase column (Alltech Assoc.) so as many as 30 samples could be analyzed daily.
Light and dark CO2 incorporation rates were determined by measuring the biological conversion of acid-labile 14C-bicarbonate into acid-stable organic 14C, as described previously (Cuhel et al., 2002). Briefly, suitable aliquots of fresh vent water or lake samples, usually 180 mL, were dispensed with minimal mixing into an acid-washed PP beaker. Enough high specific activity 14C-bicarbonate (56 mCi mmol−1, MP Biomedical) was added to give about 2 × 106 DPM mL−1. The sample was drawn into a positive displacement 50 mL repeating pipette tip, and 10 mL aliquots were gently added to acid-washed 20 mL LSV in custom machined aluminum incubator blocks. Additions (thiosulfate, inhibitors, other stimulants, etc.) had been previously loaded into the appropriate vials at 1:100 to 1:1000× dilution from concentrated stocks. Temperature was controlled by a water bath circulator through the large 64-place block for 15°C incubations with triplicates and many treatments. Photosynthesis was measured identically, except that the projector lamp light source (ELH, 4 × 300 W) was turned on and incubations spanned only 3 h.
Elevated temperature incubation (controls, +thiosulfate in duplicate only) was accomplished in electrically heated aluminum dry block incubators. In these, 10 mL of sample were heated from ambient to 80°C in about 11 min during a ∼12 h incubation, minimizing artifacts of transient temperature increases in the upper mesophilic range. All vials were capped with cone-seal caps to reduce loss of volatile species, while the atmospheric oxygen contained in 12 mL of headspace guaranteed aerobic conditions for the measurements (>4 mM dissolved oxygen equivalents). Anaerobic chemosynthesis assays were beyond the capabilities of the field laboratory. For practical reasons, most incubations were initiated in late evening after field sampling and sample processing, and terminated prior to the next morning’s ROV expedition, usually 10–14 h of incubation. Each measurement was terminated by injection of 0.25 mL 2 N H2SO4 followed by >12 h shaking (100 RPM) to remove unmetabolized 14C, addition of liquid scintillation cocktail (Hydrofluor; National Diagnostics), and storage until counting. Duplicate 10 mL aliquots for background 14C contamination were shot into acid-containing LSV immediately after dispensing block samples; in 2003 these typically yielded <500 DPM from the 2 × 107 DPM added initially. The time course measurements for Mary Bay 12 were accomplished in 250 mL tissue culture flasks with additions of supplements and 14C-bicarbonate as above but in the larger volume of sample. The flasks were placed on the large aluminum blocks set for 15°C and covered with foil. At zero time and at each successive time point, 10 mL were pipetted into an LSV containing the sulfuric acid and treated as above.
Phylogenetic analysis and sequence quality control
Forward and reverse reads were assembled and aligned in ARB (Ludwig et al., 2004). PCR primer regions were excluded from the alignment underlying phylogenetic inference. Phylogenies were inferred using neighbor-joining analysis of Jukes–Cantor distance matrix; primer regions were excluded. For bootstrap analysis, the alignments were exported in paup format, and tree topologies were checked with 1000 distance neighbor-joining bootstrap replicates in PAUP 4.0 (Swofford, 2000). By comparing phylogenetic trees based separately on the first and the second halves of the alignments, chimeric sequences were identified and removed from the dataset; indicators for chimeras included inconsistent topologies in multiple trees based on the different 16S rRNA gene subsections, combined with low or lacking bootstrap support, and sequence removal improving the bootstrap support for the branch harboring the problem sequence (Hugenholtz and Huber, 2003). The sequences have GenBank accession numbers HM446045 to HM4461154 (Table 2).
Table 2.
List of clone names, Genbank accession numbers, phylogenetic affiliation, and sample of origin.
| Clone | GenBank accession number | Phylum | Sampling sites |
|---|---|---|---|
| MB28_023 | HM446067 | Acidobacteria | Mary Bay 28 |
| MB28_029 | HM446068 | Acidobacteria | Mary Bay 28 |
| MB28_030 | HM446070 | Acidobacteria | Mary Bay 28 |
| MB12_019 | HM446046 | Actinobacteria | Mary Bay 12 |
| MB12_023 | HM446049 | Actinobacteria | Mary Bay 12 |
| MB12_066 | HM446052 | Actinobacteria | Mary Bay 12 |
| MB12_071 | HM446053 | Actinobacteria | Mary Bay 12 |
| SI72_007 | HM446084 | Actinobacteria | Stevenson Island 72 |
| SI72_016 | HM446086 | Actinobacteria | Stevenson Island 72 |
| SI72_057 | HM446094 | Actinobacteria | Stevenson Island 72 |
| SI72_059 | HM446095 | Actinobacteria | Stevenson Island 72 |
| SI72_069 | HM446096 | Actinobacteria | Stevenson Island 72 |
| SI72_071 | HM446097 | Actinobacteria | Stevenson Island 72 |
| WT98_026 | HM446105 | Actinobacteria | West Thumb 98 |
| WT98_035 | HM446107 | Actinobacteria | West Thumb 98 |
| WT98_048 | HM446110 | Actinobacteria | West Thumb 98 |
| WT98_049 | HM446111 | Actinobacteria | West Thumb 98 |
| WT98_055 | HM446112 | Actinobacteria | West Thumb 98 |
| WT98_067 | HM446115 | Actinobacteria | West Thumb 98 |
| WT98_074 | HM446118 | Actinobacteria | West Thumb 98 |
| WT98_075 | HM446119 | Actinobacteria | West Thumb 98 |
| WT98_079 | HM446120 | Actinobacteria | West Thumb 98 |
| WT98_092 | HM446122 | Actinobacteria | West Thumb 98 |
| WT98_094 | HM446123 | Actinobacteria | West Thumb 98 |
| WT129_012 | HM446128 | Actinobacteria | West Thumb 129 |
| WT129_027 | HM446133 | Actinobacteria | West Thumb 129 |
| WT129_041 | HM446135 | Actinobacteria | West Thumb 129 |
| WT129_044 | HM446137 | Actinobacteria | West Thumb 129 |
| WT129_070 | HM446146 | Actinobacteria | West Thumb 129 |
| WT129_075 | HM446148 | Actinobacteria | West Thumb 129 |
| WT129_081 | HM446151 | Actinobacteria | West Thumb 129 |
| MB12_089 | HM446063 | Alphaproteobacteria | Mary Bay 12 |
| MB28_002 | HM446076 | Alphaproteobacteria | Mary Bay 28 |
| MB28_013 | HM446069 | Aquificales | Mary Bay 28 |
| MB28_032 | HM446071 | Aquificales | Mary Bay 28 |
| SI72_008 | HM446085 | Aquificales | Stevenson Island 72 |
| SI72_055 | HM446092 | Aquificales | Stevenson Island 72 |
| WT129_036 | HM446134 | Aquificales | West Thumb 129 |
| WT129_042 | HM446136 | Aquificales | West Thumb 129 |
| WT129_051 | HM446138 | Aquificales | West Thumb 129 |
| WT129_071 | HM446147 | Aquificales | West Thumb 129 |
| WT129_076 | HM446149 | Aquificales | West Thumb 129 |
| WT129_092 | HM446154 | Aquificales | West Thumb 129 |
| MB12_009 | HM446045 | Bacteroidetes | Mary Bay 12 |
| MB12_053 | HM446051 | Bacteroidetes | Mary Bay 12 |
| MB28_005 | HM446066 | Bacteroidetes | Mary Bay 28 |
| WT129_019 | HM446130 | Bacteroidetes | West Thumb 129 |
| WT129_025 | HM446132 | Bacteroidetes | West Thumb 129 |
| WT129_063 | HM446144 | Bacteroidetes | West Thumb 129 |
| MB12_003 | HM446057 | Betaproteobacteria | Mary Bay 12 |
| MB12_010 | HM446058 | Betaproteobacteria | Mary Bay 12 |
| MB12_043 | HM446060 | Betaproteobacteria | Mary Bay 12 |
| MB12_060 | HM446061 | Betaproteobacteria | Mary Bay 12 |
| MB12_075 | HM446062 | Betaproteobacteria | Mary Bay 12 |
| MB12_106 | HM446064 | Betaproteobacteria | Mary Bay 12 |
| MB12_113 | HM446065 | Betaproteobacteria | Mary Bay 12 |
| MB28_022 | HM446072 | Betaproteobacteria | Mary Bay 28 |
| MB28_028 | HM446082 | Betaproteobacteria | Mary Bay 28 |
| SI72_039 | HM446089 | Betaproteobacteria | Stevenson Island 72 |
| SI72_082 | HM446098 | Betaproteobacteria | Stevenson Island 72 |
| SI72_096 | HM446099 | Betaproteobacteria | Stevenson Island 72 |
| WT98_014 | HM446103 | Betaproteobacteria | West Thumb 98 |
| WT98_032 | HM446106 | Betaproteobacteria | West Thumb 98 |
| WT98_058 | HM446113 | Betaproteobacteria | West Thumb 98 |
| WT98_060 | HM446114 | Betaproteobacteria | West Thumb 98 |
| WT98_071 | HM446116 | Betaproteobacteria | West Thumb 98 |
| WT98_080 | HM446121 | Betaproteobacteria | West Thumb 98 |
| WT129_001 | HM446124 | Betaproteobacteria | West Thumb 129 |
| WT129_006 | HM446126 | Betaproteobacteria | West Thumb 129 |
| WT129016 | HM446129 | Betaproteobacteria | West Thumb 129 |
| WT129_024 | HM446131 | Betaproteobacteria | West Thumb 129 |
| WT129_062 | HM446143 | Betaproteobacteria | West Thumb 129 |
| WT129_088 | HM446153 | Betaproteobacteria | West Thumb 129 |
| MB12_021 | HM446047 | Chloroflexi | Mary Bay 12 |
| MB12_022 | HM446048 | Chloroflexi | Mary Bay 12 |
| WT98_073 | HM446117 | Chloroflexi | West Thumb 98 |
| WT129_005 | HM446125 | Chlorobia | West Thumb 129 |
| WT129_056 | HM446139 | Chlorobia | West Thumb 129 |
| WT129_059 | HM446142 | Chloroflexi/Thermomicrobium | West Thumb 129 |
| WT129_064 | HM446145 | Chloroflexi | West Thumb 129 |
| WT129_084 | HM446152 | Chloroflexi | West Thumb 129 |
| MB12_042 | HM446050 | Cyanobacteria | Mary Bay 12 |
| MB12_088 | HM446055 | Cyanobacteria | Mary Bay 12 |
| SI72_036 | HM446088 | Cyanobacteria | Stevenson Island 72 |
| WT98_013 | HM446102 | Cyanobacteria | West Thumb 98 |
| MB28_034 | HM446073 | Deltaproteobacteria | Mary Bay 28 |
| MB28_001 | HM446077 | Deltaproteobacteria | Mary Bay 28 |
| MB28_011 | HM446079 | Deltaproteobacteria | Mary Bay 28 |
| MB28_016 | HM446080 | Deltaproteobacteria | Mary Bay 28 |
| MB28_031 | HM446083 | Epsilonproteobacteria | Mary Bay 28 |
| MB12_039 | HM446059 | Gammaproteobacteria | Mary Bay 12 |
| MB28_025 | HM446074 | Gammaproteobacteria | Mary Bay 28 |
| MB28_003 | HM446075 | Gammaproteobacteria | Mary Bay 28 |
| MB28_010 | HM446078 | Gammaproteobacteria | Mary Bay 28 |
| MB28_027 | HM446081 | Gammaproteobacteria | Mary Bay 28 |
| SI72_048 | HM446091 | Gammaproteobacteria | Stevenson Island 72 |
| WT98_041 | HM446109 | Gammaproteobacteria | West Thumb 98 |
| WT98_019 | HM446104 | Actinobacteria/Microthrix | West Thumb 98 |
| MB12_107 | HM446056 | OP10 | Mary Bay 12 |
| SI72_029 | HM446087 | Planctomycetes | Stevenson Island 72 |
| SI72_041 | HM446090 | Verrucomicrobia | Stevenson Island 72 |
| SI72_056 | HM446093 | Planctomycetes | Stevenson Island 72 |
| WT98_005 | HM446100 | Planctomycetes | West Thumb 98 |
| WT129_010 | HM446127 | Thermodesulfobacteria | West Thumb 129 |
| WT129_057 | HM446140 | Thermodesulfovibrio-related | West Thumb 129 |
| WT129_058 | HM446141 | Thermodesulfovibrio-related | West Thumb 129 |
| WT129_078 | HM446150 | Thermodesulfovibrio-related | West Thumb 129 |
| MB12_076 | HM446054 | Verrucomicrobia | Mary Bay 12 |
| WT98_006 | HM446101 | Verrucomicrobia | West Thumb 98 |
| WT98_037 | HM446108 | Verrucomicrobia | West Thumb 98 |
Principal component analysis
The 16S rRNA gene clone libraries were examined for statistical correlations to each other by principal component analysis, based on the number of shared vs separate phylotypes (operational taxonomic units, OTUs) between different clone libraries, and weighted for abundance. The analysis was performed online at the Unifrac website (http://128.138.212.43/unifrac/index.psp; Lozupone et al., 2006). OTU definition was based on more than 3% 16S rRNA sequence difference over a shared alignment length of approx. 500 nucleotides (E. coli positions 28–537).
Results
Chemical composition of hydrothermal vent waters
The chemical composition of the five water samples from sublacustrine Yellowstone Lake vents was distinct for every sample; reduced sulfur (HS−/S= or H2S), ammonia ( or NH3) and DIC ( or CO2) are of particular significance to chemolithotrophic metabolism. Sulfide and different oxidation states of sulfur (sulfite, thiosulfate) occurred in all water samples, but especially in the warmest samples Mary Bay Canyon 28, Stevenson Island 72, and West Thumb 129 (Table 1). A non-hydrothermal surface lake water reference sample from the Northwest shore, near the Lake Village laboratory, contained only the most oxidized sulfur species, sulfate, at detectable concentrations. DIC concentrations were highest in the same three samples, Mary Bay Canyon 28, Stevenson Island 71, and West Thumb 129; also, ammonia was most abundant in these three warmest vents (Table 1). The mixture of microbial electron donors (reduced sulfur and nitrogen species) and the elevated DIC concentration are compatible with high chemosynthetic activity (Table 1). While these chemical species likely indicate deeply sourced components of the vent fluid, a diagenetic source of sulfide, DIC, and ammonia, from decomposition of buried biomass is also possible.
In contrast, chloride is an unequivocal conservative marker for geothermal fluid because it has no diagenetic source. Silicate enrichment may also be a signal, though dissolution of diatom frustules can lead to enrichment in surficial sediments near vent orifices. Mary Bay Canyon 28 and West Thumb 129 showed chloride and silicate enrichment by factors 2 and >10 relative to lake water (Table 1), and were likely to have deep geothermal fluid sources; this interpretation is supported by previous porewater analyses from hydrothermally active sediments of Mary Bay and West Thumb that had yielded very high chloride concentrations, near 4–9 mM at 30 cm sediment depth (Aguilar et al., 2002). Thus, the localized high concentrations of ammonia, DIC, and sulfide at these two vents at least in part represented deeply sourced chemical species. Interestingly, the deep and hot Stevenson Island 72 sample showed no selective chloride or silicate enrichment that unequivocally distinguishes deep geothermal fluid sources. The high concentrations of sulfide, DIC, and ammonia may arise through transport in subsurface conduits as gases in the steam phase rather than as dissolved ionic species.
The samples Mary Bay West 12 and West Thumb 98 showed the lowest sulfide, DIC, ammonia, and nitrate concentrations, and no conservative marker signature (chloride, silicate) of deeply sourced geothermal fluids; the shallow West Thumb 98 sample showed no chemical enrichment of any parameter besides sulfite (Table 1). Dissolved organic carbon (DOC) was poorly related to physical–chemical variables and likely derived from lake water admixture at vent orifices.
Nitrate, an important substrate for anaerobic respiration, was virtually absent in Yellowstone Lake waters. During the 2003 season, 133 water samples, including 69 from hydrothermal vents, were analyzed for nitrate. Only 13 samples, 10 of these in the Stevenson Island domain, had nitrate concentrations above 0.5 μM; none exceeded 2 μM.
Chemosynthetic activity
The vent water samples showed distinct, temperature-dependent patterns of dark CO2 fixation (Figure 1). The addition of the bacterial protein synthesis inhibitor chloramphenicol reduced chemosynthetic activity, whereas the eukaryotic protein synthesis inhibitor cycloheximide did not affect chemosynthesis rates (data not shown). Biomass nutrient elements N and P were sufficient to support chemosynthesis at the level of electron donors available in these measurements.
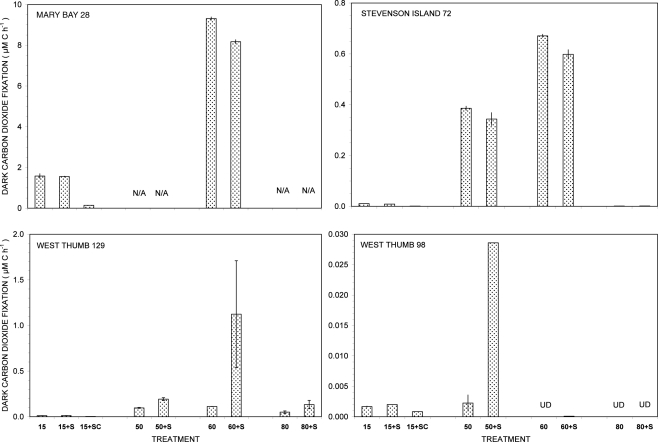
Figure 1.
Dark carbon dioxide fixation by Yellowstone Lake hydrothermal vent samples measured in triplicate (15°C) or duplicate incubations (50, 60, and 80°C) with (+S) or without addition of 5 mM thiosulfate. Biological fixation controls with added S + 20 μg/mL chloramphenicol (SC) were tested at 15°C only. UD, undetectable (<0.0005 μM C h−1); N/A, not available.
Of all dark CO2 fixation measurements at cool temperatures (15°C), Mary Bay Canyon 28 showed the highest activity near 1.7 μM C h−1 (Figure 1), even higher than the persistently active shallow bubbler at Mary Bay 12 (1.08 μM C h−1). Addition of 5 mM thiosulfate did not stimulate dark CO2 fixation rates at Mary Bay Canyon 28, but was extremely stimulatory over the 12- to 14-h incubation period at Mary Bay 12 (6.46 μM h−1) in a manner indicative of population growth. The conspicuous difference in thiosulfate stimulation at 15°C rates between Mary Bay 12 and 28 could be influenced by their different setting: Mary Bay West 12 vent fluids emerge into shallow, well-mixed open water, subject to immediate dilution, whereas Mary Bay West 28 vent fluids remain confined within a deep, narrow canyon. The highest chemosynthetic rates in unamended samples (around 8.6 μM C h−1) were commonly found in the thermophilic range at 60°C, at Mary Bay Canyon 28 (Figure 1); high temperature rates are not available for Mary Bay West 12.
Thiosulfate stimulation was only evident in the West Thumb samples (Figure 1). In West Thumb Canyon 129, stimulation was unequivocal (>10×) at 50°C but was associated with very high replicate error for 60°C incubations. Because the replicates for almost all other incubations agreed to a few percent, it is likely that the West Thumb 129 sample contained bacteria either clumped or attached to particles with patchy distribution. The positive dark fixation results at 50 and/or 60°C in Mary Bay, West Thumb 129, and Stevenson Island 72 strongly implicate chemosynthetic activity by thermophilic autotrophs in these samples. Chemosynthetic activity in the mesophilic range at 15°C was low at West Thumb 129 and Stevenson Island 72 (below 0.02 μM C h−1), indicating mesophilic autotrophs in reduced numbers or activity compared to the Mary Bay samples. West Thumb 129 was the only sample that showed low but detectable chemosynthetic activity at 80°C, indicating the existence of hyperthermophiles.
The West Thumb 98 sample showed very low dark CO2 fixation rates close to the detection limit and the lake water background, generally below 0.002 μM C h−1 for all temperatures; a low but detectable rate near 0.028 μM C h−1 was obtained for thiosulfate-amended water at 50°C (Figure 1).
The distinct temperature effects on chemosynthetic activity suggest the existence of two groups of chemosynthetic microorganisms: chemosynthetic mesophiles in the two Mary Bay Samples, and thermophiles in the Mary Bay Canyon 28, West Thumb Canyon 129, and Stevenson Island 72 samples. The Mary Bay Canyon 28 sample likely contained both types of chemosynthetic communities. The nearly inactive West Thumb 98 sample should contain few detectable chemoautotrophs, although seed populations might be present that account for low-level thermophilic sulfur stimulation.
These inferences are consistent with the results of previous sampling surveys. During a previous 3-year sampling period of 1997, 1998, and 1999, Mary Bay water samples showed the highest sulfide concentrations (max. 0.7–0.8 mM), the highest DIC concentrations (max. 16–17 mM), and also the highest dark CO2 fixation rates (max. 0.3–0.6 μM C h−1) at mesophilic temperatures, in comparison to significantly lower concentrations and rates found in Stevenson Island and West Thumb water samples (Cuhel et al., 2002). Thus, previous studies also suggest the existence of mesophilic, chemosynthetic microbial communities in Mary Bay.
Photosynthetic activity
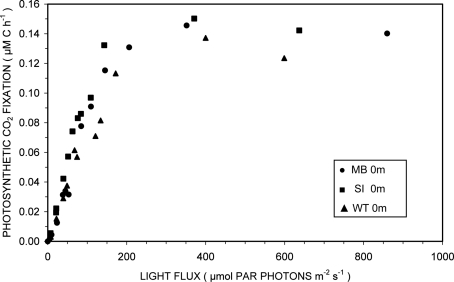
Photosynthesis-irradiance curves for surface samples from the three domains of Mary Bay, Stevenson Island, and West Thumb demonstrated light-saturable photosynthetic CO2 fixation (Figure 2). Light stimulated photosynthetic rates to 200 μmol PAR photons m−2 s−1, about 10% of full sunlight, above which rates remained constant at 0.14 ± 0.01 μM C h−1. Because this discussion concerns volumetric carbon fixation, the data were not normalized for varying algal biomass as chlorophyll a and are presented in the same units as chemosynthesis for direct comparison.
Figure 2.
Photosynthesis-irradiance relationships at 15°C for surface waters overlying hydrothermal vent fields in Mary Bay (●), Stevenson Island (■), and West Thumb (▲).
Microbial community composition
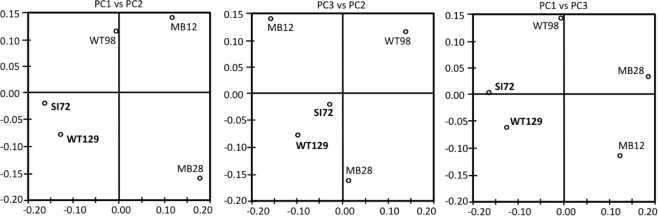
Consistent with the working hypotheses based on temperature-sensitive chemosynthetic rates, the clone libraries from the five sampling locations show distinct microbial community compositions (Table 1). The clone libraries from the two samples with the highest temperatures, West Thumb Canyon 129 and Stevenson Island 72, consistently cluster together in Principal Component analysis based on shared 16S rRNA phylotypes (Figure 3). The clone libraries for the other sites did not show any correlations, a possible consequence of data set size but also evident from deeply divergent microbial community composition (Table 1).
Figure 3.
Principal component analysis plots showing the clustering of 16S rRNA clone libraries based on shared OTUs, weighted for abundance. Each axis indicates the fraction of the variance in the data that the axis accounts for. The first and the second Principal Component explain most of the data (P1 = 41.4, P2 = 30, P3 = 17.8%).
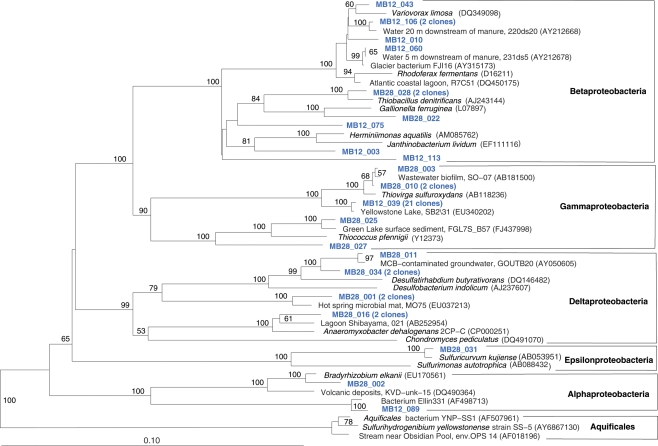
The Mary Bay West 12 sample, from an active bubbling vent near the lake shore, was dominated by gammaproteobacterial phylotypes related to the mesophilic sulfur oxidizer Thiovirga sulfuroxydans; this bacterium has an optimal growth temperature of 30–34°C (total growth range 15–42°C), and grows as an obligate chemolithoautotroph by oxidizing sulfur, sulfide, and thiosulfate aerobically; its 16S rRNA gene sequence forms a monophyletic lineage with very similar (at least 97% similarity) phylotypes from sulfidic and geothermal waters (Ito et al., 2005). The Mary Bay West 12 sample also yielded clones related to a betaproteobacterial cluster from diverse freshwater environments that included the freshwater bacteria Variovorax and Rhodoferax (Table 1; Figure 4). This betaproteobacterial cluster is generally found in freshwater environments, and has a cosmopolitan distribution (Zwart et al., 2002). The Mary Bay Canyon 28 sample harbored Thiovirga-related clones as well, but its bacterial community appeared to be differently structured and consisted at least in part of clones related to anaerobic bacteria (Table 1); no PCA clustering of the two Mary Bay samples was observed (Figure 3). Several clones were affiliated with anaerobic Deltaproteobacteria that usually occur in aquatic sediments but not in the water column; these include sulfate-reducing bacteria of the Desulfobacteraceae, with the butyrate oxidizer Desulfatirhabdium butyrativorans as the closest match (Balk et al., 2008); the heterotrophic halorespiring bacterium Anaeromyxobacter dehalogenans (Sanford et al., 2002); and uncultured bacteria from the sediment of an eutrophic lake. No other water sample yielded deltaproteobacterial clones. Also, Mary Bay West 28 contained close relatives of the anaerobic, nitrate-reducing and sulfur-oxidizing autotrophic bacterium Thiobacillus denitrificans within the Betaproteobacteria, and of the facultatively anaerobic, microaerophilic, or nitrate-reducing sulfur oxidizer Sulfuricurvum kujiense within the Epsilonproteobacteria (Kodama and Watanabe, 2004; Figure 4). This distinct microbial community signature corresponded to a different setting. Mary Bay 12 was a bubbler in shallow open water at low temperature (15°C) and high mixing rate, which never exceeded 20°C and was near freezing for 6 months of the year. In contrast, the hot Mary Bay Canyon 28 sample (49°C at sampling), the most confined of all the sample locations, came from the bottom of a canyon-like crater at ca. 56 m depth, with nearby sediments hot enough to melt a core liner. With a sill depth of 20 m, these steep-walled hydrothermal explosion craters are characteristic for Mary Bay (Morgan et al., 2003b) and provide more geochemically favorable conditions for anaerobic, especially heterotrophic bacterial growth. The conspicuous sequence signature of obligately or facultatively anaerobic bacteria may indicate deep-growing populations from hydrothermal sediments at the bottom of the crater that undergo upward advection and suspension within into the deep-water layers of this enclosed basin.
Figure 4.
Phylogeny of Mary Bay West 12 and Mary Bay Canyon 28 proteobacterial clones, based on E. coli 16S rRNA gene nucleotide positions 723–1491.
In addition, Mary Bay Canyon 28 harbored a clone most closely related to the autotrophic iron-oxidizing bacterium Gallionella ferruginea, and two clones of the thermophilic, chemosynthetic phylum Aquificales, specifically the hydrogen- and sulfur-oxidizing genus Sulfurihydrogenibium (Figure 4). Thus, Mary Bay Canyon 28 contains an unusually multifaceted microbial community, consisting of mesophilic and thermophilic chemosynthetic bacteria within the Beta-, Gamma-, and Epsilonproteobacteria and the Aquificales (consistent with high chemosynthetic rates at both temperature regimes), as well as nitrate- and sulfate-reducing anaerobes.
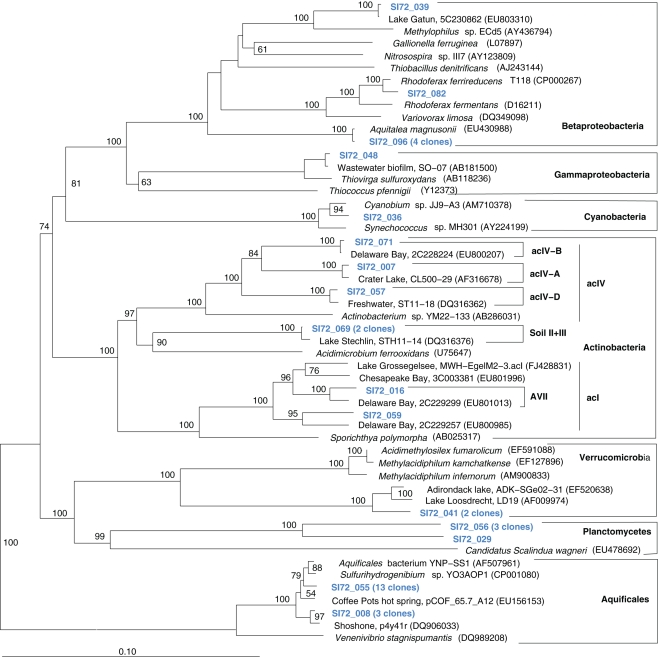
The microbial community of the deep, hot Stevenson Island 72 sample was distinct from those of both Mary Bay samples. Almost half of the cloned 16S rRNA gene sequences grouped with the Aquificales (Table 1) and are most closely related to clones from other Yellowstone Park locations (Figure 5) and the cultured strain Sulfurihydrogenibium spp. YO3AOP1 from Obsidian Pool in Yellowstone National Park (Reysenbach et al., 2009). These populations were likely from the vent throat or orifice near deep lake water; the thermistor on the sampling arm of the ROV gave 110°C, indicating source fluid temperatures possibly above 125°C. The temperature optimum of cultured Sulfurihydrogenibium strains and species is between 65 and 70°C; depending on the species, growth is possible between 40 and 78°C (Nakagawa et al., 2005). The species most closely related to the Yellowstone Lake clones, Sulfurihydrogenibium yellowstonense strain SS-5, has a growth temperature range of 55–78°C, 5–8°C more tolerant of high temperatures than other described species (Nakagawa et al., 2005); it uses sulfur and thiosulfate but not hydrogen as electron donors, and is obligately aerobic; it assimilates organic carbon compounds for biosynthetic purposes, but is unable to use them as energy sources (Nakagawa et al., 2005). Other phylum-level bacterial groups in the Stevenson Island sample included Betaproteobacteria, Actinobacteria, Planctomycetales, and Verrucomicrobia, plus a single Thiovirga-related gammaproteobacterial clone (Table 1).
Figure 5.
Phylogeny of Stevenson Island 72 bacterial clones, based on E. coli 16S rRNA gene nucleotide positions 25–1491.
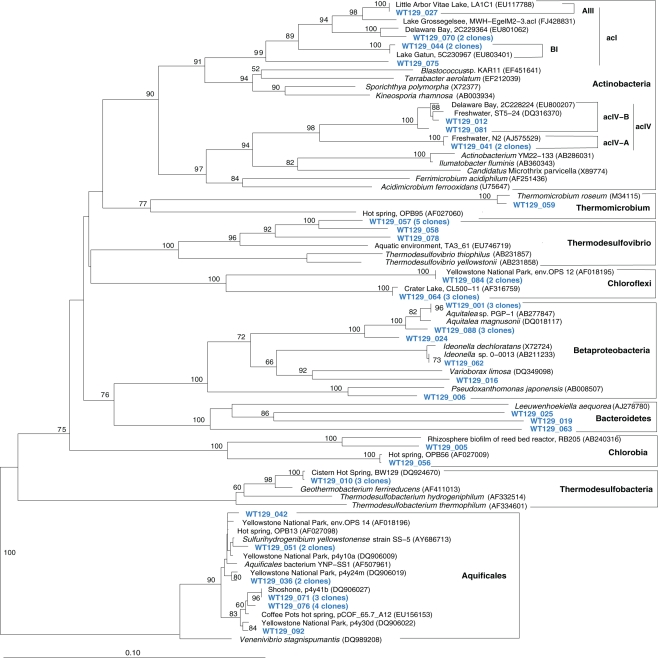
The microbial communities of the Stevenson Island 72 and West Thumb Canyon 129 resembled each other, as confirmed by clustering in PCA analysis (Figure 3). West Thumb Canyon 129 had the next-highest temperature (77°C) after the Stevenson Island sample; both samples shared abundant Aquificales clones specifically related to the cultured species of the genus Sulfurihydrogenibium, and to Sulfurihydrogenibium-related clones from different locations within Yellowstone National Park (Figure 6). The abundance of Sulfurihydrogenibium clones is consistent with the high sulfur-stimulated chemosynthetic rates at 60°C, and the lower but still substantial rates at 50°C in these two samples (Figure 1). Since chemosynthesis rates and sulfur stimulation persisted at 80°C at West Thumb Canyon 129 (Figure 1), it is possible that this sample contained high-temperature-adapted strains of Sulfurihydrogenibium, or different types of thermophiles and hyperthermophiles that remain to be found. Next to the Aquificales, the most frequently recovered clones represent Betaproteobacteria and Actinobacteria, which occur in the West Thumb Canyon 129, Stevenson Island 72, Mary Bay West 12, and (to a lesser extent, only Betaproteobacteria) the Mary Bay Canyon 28 samples (Table 1).
Figure 6.
Phylogeny of West Thumb Canyon 129 bacterial clones, based on E. coli gene nucleotide positions 25–573.
The West Thumb Canyon 129 sample also contained, in similar abundance, phylotypes of the Chloroflexi and the Thermodesulfovibrio lineages; the latter was not found in other clone libraries (Table 1), potentially a result of terrestrial spring influx limited to the West Thumb region. Within the Thermodesulfovibrio lineage, the phylotypes from West Thumb Canyon 129 were most closely related to the autotrophic, hydrogen-oxidizing, iron-reducing bacterium Geothermobacterium ferrireducens (Figure 6) previously isolated from hot spring sediments in Yellowstone National Park (Kashefi et al., 2002). The West Thumb area harbors a suitable biogeochemical niche for this bacterium: iron-manganese oxide crusts were found on the hydrothermally active lake bottom, and the upper sediment layers contain porewater concentrations of 20–40 μM dissolved, reduced iron (Aguilar et al., 2002).
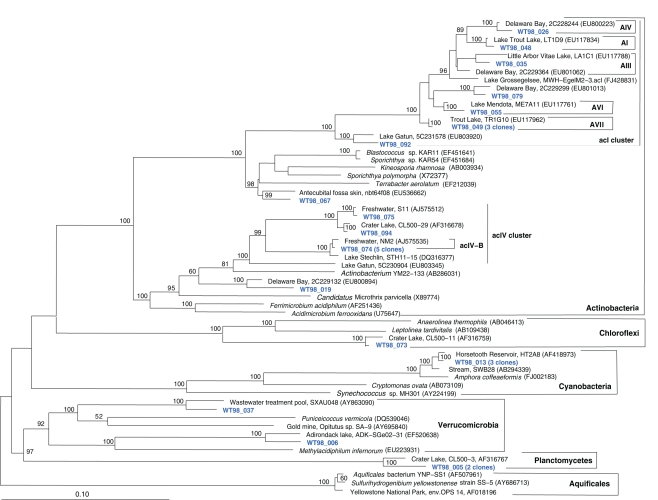
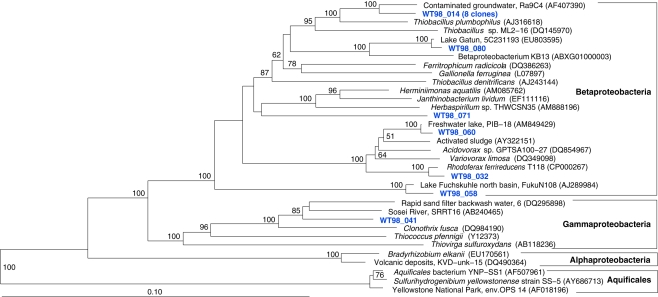
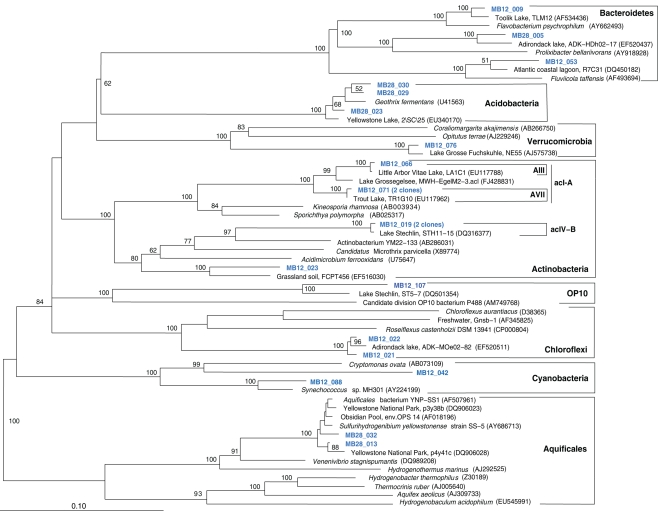
The fifth sample, a relatively cool West Thumb 98 sample with low chemosynthetic activity, yielded no Aquificales or other chemosynthetic bacterial populations, but predominantly members of the Actinobacteria (Figure 7) and the Betaproteobacteria (Figure 8), the bacterial community components that occur to some extent in all Yellowstone Lake water samples (Table 1). The actinobacterial clones in this and other Yellowstone Lake samples are members of uncultured Actinobacterial freshwater lineages with a cosmopolitan distribution, the acI and acIV clusters (Warnecke et al., 2004). In subsequent sequencing surveys of freshwater lakes, 16S rRNA genes of these clusters were found very frequently and in high diversity. The clades were further subdivided into clades AI to AVII for the acI linage (Newton et al., 2007) and clades acIV-A to acIV-E for the acIV lineage (Warnecke et al., 2004; Holmfeldt et al., 2009); this nomenclature is followed in phylogenetic trees in this study. Actinobacterial acI or acIV clades were found in almost every water sample, Mary Bay 12 (Figure 9), Stevenson Island 72 (Figure 5), West Thumb Canyon 129 (Figure 6), and West Thumb 98 (Figure 7), and most likely represent the admixture of Yellowstone Lake water and its indigenous microbial community to the thermal water samples. Since acI and acIV strains have not been cultured yet, a physiological rationale for the occurrence of these bacteria in Yellowstone Lake water is hard to infer. Interestingly, comparative quantifications of the acI clade in high mountains lakes have identified UV irradiation as a potential factor selecting for acI actinobacteria (Warnecke et al., 2005).
Figure 7.
Phylogeny of West Thumb 98 bacteria except Proteobacteria, based on E. coli 16S rRNA gene nucleotide positions 25–1491.
Figure 8.
Phylogeny of West Thumb 98 proteobacterial clones, based on E. coli 16S rRNA gene nucleotide positions 25–1491.
Figure 9.
Phylogeny of Mary Bay West 12 and Mary Bay Canyon 28 bacterial clones, except Proteobacteria, based on E. coli 16S rRNA gene nucleotide positions 723–1491.
Discussion
Hydrogen- and sulfur-based chemosynthesis
The results from the Yellowstone Lake hydrothermal vents form an interesting contrast to previous results from multiple aerial hot springs in Yellowstone National Park (Spear et al., 2005). By combining chemical analysis of spring waters for electron donors including sulfide and hydrogen, thermodynamic modeling of energy yields from the aerobic oxidation of these electron donors, and extensive 16S rRNA gene sequencing, aerobic hydrogen oxidation was identified as the favored energy source in Yellowstone hot springs, consistent with 16S rRNA gene clone libraries dominated by hydrogenotrophic genera of the Aquificales, such as Hydrogenothermus, Hydrogenobacter, and Hydrogenobaculum (Spear et al., 2005). Close relatives of mesophilic sulfur-oxidizing bacteria (Thiovirga spp.) occur in some locations, for example in the Mary Bay sampling sites, and most likely contribute to dark CO2 fixation rates measured at cool temperatures. Consistent with these results, previous studies have shown that dark CO2 fixation rates at Mary Bay locations are strongly stimulated by thiosulfate enrichment (Cuhel et al., 2002). In contrast, clone library results from the hot Stevenson Island and West Thumb Canyon vents, show that mesophilic sulfur oxidizers such as Thiovirga spp. are not detected in these habitats, or occur only as a minority. Here, the spectrum of clones changes toward phylotypes that are most closely related to thermophilic and hyperthermophilic chemolithoautotrophs of the genus Sulfurihydrogenibium that use sulfur and hydrogen as electron donors. As a caveat, S. yellowstonense, the closest relative to the Yellowstone Lake clones in this dataset, oxidizes sulfur and thiosulfate, but not hydrogen.
Since chemolithoautotrophic microorganisms may use one or several different electron donors, including reduced sulfur compounds such as H2S, polythionates, Fe(II), Mn(II), NH3, or H2, our results have obvious limitations. Here, thiosulfate was chosen as model reduced sulfur stimulant of chemosynthesis because it does not as easily react with other substrates as might have happened with H2S as a supplement. Since thiosulfate was rarely found in quantity even when H2S was abundant (Table 1), it was most likely utilized rapidly upon (a)biotic production from sulfide. Thiosulfate-stimulated autotrophic CO2 fixation implicated the presence of bacteria capable of using it, and possibly other reduced sulfur compounds, as electron donors. A lack of thiosulfate stimulation would imply either electron donor saturation, or the absence of thiosulfate-oxidizing bacteria. Interestingly, thermophilic dark CO2 fixation rates at Mary Bay Cayon 28 and Stevenson Island 72 were reduced by ca. 10% after thiosulfate addition (Figure 1); reactions of the added thiosulfate (or its disproportionation products) with other electron donors such as Fe(II), might reduce their availability, and thus reduce the autotrophic contribution of non-sulfur lithotrophs.
Nitrate reduction and metal reduction
The chemosynthesis rates in this study were measured aerobically, assuming that chemosynthetic microbial communities venting into a well-oxygenated oligotrophic lake would respire with oxygen. The prevalence of 16S rRNA gene sequences related to aerobic chemosynthetic bacteria (Thiovirga, Aquificales) supports this assumption. Niches certainly exist for anaerobic, nitrate- and metal-reducing chemosynthesis in Lake Yellowstone, for example anaerobic vent fluid, sediments or hydrothermal mineral deposits. Very low oxygen solubility at high temperature, and the presence of reducing compounds including Fe(II) and H2S, might facilitate anoxic conditions in vent source waters and possibly during channelized flow toward the lake-bottom orifice. As a result, phylotypes related to anaerobic chemosynthetic bacteria, such as the nitrate-reducing sulfur oxidizers Thiobacillus denitrificans and Sulfuricurvum kujiense, or the iron-reducing hydrogenotroph Geothermobacterium ferrireducens, were detected as well. Because 16S rRNA genes related to nitrate-reducing sulfur oxidizers were detected in Mary Bay 28, this deep, stratified canyon represents a likely habitat where nitrate-respiring vent or sediment populations accumulate to sufficient densities to be detected in clone libraries; such microbial nitrate sinks could compound the functional absence of nitrate (usually <0.2 μM) in any of the lake and vent water habitats sampled. Authigenic iron and manganese oxide crusts on the lake bottom, and bottom sediments with 20–50 μM porewater concentrations of Fe(II) (Aguilar et al., 2002) could provide a niche for hyperthermophilic iron-reducing hydrogenotrophs such as Geothermobacterium ferrireducens. Hydrothermal sediment cores show porewater chemical profiles consistent with sulfide oxidation and sulfate production, ammonia oxidation, and DIC consumption, from 30 cm depth toward the sediment surface (Aguilar et al., 2002). To summarize, anaerobic chemosynthesis remains to be systematically investigated by quantitative microbial sampling and measurements of anaerobic respiration rates.
New discoveries at West Thumb
In previous studies of Yellowstone Lake water column and vent fluids, samples could be grouped by geochemical and chemosynthetic characteristics into functional domains (Cuhel et al., 2002): West Thumb was characterized by low concentrations of electron donors, and high levels of chloride and silicate; Stevenson Island and Mary Bay sites were characterized by high electron donor and low chloride concentrations; Sedge Bay showed reducing gases in fumarole-dominated emanations in combination with low chloride (Konkol et al., 2010); and South Arm served as a control area outside of caldera influence. The latter two are not presented here. In pre-2002 surveys, West Thumb vents rarely exceeded 0.02 μM C h−1 in chemosynthesis and did not display thiosulfate stimulation, while different Mary Bay sites and to a more sporadic extent Stevenson Island vents frequently reached 10 times that value (Cuhel et al., 2002) and were often stimulated by thiosulfate addition. The present work adds the chemosynthetically active site West Thumb Canyon 129, the result of more sophisticated ROV-based sampling and continuous search for new vent fields. While the very low rates of chemosynthesis at ambient temperature (ca. 15°C) in the West Thumb Canyon 129 samples are consistent with past results in the wider West Thumb area (Cuhel et al., 2002), the persistent and apparently reduced sulfur-stimulated dark CO2 fixation at 50–80°C in the West Thumb Canyon 129 vent fluids may indicate hitherto undiscovered communities that function deeply enough in the vent conduits to both completely consume electron donors and to cool to unresponsive conditions after collection by previous sampling technologies.
Caveats on linking vent fluid chemistry, chemosynthesis rates and community structure
In this study, linkages between the 16S rRNA sequence data and the chemosynthetic rates are indirect: the chemosynthetic bacteria whose activity is measured in the rates may appear in the clone libraries, but do not constitute them – the clone libraries contain other bacteria as well, and are subject to the methodological problems of DNA extraction, PCR and cloning bias. Vice versa, the dark CO2 fixation rates as measured (aerobic and thiosulfate-stimulated rates) provide a window on the overall chemosynthetic potential, but cannot be equated with it. Other chemosynthetic bacteria that require different testing conditions might be present, for example the phylotypes related to nitrate- and iron-reducing bacteria. Chemosynthesis rates and 16S rRNA sequence data could be linked more effectively by cultivation, quantification, and dark CO2 fixation rate measurements of key species, such as the Yellowstone Lake strains of Thiovirga and Sulfurihydrogenibium.
Chemosynthetic activity is not only determined by the presence or absence of inorganic electron donors, but also by the residence time of vent fluids, a function of admixture of sediment porewater or lake water. Although vent flow rates were not quantified in this study, efflux of water from vent orifices into the cold Yellowstone Lake water column was often clearly visible, and suggested that the residence time of vent fluids recovered by the sipper system might range from minutes to hours in the vicinity of the vent orifice. These turbulent mixtures of hot, reducing vent fluids and cold, oxygenated, oligotrophic Lake water would select for mesophilic, neutrophilic, and chemolithoautotrophic sulfide and hydrogen oxidizers with fast specific growth rates, exemplified by the genera Thiovirga (Ito et al., 2005), as detected here, and by Thiomicrospira (Jannasch et al., 1985; Brinkhoff et al., 1999). Within the very hot and chemically distinctive fluids, clones are more likely to reflect locally active populations than in bulk lake water. Here, high chemosynthesis rates may stimulate growth of heterotrophic consumers, who – depending on growth rate and abundance relative to the in situ chemosynthetic community – might escape phylogenetic detection. In vents with low content of reduced minerals, bacterial populations growing in the throat of the conduit may have consumed electron donors prior to sampling, yielding phylotypes in apparent absence of their energy source. Thus, the expected correlations between community structure and chemosynthetic rates will always be impacted by hydrothermal fluid concentration, temperature, fluid flow, and mixing rates; controlling for these variables over time requires long-term in situ chemical, physical, and microbiological monitoring efforts similar to those at deep-sea hydrothermal vents (Huber et al., 2002; Page et al., 2008).
Chemosynthetic contribution to Yellowstone Lake productivity
The contribution of microbial chemosynthesis to overall biological productivity of Yellowstone Lake is hard to quantify, due to a lack of spatially integrated data for dark CO2 fixation. Yet, chemosynthetic rate measurements at different sites in Yellowstone Lake since 1994 have consistently shown that maximum volume-based dark CO2 fixation rates of hydrothermal water samples, mostly from Mary Bay, are at least in the same order of magnitude (0.15–0.75 μM C h−1) as volume-based maximum photosynthesis rates at the lake surface (Figure 2; Aguilar et al., 2002). Further, chemosynthesis rates at specific sites can be significantly higher, such as the Mary Bay samples surveyed in 2003, and increase further with added reduced S. Some sections of the lake, for example the Mary Bay domain with its exceptionally high geothermal heatflow and abundance of active vents (Morgan et al., 2003b), are likely to have a substantial chemosynthetic contribution to total biomass production. This assumption is supported by observations of localized biological oases supported by Yellowstone Lake vents, for example extensive microbial mats covering sediments and macrophytes in Sedge Bay (Remsen et al., 2002; Konkol et al., 2010), or a warm DIC-rich vent in the West Thumb region that sustained an aquatic moss colony inhabited by crustaceans, segmented worms and flatworms (Lovalvo et al., 2010).
This study has shown that sublacustrine vents of Yellowstone Lake provide fascinating and novel opportunities to explore microbial community structure and function of chemosynthetically supported aquatic ecosystems. The combination of biogeochemical ecophysiology and phylogenetic analysis enables a deeper understanding of extremophilic microbiology in Yellowstone Lake hydrothermal vent habitats.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the National Science Foundation Research Experience for Undergraduates (REU, OCE 0097622), Life in Extreme Environments (LExEn, NSF-EAR 0085515), and Environmental Geochemistry and Biogeochemistry (EGB, NSF-EAR 9708501) programs awarded to Cuhel, Aguilar and Maki (Marquette University), and the Great Lakes WATER Institute at the University of Wisconsin – Milwaukee for supporting annual Yellowstone Lake expeditions and sampling opportunities. We truly appreciate David Lovalvo for extraordinarily skilled ROV operations. We are indebted to National Park Service leaders John Varley, John Lounsbury, and especially Todd Koel and the Fisheries and Aquatic Sciences Program and personnel YNP, WY for sampling access and accommodation of the project. Appreciation is extended to the 2003 REU undergraduates Jamie Becker, Erin Howard, and Lynn Killberg for their contribution to the extensive chemical analyses. Andreas Teske and Shawn Lyons were supported by the NASA Astrobiology Institute “Environmental Genomes.” Tingting Yang was supported by NSF grant No. 0727175.
References
- Aguilar C., Cuhel R. L., Klump J. V. (2002). “Porewater and hydrothermal vent water input into Yellowstone Lake, Wyoming,” in Yellowstone Lake: Hotbed of Chaos or Reservoir of Resilience? Proceedings of the 6th Biennial Scientific Conference on the Greater Yellowstone Ecosystem, eds Anderson R. J., Harmon D. (Yellowstone National Park, WY: Yellowstone Center for Resources and George Wright Society; ), 1–18 [Google Scholar]
- APHA (1992). Standard Methods for the Examination of Water and Wastewater, 18th Edn. Washington, DC: American Public Health Association [Google Scholar]
- Balk M., Altinbas W.M., Rijpstra I. C., Sinninghe Damste J. S., Stams A. J. M. (2008). Desulfatirhabdium butyrativorans gen. nov., sp. nov., a butyrate-oxidizing, sulfate-reducing bacterium isolated from an anaerobic bioreactor. Int. J. Syst. Evol. Microbiol. 58, 110–115 [DOI] [PubMed] [Google Scholar]
- Brinkhoff T., Muyzer G., Wirsen C. O., Küver J. (1999). Thiomicrospira chilensis sp. nov, a mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacterium isolated from a Thioploca mat. Int. J. Syst. Bacteriol. 49, 875–879 [DOI] [PubMed] [Google Scholar]
- Cuhel R. L., Aguilar C., Anderson P. D., Maki J. S., Paddock R. W., Remsen C. C., Klump J. V., Lovalvo D. (2002). “Underwater domains in Yellowstone Lake: hydrothermal geochemistry and bacterial chemosynthesis,” in Yellowstone Lake: Hotbed of Chaos or Reservoir of Resilience? Proceedings of the 6th Biennial Scientific Conference on the Greater Yellowstone Ecosystem, eds Anderson R. J., Harmon D. (Yellowstone National Park, WY: Yellowstone Center for Resources and George Wright Society), 27–53 [Google Scholar]
- Cuhel R. L., Aguilar C., Remsen C. C., Maki J. S., Lovalvo D., Klump J. V., Paddock R. W. (2004). The bridge bay spires: collection and preparation of a scientific specimen and museum piece. Yellowstone Sci. 12, 35–40 [Google Scholar]
- Hall P. O. J., Aller R. C. (1992). Rapid, small-volume, flow injection analysis for CO2 and NH4+ in marine, and freshwaters. Limnol. Oceanogr. 37, 1113–1119 [Google Scholar]
- Henry E. A., Devereux R., Maki J. S., Gilmour C. C., Woese C. R., Mandelco L., Schauder R., Remsen C. C., Mitchell R. (1994). Characterization of a new thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol. 161, 62–69 [PubMed] [Google Scholar]
- Holmfeldt K., Dziallas C., Titelman J., Pohlmann K., Grossart H.-P., Riemann L. (2009). Diversity and abundance of freshwater Actinobacteria along environmental gradients in the brackish northern Baltic Sea. Environ. Microbiol. 11, 2042–2054 [DOI] [PubMed] [Google Scholar]
- Huber J. A., Butterfield D. A., Baross J. A. (2002). Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P., Huber T. (2003). Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53, 289–293 [DOI] [PubMed] [Google Scholar]
- Ito T., Sugita K., Yumoto I., Nodasaka Y., Okabe S. (2005). Thiovirga sulfuroxydans gen. nov., sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium isolated from a microaerobic waste-water biofilm. Int. J. Syst. Evol. Microbiol. 55, 1059–1064 [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Wirsen C. O., Nelson D. C., Robertson L. A. (1985). Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35, 422–424 [Google Scholar]
- Kashefi K., Holmes D. E., Reysenbach A.-L., Lovley D. R. (2002). Use of Fe(III) as an electron acceptor to recover previously uncultured hyperthermophiles: isolation and characterization of Geothermobacterium ferrireducens gen. nov., sp. nov. Appl. Environ. Microbiol. 68, 1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump J. V., Remsen C. C., Kaster J. L. (1988). “The presence and potential impact of geothermal activity on the chemistry and biology of Yellowstone Lake, Wyoming,” in NOAA Symposium Series for Undersea Research, Vol. 6 (Rockville, MD: NOAA, Oceanic and Atmospheric Research, Office of Undersea Research), 81–97 [Google Scholar]
- Klump J. V., Remsen C. C., Lovalvo D., Anderson P. D., Cuhel R. L., Kaplinski M., Kaster J. L., Maki J. S., Paddock R. W. (1995). 20,000 Leagues under Yellowstone Lake: strangeness and beauty in the hidden deeps. Yellowstone Sci. 3, 10–14 [Google Scholar]
- Kodama Y., Watanabe K. (2004). Sulfuricurvum kujiense gen. nov. sp. nov., a facultatively anaerobic, chemolithoautotrophic sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54, 2297–2300 [DOI] [PubMed] [Google Scholar]
- Konkol N. R., Bruckner J. C., Aguilar C., Lovalvo D., Maki J. S. (2010). Dominance of epithytic filamentous Thiothrix spp. on an aquatic macrophyte in a hydrothermal vent flume in Sedge Bay, Yellowstone Lake, Wyoming. Microb. Ecol. 60, 528–538 [DOI] [PubMed] [Google Scholar]
- Lovalvo D., Clingelpeel S. R., McGinnis S., Macur R. E., Varley J. D., Inskeep W. P., Glime J., Nealson K., McDermott T. R. (2010). A geothermal-linked biological oasis in Yellowstone Lake, Yellowstone National Park, Wyoming. Geobiology 8, 327–336 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Hamady M., Knight R. (2006). UniFrac – an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7, 371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhu K., Buchner A., Lai T., Steppi S., Jobb G., Förster W., Brettske I., Gerber S., Ginhart A. W., Gross O., Grumann S., Hermann S., Jost R., König A., Liss T., Lüßmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., Schleifer K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. A., Shanks P., Lovalvo D., Pierce K., Lee G., Webring M., Stephenson W., Johnson S., Finn C., Schulze B., Harlan S. (2003a). The floor of Yellowstone Lake is anything but quiet. Yellowstone Sci. 11, 15–30 [Google Scholar]
- Morgan L. A., Shanks W. C., III, Lovalvo D. A., Johnson S. Y., Stephenson W. J., Pierce K. L., Harlan S. S., Finn C. A., Lee G., Webring M., Schulze B., Dühn J., Sweeney R., Balistrieri L. (2003b). Exploration and discovery in Yellowstone Lake: results from high-resolution sonar imaging, seismic reflection profiling, and submersible studies. J. Volcanol. Geoth. Res. 122, 221–242 [Google Scholar]
- Nakagawa S., Shtaih Z., Banta A., Beveridge T. J., Sako Y., Reysenbach A.-L. (2005). Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfur- oxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int. J. Syst. Evol. Microbiol. 55, 2263–2268 [DOI] [PubMed] [Google Scholar]
- Newton R. J., Jones S. E., Helmus M. R., McMahon K. D. (2007). Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl. Environ. Microbiol. 73, 7169–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A., Tivey M. K., Stakes D., Reysenbach A.-L. (2008). Temporal and spatial archaeal colonization of hydrothermal vent deposits. Environ. Microbiol. 10, 874–884 [DOI] [PubMed] [Google Scholar]
- Remsen C. C., Klump J. V., Kaster J. L., Paddock R. W., Anderson P. D. (1990). Hydrothermal springs and gas fumaroles in Yellowstone Lake, Yellowstone National Park, Wyoming. Natl. Geogr. Res. 6, 509–515 [Google Scholar]
- Remsen C. C., Maki J. S., Klump J. V., Aguilar C., Anderson P. D., Buchholz L., Cuhel R. L., Lovalvo D., Paddock R. W., Waples J., Bruckner J. C., Schroeder C. M. (2002). “Sublacustrine geothermal activity in Yellowstone Lake: studies past and present,” in Yellowstone Lake: Hotbed of Chaos or Reservoir of Resilience? Proceedings of the 6th Biennial Scientific Conference on the Greater Yellowstone Ecosystem, eds Anderson R. J., Harmon D. (Yellowstone National Park, WY: Yellowstone Center for Resources and George Wright Society), 192–212 [Google Scholar]
- Reysenbach A.-L., Hamamura N., Podar M., Griffiths E., Ferreira S., Hochstein R., Heidelberg J., Johnson J., Mead D., Pohorille A., Sarmiento M., Schweighofer K., Seshadri R., Voytek M. A. (2009). Complete and draft genome sequences of six members of the Aquificales. J. Bacteriol. 191, 1992–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford A. R., Cole J. R., Tiedje J. M. (2002). Characterization and description of Anaeromyxobacter dehalogenans gen. nov, sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear J. R., Walker J. J., McCollom T. M., Pace N. R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. U.S.A. 102, 2555–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. (2000). PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4. Sunderland, MA: Sinauer Associates [Google Scholar]
- Teske A., Hinrichs K.-U., Edgcomb V., de Vera Gomez A., Kysela D., Sylva S. P., Sogin M. L., Jannasch H. W. (2002). Microbial diversity in hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A., Wawer C., Muyzer G., Ramsing N. B. (1996). Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62, 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairavamurthy A., Mopper K. (1990). Determination of sulfite and thiosulfate in aqueous samples including anoxic seawater by liquid chromatography after derivatization with 2,2′-dithiobis (5-nitropyridine). Environ. Sci. Technol. 24, 333–337 [Google Scholar]
- Ward D. M., Ferris M. J., Nold S. C., Bateson M. M. (1998). A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62, 1353–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F., Amann R., Pernthaler J. (2004). Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6, 242–253 [DOI] [PubMed] [Google Scholar]
- Warnecke F., Sommaruga R., Sekar R., Hofer J. S., Pernthaler J. (2005). Abundances, identity, and growth state of actinobacteria in mountain lakes of different UV transparency. Appl. Environ. Microbiol. 71, 5551–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart G., Crump B., Kamst-van Agterveld M. P., Hagen F., Han S.-K. (2002). Typical freshwater water: an analysis of available 15S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28, 141–155 [Google Scholar]