SUMMARY
RfaH, a paralog of the general transcription factor NusG, is recruited to elongating RNA polymerase at specific regulatory sites. The X-ray structure of Escherichia coli RfaH reported here reveals two domains. The N-terminal domain displays high similarity to that of NusG. In contrast, the α-helical coiled-coil C domain, while retaining sequence similarity, is strikingly different from the β barrel of NusG. To our knowledge, such an all-β to all-α transition of the entire domain is the most extreme example of protein fold evolution known to date. Both N domains possess a vast hydrophobic cavity that is buried by the C domain in RfaH but is exposed in NusG. We propose that this cavity constitutes the RNA polymerase-binding site, which becomes unmasked in RfaH only upon sequence-specific binding to the nontemplate DNA strand that triggers domain dissociation. Finally, we argue that RfaH binds to the β′ subunit coiled coil, the major target site for the initiation σ factors.
INTRODUCTION
In bacteria, a single core RNA polymerase species (RNAP, subunit composition α2ββ′ω) transcribes all genes. RNAP is an amazingly efficient molecular motor that synthesizes RNA chains reaching tens of thousands of nucleotides in length; however, it has a limited ability to respond to environmental and cellular cues and is incapable of promoter-specific initiation. Numerous regulators bind to RNAP during initiation, elongation, and termination stages of transcription to adjust patterns of gene expression to changing conditions. The classic paradigm of regulation is σ competition, wherein multiple σ initiation factors compete for binding to core RNAP to direct it to a subset of bacterial promoters (Gruber and Gross, 2003). Following promoter escape, RNAP becomes a target for elongation and termination factors whose action can also lead to dramatic changes in gene expression (Mooney et al., 1998). Even though many elongation factors may be present in the cell at the same time, their binding to RNAP is not typically viewed as competitive, primarily because their effects on transcription are diverse and target sites on RNAP are unknown. The regulatory circuits are even more complex in eukaryotic cells where many more proteins compete for binding to three different RNAPs (Sims et al., 2004).
Gene duplication, followed by neofunctionalization (Koonin, 2005), allows the cell to expand the repertoire of regulators. For example, alternative σ factors that play major roles in reprogramming transcription are structurally related (Gruber and Gross, 2003) and the differences among them are largely limited to sequences/structures of their promoter targets, whereas their mechanisms of action, in recruiting RNAP to a promoter, are largely identical—such specialization can be achieved through minor changes in protein sequence. In contrast, more drastic alterations of the primary and tertiary structures may underlie diversification of other paralogs, such as RfaH and NusG, which have rapidly evolved to mediate different and in some instances opposite effects on gene expression (Bailey et al., 1997).
NusG and RfaH regulate transcriptional pausing and termination. Although they diverged rather recently (about the time of the branching of proteobacterial lineage), their functions are distinct, and only limited sequence identity remains (Artsimovitch and Landick, 2002). The extant NusG is likely similar to the common ancestor: NusG is present in all bacteria with the exception of a few intracellular parasites, and its rate of evolution is dramatically slower than that of RfaH. In E. coli, NusG is essential and acts as a sequence-independent elongation factor that both suppresses pausing and increases Rho-dependent termination. In contrast, RfaH is an operon-specific regulator, which suppresses polarity in long operons encoding virulence and fertility determinants (Bailey et al., 1997); RfaH is dispensable for cell viability in rich media, but it is essential for virulence in animal models (Nagy et al., 2006).
The key difference between NusG and RfaH is that RfaH action depends on the ops site in vivo and in vitro. The ops site is thought to play three roles during RfaH recruitment: it slows RNAP down to allow more time for RfaH recruitment (Artsimovitch and Landick, 2002; Leeds and Welch, 1997), mediates sequence-specific binding of RfaH to the nontemplate DNA strand exposed on the surface of the transcription elongation complex (TEC) (Artsimovitch and Landick, 2002), and induces TEC isomerization into a structurally distinct paused state (Artsimovitch and Landick, 2000) that may be necessary for productive recruitment of RfaH. Similarly to the phage λ antiterminators, following its recruitment RfaH presumably becomes an integrated part of the TEC and reduces pausing and termination to allow for complete synthesis of long RNA chains (Artsimovitch and Landick, 2002; Carter et al., 2004). While overlapping effects on transcription and common origin suggest similar mechanisms of action and targets on RNAP, the differences between RfaH and NusG that lead to different regulatory outcomes are particularly fascinating. To understand the transformation of a sequence-independent, essential transcription factor into a sequence-specific regulator, we determined the structure of RfaH and studied its molecular mechanism.
RESULTS AND DISCUSSION
Structure of the RfaH Protein
The structure of RfaH was determined by the single-anomalous wavelength diffraction (SAD) technique using the SeMet-substituted protein (Table 1). The atomic model (Figure 1) was built into the SAD electron density map and was then refined against the high-resolution (2.1 Å) data set obtained from the native crystals to the final R factor/Rfree = 0.238/0.273. The asymmetric unit contains two molecules of RfaH (see Figure S1 in the Supplemental Data available with this article online); each protomer consists of two domains (N and C terminal) connected by a 14 residue-long flexible linker that lacks electron density and thus appears to be disordered in the crystal (Figure 1A). The globular N domain (residues 2–100) possesses an α + β fold comprising the central four-stranded antiparallel β sheet flanked by two α helices on one side and one α helix on the other. A long hairpin loop (residues 31–52) inserts between the two β strands of the N domain; its conformation and orientation are likely stabilized through interactions with the C domain. The C domain (residues 115–156) consists of two long, antiparallel α helices that form a coiled coil (CC).
Table 1.
Data Collection and Refinement Statistics
| Space Group | P6522 | ||
| Unit cell parameters (Å) | a = b = 45.15, c = 600.16 | ||
| Synchrotron beamline | SERCAT (APS, Argonne, USA) | ||
| Data Collection | |||
| Anomalous scatter | None | Se | |
| Data set | Native | Peak | |
| Resolution (Å) | 50–2.10 (2.18–2.10)a | 50–2.40 (2.49–2.40) | |
| Reflections (unique) | 22,156 | 15,331 | |
| Redundancy | 4.7 (2.6) | 4.1 (2.9) | |
| I/σ(I) | 27.6 (2.4) | 13.7 (2.1) | |
| Rmerge (%) | 5.1 (49.3) | 7.1 (37.3) | |
| Completeness (%) | 93.8 (76.7) | 95.1 (89.9) | |
| Phasing Statistics | |||
| Space group | P6522 | ||
| Resolution (Å) | 40.0–3.0 | ||
| Number of Se atoms | 4 | ||
| Figure of merit | 0.47 | ||
| Refinement | Model Quality | ||
| Space group | P65 | Rmsd bond length (Å) | 0.018 |
| Resolution (Å) | 30.0–2.1 (2.18–2.10) | Rmsd bond angles (°) | 1.90 |
| Reflections used | 38,387 | Rmsd improper angles (°) | 1.06 |
| R factor (%) | 23.8 (33.2) | ||
| Rfree (%) | 27.3 (33.6) | Ramachadran Plot (Regions) | Number of Residues (%) |
| Most favorable | 85.6 | ||
| Overall B factor/rmsd (Å2) | 43.3/1.5 | Allowed | 14.4 |
| Number of protein atoms | 4512 | Generously allowed | 0.0 |
| Number of water molecules | 477 | Disallowed | 0.0 |
Rmerge = ΣhklΣj | Ij(hkl) − <I(hkl)> | / ΣhklΣj <I(hkl)>, where Ij(hkl) and <I(hkl)> are the intensity of measurement j and the mean intensity for the reflection with indices hkl, respectively. R factor, Rfree = Σhkl||Fcalc(hkl) | − |Fobs(hkl) || / Σhkl |Fobs|, where the crystallographic R factor is calculated including and excluding reflections in the refinement. The free reflections constituted 5% of the total number of reflections. Rmsd, root-mean-square deviation. I/σ(I), ratio of mean intensity to a mean standard deviation of intensity.
The data for the highest resolution shell are shown in parentheses.
Figure 1. Structure of the RfaH Protein.
(A) The overall structure.
(B) Stereo view of the RfaH interdomain interface. All structural figures were prepared using the programs MOLSCRIPT, BOBSCRIPT, and Raster3D (Esnouf, 1999; Kraulis, 1991; Merrit and Bacon, 1997).
Both the domain architecture of RfaH and the positioning of two acidic residues at the tip of the C domain (Glu132 and Asp134) are reminiscent of several transcriptional regulators (GreA, GreB, Gfh1, and DksA), in which similarly positioned conserved acidic residues are thought to stabilize Mg2+ ions near the active site, thereby altering RNAP catalytic properties (Lamour et al., 2006; Laptenko et al., 2003, 2006; Opalka et al., 2003; Perederina et al., 2004; Sosunova et al., 2003; Symersky et al., 2006). Similarly to RfaH, Gre factors reduce transcriptional pausing (Marr and Roberts, 2000). However, the C domain of RfaH is packed against the globular N domain along its entire length, whereas the CC domains in the Gre-like proteins make only a few interactions with, and protrude away from, their globular domains. Underscoring these differences, substitutions of Glu132 and/or Asp134 for alanine do not alter RfaH effects on transcription in vitro and, unlike all other CC regulators, RfaH is not cleaved by Fe2+ bound in place of the catalytic Mg2+ ion in an assay that is routinely used to detect protein proximity to the active site (data not shown).
In the crystal structure, the C domain acts as a cap that closes over the vast hydrophobic cavity on the surface of the N domain. The interdomain interface is highly (>80%) nonpolar; nine hydrophobic residues donated by both α helices of the C domain complement the 14 hydrophobic side chains lining the cavity of the N domain to form an extensive hydrophobic core (Figure 1B). Arg138, the only polar residue in the C domain involved in interdomain interactions, makes numerous interactions that are likely essential for the association of the RfaH domains: its methylene backbone contributes to the predominantly hydrophobic interdomain interactions, whereas its guanidinium group forms multiple hydrogen bonds with the main-chain oxygens from both domains, as well as with the side chain of Glu48 from the N domain (Figure 1B).
Structures of RfaH and NusG: Similarities and Differences
RfaH and NusG share low but statistically significant sequence similarity throughout their polypeptide chains (Artsimovitch and Landick, 2002), and sequence identity is about the same in both domains (17%; Figure 2). Three NusG structures (Knowlton et al., 2003; Reay et al., 2004; Steiner et al., 2002) show a common two-domain architecture, with both domains potentially capable of making contacts to RNAP (Li et al., 1992), Rho (Li et al., 1993), and nucleic acids (Nehrke and Platt, 1994; Steiner et al., 2002). Perhaps not surprisingly, the structure of the RfaH N domain closely resembles (rmsd = 1.8 Å over the 82 Cα positions; Figure 2A) that of the Aquifex aeolicus NusG (Steiner et al., 2002). In A. aeolicus NusG, a β sandwich domain is inserted in place of the β hairpin. The β sandwich insert is, however, absent in most species; and in E. coli NusG (Steiner et al., 2002) this segment was modeled in a conformation similar to that reported here for RfaH. The overall structural similarity between RfaH and NusG might be extended to the long flexible linkers connecting the C and N domains.
Figure 2. Comparison of the RfaH and NusG Structures.
(A) Superposition of the RfaH and NusG N domains.
(B) The overall structure of the NusG C domain.
(C) Sequence alignment of the RfaH and NusG proteins built with DNA Star; the bar height indicates residue conservation. The conserved hydrophobic residues are highlighted in yellow; those lining the hydrophobic cavities of the N domains are additionally boxed. The secondary structure elements in A. aeolicus NusG and E. coli RfaH are shown above and below the alignment, respectively, and colored as in (A) and (B). The vertical black arrow indicates the position of a large insertion in A. aeolicus NusG.
In contrast, the structures of the C domains are strikingly distinct: the NusG C domain consists exclusively of β strands that fold into a compact globular four-stranded β barrel (SH3 domain-like, KOW motif, Figure 2B), while the C domain of RfaH forms an α-helical CC. In solution, the NusG globular C domain, whose fold is dictated by the residues forming its hydrophobic core, should be more stable than the RfaH C domain, which is stabilized by a mixture of the polar and hydrophobic interactions distributed along the entire length of the α helices and possesses substantially larger solvent-exposed surface rich in hydrophobic side chains. One might expect that RfaH forms a CC because key residues in the hydrophobic core of the NusG C domain have been exchanged during evolution for the polar/charged ones in RfaH. Surprisingly, however, this did not occur: all key hydrophobic residues are highly conserved between NusG and RfaH (Figure 2C). Moreover, homology modeling demonstrated that the RfaH C-domain sequence could be easily integrated (as is, without energy minimization) into the NusG β barrel structure, while maintaining the hydrophobic core and producing neither steric clashes nor unfavorable contacts (Figure S2). The change in the domain fold observed in RfaH rearranges many of these conserved hydrophobic residues from the core of the ancestral β barrel onto the surface of the α-helical CC where they become available for interactions with the hydrophobic cleft of the N domain (Figure 1B).
From Structure to Function I: RfaH Activation
RfaH likely contains at least two separate binding sites: one to mediate sequence-specific recognition of the non-template DNA at ops site, another to maintain interactions with RNAP following the escape from the ops site. However, in the crystal structure, RfaH protomer lacks any cavities and/or grooves that might accommodate the nucleotide bases of the nontemplate strand. The groove at the interface of the monomers in the crystal (Figure S1) initially suggested that dimer might be the functional oligomeric state of RfaH. However, both the gel filtration analysis and the absence of specific crosslinking of the cysteine side chains introduced at position 32 (in place of a methionine residue located at the center of dimer symmetry) indicate that RfaH is a monomer in solution (data not shown).
In spite of their functional differences, RfaH and NusG both increase the rate of elongation (Artsimovitch and Landick, 2002; Burova et al., 1995) and compete following their recruitment to the TEC in vitro (I.A., V.S., R. Landick, and R. Mooney, unpublished data), suggesting that RfaH and NusG utilize their structurally similar N domains to bind to the same site on RNAP. However, their limited sequence identity suggests that binding to RNAP is largely determined by van der Waals interactions, for which the hydrophobic nature, rather than the identity of the side chains, is of central importance. This hypothesis predicts that the best candidate for a conserved RNAP-binding site on NusG and RfaH is the large hydrophobic cavity found in the N domain of both proteins. In RfaH, this cavity is masked by interactions with the C domain (Figure 1B), whereas it is completely exposed in one of the crystal forms of NusG (in which the C domain is flexible) and only partially protected in the second crystal form by the hydrophobic residues from the C domain of the symmetry-related molecule (Knowlton et al., 2003; Steiner et al., 2002).
The latter observation suggested a hypothetical “spring-loaded” state for NusG (Knowlton et al., 2003) that is analogous to the inactive state of the RfaH protomer (see below). However, analysis of the RfaH structure indicates that interface buried between the C and N domains is vast (~1800 Å2) and, more importantly, is more than 80% hydrophobic (see also Figure 1B), whereas the interface in the spring-loaded state of NusG (~1000 Å2; Knowlton et al., 2003) is less than 55% hydrophobic, casting doubt on the existence of the spring-loaded state in solution in the absence of the crystal-packing forces. Indeed, the intramolecular interaction of the NusG domains (as opposed to the domain-swapped dimer) has not been demonstrated by Knowlton et al., 2003, whereas the solution structure and subsequent biophysical analysis of Thermus thermophilus NusG failed to reveal interdomain interactions (Reay et al., 2004). Thus, existence of any other than observed extended conformation of NusG still awaits a rigorous experimental demonstration (see the Supplemental Data for the detailed discussion).
Altogether, these structural considerations lead to several testable predictions. First, upon binding to its target in the TEC, the domains of RfaH must dissociate to expose the putative RNAP-binding site located on the open hydrophobic cavity of the N domain. Second, the N domain alone should confer the ability to accelerate transcription, a property that RfaH and NusG share. Third, if binding to the ops DNA is required solely to trigger the domain opening, the N domain in isolation should act in a sequence-independent fashion. Below we show that these predictions are fulfilled.
Domain Opening Is Required for RfaH Action
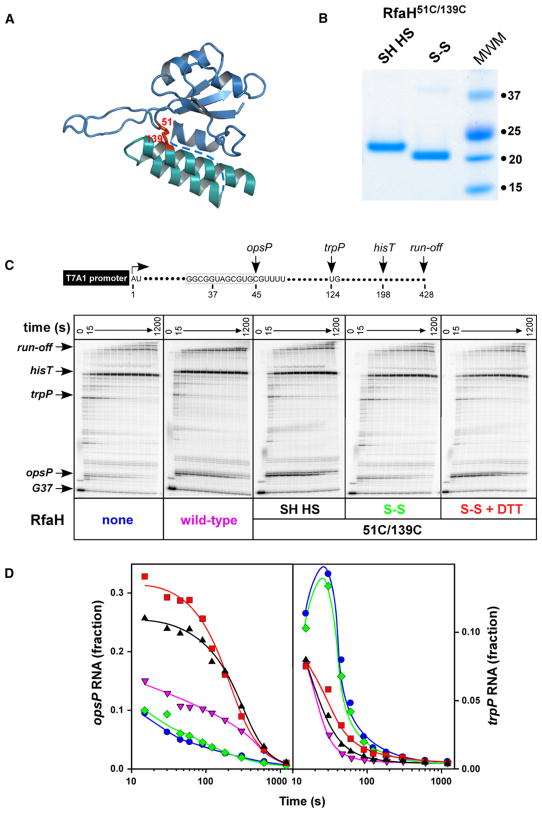
To test the first prediction, we engineered a covalent bond at the interface between the N and C domains. We introduced two cysteine residues in place of Phe51 and Ser139 (Figure 3A); the altered RfaH remained transcriptionally active in reducing conditions (see below). Formation of an S-S covalent bond between these two residues should not only prevent the domain opening but also change protein mobility in a denaturing gel, as the molecule will not be able to unfold completely. Indeed, upon oxidation the mobility of RfaH51C,139C was increased (Figure 3B), and the protein appeared fully crosslinked. We then tested crosslinked RfaH51C,139C in a standard single-round transcription assay (Figure 3C) using the template with a strong T7A1 promoter followed by an initial transcribed region that allows for formation of radiolabeled TECs stalled after incorporation of a G residue at position 37 (G37) when transcription is initiated in the absence of UTP. The template also encodes the ops pause (opsP) signal, the trp pause site (trpP), and the his terminator (hisT). On ops-containing templates RfaH increases RNAP pausing at the ops site (at C45 position on a template shown in Figure 3) but accelerates transcription downstream, reducing both the efficiency and the longevity of pauses (Artsimovitch and Landick, 2002). The delay at the ops site is the consequence rather than the requirement for RfaH recruitment: both the RNAP that is delayed at C45 and the one that is not are accelerated by RfaH. Pausing at the ops site is an excellent reporter of RfaH recruitment and is used throughout the manuscript largely as a qualitative parameter.
Figure 3. Domain Opening Is Required for RfaH Function.
(A) A disulfide bond engineered between the N- and C-domain residues 51 and 139; the interdomain linker absent from the structure is shown by a dashed line.
(B) A 12% denaturing protein gel loaded with reduced (left) and oxidized (right) forms of RfaH51C,139C (see the Experimental Procedures for details); the sizes of molecular weight markers are shown on the right.
(C) Transcriptional phenotypes of RfaH51C,139C. (Top) Transcript generated from the T7A1 promoter on a linear pIA449 template; transcription start site (+1), positions of ops and trp pause sites, his terminator, and run-off are indicated. (Bottom) Halted radiolabeled G37 TECs were preincubated with RfaH51C,139 in reduced (SH HS), oxidized (S-S), oxidized state reduced with DTT (S-S + DTT), or storage buffer (none) for 5 min at 37°C, and then challenged with rifampicin at 25 μg/ml and NTPs (10 μM GTP and 150 μM ATP, CTP, and UTP). Aliquots were withdrawn at times ranging from 15 to 1200 s, followed by the 200 μM GTP chase for 300 s, and analyzed on 8% denaturing gels.
(D) Quantification of the fraction of RNA at the ops (transcript position C45, left) and the trp (transcript position U124, right) pause sites from the data shown in (C). Fraction refers to the ratio of product at pause site to the sum of all products resulting from elongation of TEC halted at G37. The symbols are color-coded as in (C).
Upon addition of all four NTP substrates and rifapentin (to block reinitiation), RNAP elongated the nascent RNA at a characteristic rate, pausing at the opsP and the trpP sites (Figure 3C). Similarly to the wild-type RfaH, RfaH51C,139C in a reduced form (SH HS) significantly delayed the escape of RNAP from the ops site and reduced pausing at the downstream trpP site. In contrast, the crosslinked RfaH51C,139C (S-S) did not affect RNAP pausing at either site; this defect was fully reversed upon addition of dithiothreitol.
These results, represented graphically in Figure 3D, demonstrate that the covalent bond which effectively locks RfaH in a closed state interferes with its function. Thus, we conclude that domain dissociation is a necessary component of the RfaH mechanism. Given the apparently specific and stable association of the RfaH domains, the free energy cost of their dissociation would have to be offset by forming interactions with the TEC. Sequence-specific recognition of the nontemplate DNA strand (an essential feature of the RfaH, but not the NusG, mechanism) might play such a role.
The N Domain Retains Full Activity but Lacks Sequence Specificity of RfaH
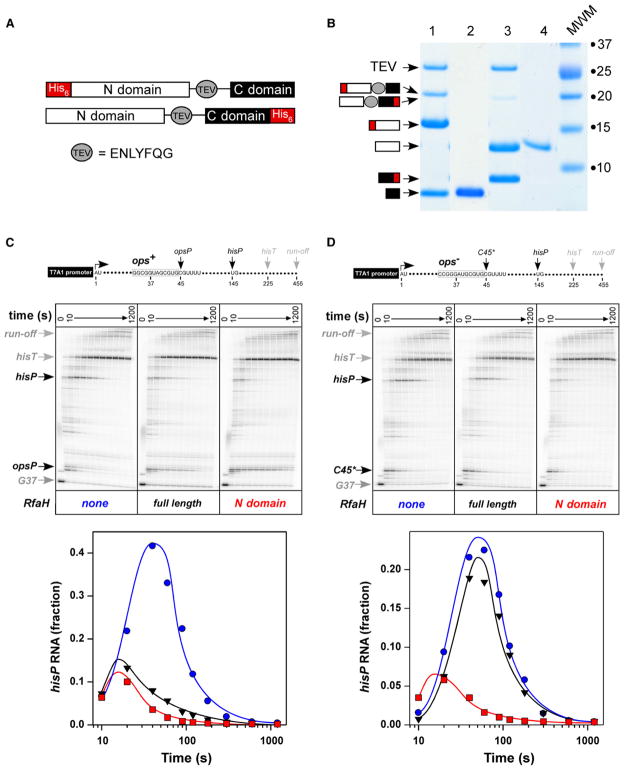
When expressed separately, the RfaH N domain was insoluble. To circumvent this problem, we introduced a TEV protease cleavage site into the linker between the N and C domains. We made two versions of N-TEV-C constructs with the His6 tag either at the N- or the C-terminal end of RfaH (Figure 4A); both remained fully active (data not shown). We then utilized the His6-tagged TEV protease to cleave the purified proteins and removed both the protease and one of the domains by absorption to the Ni-Sepharose resin (Figure 4B). The C domain by itself was transcriptionally inert (data not shown), whereas the N domain acted similarly to the full-length RfaH on an ops-containing template (Figure 4C): both proteins delayed RNAP at the opsP site and reduced pausing at the hisP site; the pause efficiency (estimated from semilogarithmic plots of escape kinetics; Landick et al., 1996) was increased by 3-fold at opsP (Figure S4) and reduced by 3- to 4-fold at hisP (from ~0.8 to ~0.2; Figure 4C, bottom).
Figure 4. The N Domain Mediates the RfaH Effect on Elongation.
(A) The pIA750-encoded protein designed to purify the C domain (black rectangle) of RfaH contained the TEV protease site (gray oval) in the linker between the two domains and the His6 tag (red rectangle) fused to the N domain, whereas pIA777-encoded protein that was used to purify the N domain (white rectangle) had His6 tag fused to the C domain.
(B) SDS-PAGE gel of 750 and 777 proteins after TEV protease digestion (1 and 3, respectively) and absorption to Ni-Sepharose (2 and 4, respectively). The digested products are presented schematically as in (A).
(C and D) In vitro transcription assays were performed similarly to those described in the Figure 3 legend on templates containing canonical (C) or scrambled (D) opsP pause sites (Artsimovitch and Landick, 2002) and a hairpin-dependent pause site hisP in the absence of RfaH (left panels), in the presence of full-length RfaH (middle panels) and purified N domain (right panels). Quantification of the fraction of RNA at the hisP site is shown below with the symbols color-coded to match labels for each gel panel; quantification of the opsP RNA is shown in Figure S4.
We next tested whether the ops element was still required for the N-domain action using a template with the “scrambled” ops site, which induces pausing similarly to the canonical ops element but fails to respond to RfaH (Artsimovitch and Landick, 2002), followed by the hisP site (Figure 4D). On this template, neither the N domain nor the full-length RfaH delayed RNAP at the C45 site, positioned similarly to the opsP site (Figure S4); yet, in stark contrast to the full-length RfaH, the N domain was fully active in reducing pausing at the hisP site (by 4-fold; Figure 4D, bottom). Similar effect was observed on other ops-less templates (data not shown). We conclude that the ops sequence is required during the initial recruitment of RfaH and becomes dispensable once the N and C domains are separated, allowing for the stable binding of RfaH to the TEC.
From Structure to Function II: RfaH Target Site on RNAP
These results support a model in which an exposed hydrophobic cavity on RfaH mediates its binding to RNAP. Where is the RfaH-binding site on RNAP? Such a binding site should satisfy three major criteria. First, it should be located on a protruding (or at least convex) structural segment. Second, it should be positioned near the nontemplate strand to allow for RfaH binding to the ops element. Third, it should be abundant in the hydrophobic side chains to complement those in the RfaH hydrophobic cleft.
The N-terminal CC of the β′ subunit (β′CC, E. coli residues 265–310), which is thought to constitute the major binding site for the initiation σ factors (Arthur et al., 2000), meets all these criteria. First, the β′CC protrudes from the RNAP surface and thus might be inserted into the RfaH cavity. Second, in the RNAP holoenzyme structure (Vassylyev et al., 2002) σ regions 2.1- to 2.4-fold around β′CC and σ2.4 binds specifically to the nontemplate strand of the −10 promoter element (Marr and Roberts, 1997), placing β′CC near the unwound nontemplate strand. Finally, the tip of β′CC contains a set of highly conserved exposed hydrophobic residues (Figure S3).
If this hypothesis were correct, disrupting the hydrophobic interactions between RfaH and β′CC should abolish RfaH effects on transcription. Assuming that the modes of binding of the σ factors (Vassylyev et al., 2002) and RfaH to the β′CC are likely distinct, we designed a deletion-substitution variant of the E. coli RNAP in which an 11 residue segment at the β′CC tip was substituted by Gly4 linker (Figure 5). As predicted, the altered RNAP was transcriptionally competent and did not behave any differently from the wild-type enzyme in all the elongation assays tested (Figure 5 and data not shown). However, this enzyme completely lost the ability to respond to either full-length RfaH or the N domain alone. We conclude that the β′CC likely constitutes the major binding site for RfaH on RNAP and that this interaction is necessary for RfaH action even if the ops-mediated recruitment is bypassed.
Figure 5. The Tip of the β′CC Is Required for RfaH Action.
In vitro analysis of transcript elongation by the altered RNAP enzyme, in which the residues within the gray box were substituted with four glycines, on the template shown in Figure 4C. The assays were done in the absence of RfaH (left panel), in the presence of full-length RfaH (middle panel), and with the purified N domain (right panel). Quantification and comparison with the wild-type RNAP are presented in Figure S5.
We argue that the elongation factor RfaH (and its paralog NusG) binds to the N-terminal β′CC, the major target site for the σ factors; competition for this site between elongation factors and σ may favor σ release during transition to elongation (Mooney et al., 2005). On the other hand, reports that σ may bind to the TEC at sites where the nontemplate strand exhibits similarity with the promoter −10 region (Mooney and Landick, 2003; Roberts et al., 1998) suggest that the σ factors may play a role in regulation of transcript elongation in a similar fashion to that of RfaH/NusG.
Modeling of the TEC/RfaH Complex
We next constructed a model of RfaH recruited to the TEC (Figure 6A). First, the downstream DNA and the RNA/DNA hybrid from the yeast TEC crystal structure (Kettenberger et al., 2004) were built into the RNAP core enzyme extracted from the T. thermophilus holoenzyme (Vassylyev et al., 2002) upon simple superposition of the protein structures. We next added the nontemplate strand and the upstream double-stranded DNA to the model to complete the transcription bubble. Though this portion of the model was built ab initio and thus by no means represents the detailed and precise atomic model, its building was substantially restrained through the feasible location of the nucleic acids in the bubble, the fixed length of the non-template strand, and the requirement to maintain protein-nucleic acid contacts while avoiding steric hindrance. The present model provides a good estimate of the spatial arrangement of all the TEC components and agrees well with the semiexperimental TEC model (Korzheva et al., 2000), and therefore was used as a reference for modeling of the TEC/RfaH complex.
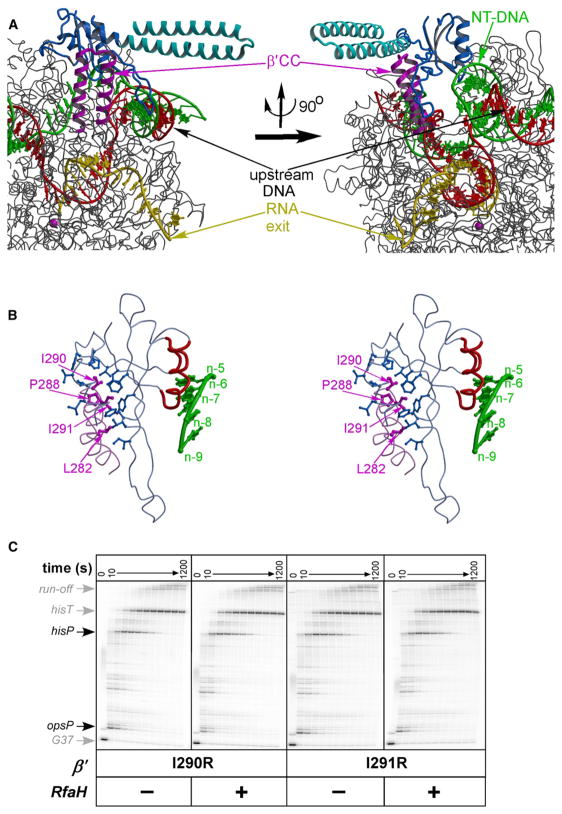
Figure 6. The Model of RfaH Recruited to the TEC.
(A) Overall model is presented in two distinct orientations. The RNAP is shown in gray with the β′CC (the RfaH-binding site) highlighted in magenta. The color scheme for the RfaH N and C domains is the same as in Figure 1. The RNAP active site is marked by the Mg2+ ion (magenta sphere). The template DNA, the nontemplate DNA, and the RNA transcript are colored in red, green, and yellow, respectively.
(B) The close-up stereo view of RfaH bound to the β′CC. The side chains forming the intermolecular hydrophobic core are represented by the balls-and-sticks model (blue and magenta for the RfaH and β′CC, respectively). The nontemplate strand nucleotides adjacent to RfaH in the TEC model are shown in green and are numbered with respect to the position of the acceptor template base (register n). The cluster of the RfaH residues that confer defects in the ops DNA binding is colored in red.
(C) Transcript elongation on pIA349 template (Figure 4C) by RNAPs containing I290R or I291R substitutions at the tip of the β′CC in the absence (left panels) or in the presence (right panels) of full-length RfaH. Quantification is presented in Figure S6.
The following criteria were applied to the model building of RfaH bound to the TEC: first, the N and C domains should be separated. Second, the tip of the β′CC should be inserted into the hydrophobic cavity of the RfaH N domain. Third, the model should possess the most extensive interaction surface between RfaH and RNAP while avoiding any steric clashes. Fourth, no polar side chains should appear within the hydrophobic core formed at the RfaH/RNAP interface. We first carried out manual docking of the N domain to the β′CC that resulted in a unique model matching all of the above criteria. The C domain was then added through extension of the N-domain C-terminal α helix, with the interdomain linker (disordered in the crystals) modeled in an α-helical conformation. This modeling indicates that the C domain may extend from the complex surface forming no interactions with RNAP and/ or nucleic acids, in agreement with our finding that the isolated N domain is fully active (Figure 4).
This model is built from individual parts coming from different systems (nucleic acids from yeast TEC, T. thermophilus RNAP, and E. coli RfaH) using rigid-body docking and obviously does not reflect the fine details of the RfaH/TEC contacts; however, its feasibility was additionally justified through the model-guided biochemical experiments. First, among all hydrophobic residues at the β′CC tip buried inside the hydrophobic cleft of the RfaH N domain in the model, two highly conserved residues, Ile290 and Ile291 (Figure 6B and Figure S3), are located in the middle of the N-domain cavity and make multiple van der Waals contacts with RfaH hydrophobic side chains. Single substitution of each of these residues for arginine completely abolished response to RfaH in vitro (Figure 6C and Figure S5). In contrast, nonconservative substitutions of Leu283, Arg275, Arg278, and Arg293 (which make no contacts to RfaH in the model) had no effect on RfaH function (I.A. and G.A.B., unpublished data). Second, there is a cluster of residues located on the N-domain surface opposite of the hydrophobic cavity and near the nontemplate DNA strand in the model (Figure 6B); substitutions of these residues for alanine impair RfaH ability to bind to the nontemplate DNA (as inferred from the loss of RNAP delay at the ops site; I.A., G.A.B., and V.S., unpublished data). It is worth noting that although these residues are likely able to interact with the nontemplate strand backbone, they alone seem unlikely to constitute a sequence-specific binding site for nucleotides, given the absence of cavities/grooves that might accommodate the DNA bases. We therefore conjecture that the specific recognition of the ops element occurs in a site formed by RfaH and RNAP in a concerted manner.
Mechanism of the RfaH Action
Our data suggest the following mechanism. RfaH (presumably together with RNAP) recognizes and binds to the nontemplate DNA in the ops-paused TEC. Interactions with TEC likely induce some structural alterations in RfaH that facilitate domain dissociation. This step likely represents an RfaH-specific innovation compared to NusG, in which the N-domain hydrophobic cavity cannot be stably closed by the C-terminal β barrel and whose only populated state is an extended monomer (see above). Persistent extended (active) state versus conditional domain opening could then be the major difference between these homologous general (NusG) and operon-specific (RfaH) regulators. The subsequent exposure of the hydrophobic cavity of the N domain allows RfaH to form a stable complex with RNAP using β′CC as an anchor. Escape of the RfaH-modified TEC from the ops site is accompanied by the loss of the sequence-specific contacts with the non-template DNA; nonspecific protein/DNA contacts might still persist during transcription of the RfaH-controlled operons that can reach tens of thousands of nucleotides in length (Bailey et al., 1997). Interestingly, Carol Gross and coauthors showed that an interaction between β′CC and σ region 2.2 induces specific binding of σ70 to the nontemplate DNA (Young et al., 2001). While the direction of a σ-mediated allosteric signal is reversed as compared to that of RfaH, both findings highlight the functional importance of allosteric isomerization during recruitment of the regulators to the transcription complex.
The present mechanistic model suggests two possible pathways of RfaH-mediated acceleration of elongation. In one scenario, interactions with the nontemplate strand and/or with the adjacent domains of RNAP might trigger an allosteric signal propagating to the active site to optimize its configuration, thereby facilitating the catalytic reaction. RfaH may contact the β subunit domain (E. coli residues 456–508) that has direct connections to the rifamycin-binding site, a proposed origin of the allosteric signal (albeit inhibiting rather than facilitating transcription) transmitted to the active center (Artsimovitch and Vassylyev, 2006). In another scenario, RfaH, which binds to the TEC near the first base pair of the upstream DNA duplex, may directly facilitate DNA annealing, thereby favoring forward DNA translocation and/or stabilizing the posttranslocated state. Which of these (or some other, yet unforeseen) mechanisms are correct remains to be elucidated.
Evolutionary Transformation
We show that evolutionary transition from a NusG-like common ancestor to the specialized regulator RfaH was accompanied by a dramatic structural change of the C domain, whereas the N-domain fold remained intact. Due to significant sequence similarity in the C domains of NusG and RfaH and a clear presence of KOW motif in both, it was puzzling to find an all-helical conformation of a sequence that is expected to fold as a β barrel. Quite a few instances of structural changes between homologs have been described, and some include transformations of α helices to β strands (Cordes et al., 2003; de Chiara et al., 2005; Ikeda and Higo, 2003; Tidow et al., 2004; Yang et al., 1998). However, in all reported examples, these changes involve only parts of otherwise structurally similar domains, and RfaH/NusG is the first known to us case of all-β to all-α transition of the complete domain structure. We hypothesize that an RfaH ancestor developed a conformationally dual “chameleon” sequence (Andreeva and Murzin, 2006) in the C domain, probably because its functional role was changing, and such sequence could fold either as a β barrel or as a CC. It is possible that through subsequent accumulation of mutations, RfaH C domain has lost the chameleon sequence and exists either as α helices (while interacting with the N domain) or in an unfolded free state. However, it is tempting to speculate that this domain is still able to fold into a β barrel and can exist in two drastically different states; moreover, this conformational switch may have a functional relevance. For instance, the helical conformation may be blocking the hydrophobic cavity of the N-terminal domain, but upon the domain dissociation, the C domain adopts a sheet conformation and is able to bind to other target molecules.
The conserved fold of the N domain is consistent with our findings that this domain alone is sufficient to accelerate transcription in vitro in an ops-independent fashion, an effect that is common to RfaH and NusG. In contrast, the distinct C domains may mediate the protein-specific functions: contacts with Rho for NusG (Li et al., 1993), interactions with the translation/secretion machineries for RfaH (Bailey et al., 2000). In addition, the C domain apparently plays two unique roles in RfaH. First, it closes the extensive hydrophobic cavity of the N domain, thereby protecting the protein from aggregation. Second, it indirectly confers sequence specificity: the required for activation dissociation of the C domain triggered by the ops binding restricts RfaH effect to only a few targets—a key regulatory property that distinguishes RfaH from NusG.
EXPERIMENTAL PROCEDURES
Crystallization and Data Collection
Crystallization was carried out for the native protein and its SeMet derivative as described previously (Vassylyeva et al., 2006). The crystals belong to the P6522 space group, with unit cell dimensions a = b = 45.15 and c = 600.16 Å. The complete 2.1 Å resolution data for the native protein crystals and the 2.4 Å resolution MAD data sets for the Se-Met-derivatized RfaH crystals were collected at the three wavelengths (peak, edge, and remote) near the absorption edge for the Se atom at 100K at the SERCAT beamline at the Advanced Photon Source (Argonne, USA) using MAR-300 CCD detector. The data were processed using the HKL2000 program package (Otwinowski and Minor, 1997) (Table 1). Structure determination and refinement are described in the Supplemental Experimental Procedures.
Plasmids, Proteins, and Reagents
Plasmids, proteins, and reagents are described in the Supplemental Experimental Procedures.
Protein Crosslinking
The closure of the interdomain disulfide was accomplished by incubating 10 μM RfaH51C,139C with 2 mM CuCl2 and 6 mM 1,10-phenanthroline in 50 mM Tris-HCl (pH 6.9), 250 mM NaCl, 8% glycerol, and 50 μM EDTA for 1 hr at 20°C. The oxidized protein was supplemented with 10 mM EDTA and dialyzed against the storage buffer.
In Vitro Transcription Assays
Linear DNA template generated by PCR amplification (30 nM), holo RNAP (40 nM), ApU (100 μM), and starting NTP subsets (1 μM CTP, 5 μM ATP and GTP, 10 μCi [α32P]CTP, 3000 Ci/mmol) were mixed in 100 μl of TGA buffer (20 mM Tris-HCl, 2 mM MgCl2, 20 mM NaCl, 5% glycerol, and 0.1 mM EDTA [pH 7.9] for assays in Figure 3; 20 mM Tris-HCl, 14 mM MgCl2, 20 mM NaCl, 5% glycerol, 1 mM DTT, and 0.1 mM EDTA [pH 7.9] for assays in Figures 4 and 6C; 20 mM Tris-acetate, 20 mM Na-acetate, 2 mM Mg-acetate, 5% glycerol, 1 mM DTT, and 0.1 mM EDTA [pH 7.9] for assays in Figure 5 and Figure S5). Reactions were incubated for 15 min at 37°C. RfaH was added to 50 nM where indicated, and transcription was restarted by addition of nucleotides (10 μM GTP and 150 μM ATP, CTP, and UTP) and rifapentin to 25 μg/ml at 37°C. Samples were removed at times indicated in the figures (in a range from 10 to 1200 s) and after a final 5 min incubation with 200 μM GTP. Sample analysis and quantification were as described in Landick et al. (1996).
Supplemental Data
Supplemental Data include six figures, Supplemental Experimental Procedures, Supplemental Discussion, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/26/1/117/DC1/.
Supplementary Material
Acknowledgments
We thank Dr. A. Perederina for assistance in protein crystallization and data collection, and Dr. R. Saecker for critical reading of the manuscript and stimulating discussions. This work was supported in part by the NIH grants GM74252 and GM74840 (to D.G.V.), GM67153 and AI064819 (to I.A.), and GM67165 (to N.V.G.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research under contract number W-31-109-Eng-38.
Footnotes
Accession Numbers
The PDB ID for the structure of E. coli RfaH reported in this paper is 2OUG.
References
- Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Arthur TM, Anthony LC, Burgess RR. Mutational analysis of beta′260–309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. J Biol Chem. 2000;275:23113–23119. doi: 10.1074/jbc.M002040200. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Vassylyev DG. Is it easy to stop RNA polymerase? Cell Cycle. 2006;5:399–404. doi: 10.4161/cc.5.4.2466. [DOI] [PubMed] [Google Scholar]
- Bailey M, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Hughes C, Koronakis V. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol Gen Genet. 2000;262:1052–1059. doi: 10.1007/pl00008648. [DOI] [PubMed] [Google Scholar]
- Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HD, Svetlov V, Artsimovitch I. Highly divergent RfaH orthologs from pathogenic proteobacteria can substitute for Escherichia coli RfaH both in vivo and in vitro. J Bacteriol. 2004;186:2829–2840. doi: 10.1128/JB.186.9.2829-2840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes MH, Walsh NP, McKnight CJ, Sauer RT. Solution structure of switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon. J Mol Biol. 2003;326:899–909. doi: 10.1016/s0022-2836(02)01425-0. [DOI] [PubMed] [Google Scholar]
- de Chiara C, Menon RP, Adinolfi S, de Boer J, Ktistaki E, Kelly G, Calder L, Kioussis D, Pastore A. The AXH domain adopts alternative folds: the solution structure of HBP1 AXH. Structure. 2005;13:743–753. doi: 10.1016/j.str.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Higo J. Free-energy landscape of a chameleon sequence in explicit water and its inherent alpha/beta bifacial property. Protein Sci. 2003;12:2542–2548. doi: 10.1110/ps.03143803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Lamour V, Hogan BP, Erie DA, Darst SA. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J Mol Biol. 2006;356:179–188. doi: 10.1016/j.jmb.2005.10.083. [DOI] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan C. Quantitative analysis of transcriptional pausing by RNA polymerase: the his leader pause site as a paradigm. Methods Enzymol. 1996;274:334–352. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, Berenguer J, Kong XP, Borukhov S. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25:2131–2141. doi: 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds JA, Welch RA. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J Bacteriol. 1997;179:3519–3527. doi: 10.1128/jb.179.11.3519-3527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and anti-termination of transcription. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- Merrit EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Mooney RA, Landick R. Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev. 2003;17:2839–2851. doi: 10.1101/gad.1142203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L, Hinton JC, Hacker J. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun. 2006;74:5914–5925. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke KW, Platt T. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J Mol Biol. 1994;243:830–839. doi: 10.1006/jmbi.1994.1685. [DOI] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Reay P, Yamasaki K, Terada T, Kuramitsu S, Shirouzu M, Yokoyama S. Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins. 2004;56:40–51. doi: 10.1002/prot.20054. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky J, Perederina A, Vassylyeva MN, Svetlov V, Artsimovitch I, Vassylyev DG. Regulation through the RNA polymerase secondary channel. Structural and functional variability of the coiled-coil transcription factors. J Biol Chem. 2006;281:1309–1312. doi: 10.1074/jbc.C500405200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidow H, Lauber T, Vitzithum K, Sommerhoff CP, Rosch P, Marx UC. The solution structure of a chimeric LEKTI domain reveals a chameleon sequence. Biochemistry. 2004;43:11238–11247. doi: 10.1021/bi0492399. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vassylyeva MN, Svetlov V, Klyuyev S, Devedjiev YD, Artsimovitch I, Vassylyev DG. Crystallization and preliminary crystallographic analysis of the transcriptional regulator RfaH from Escherichia coli and its complex with ops DNA. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:1027–1030. doi: 10.1107/S174430910603658X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WZ, Ko TP, Corselli L, Johnson RC, Yuan HS. Conversion of a beta-strand to an alpha-helix induced by a single-site mutation observed in the crystal structure of Fis mutant Pro26Ala. Protein Sci. 1998;7:1875–1883. doi: 10.1002/pro.5560070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BA, Anthony LC, Gruber TM, Arthur TM, Heyduk E, Lu CZ, Sharp MM, Heyduk T, Burgess RR, Gross CA. A coiled-coil from the RNA polymerase beta′ subunit allosterically induces selective nontemplate strand binding by sigma(70) Cell. 2001;105:935–944. doi: 10.1016/s0092-8674(01)00398-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.