Abstract
The current epidemic of the mountain pine beetle (MPB), an indigenous pest of western North American pine, has resulted in significant losses of lodgepole pine. The leading edge has reached Alberta where forest composition shifts from lodgepole to jack pine through a hybrid zone. The susceptibility of jack pine to MPB is a major concern, but there has been no evidence of host-range expansion, in part due to the difficulty in distinguishing the parentals and their hybrids. We tested the utility of a panel of microsatellite loci optimized for both species to classify lodgepole pine, jack pine and their hybrids using simulated data. We were able to accurately classify simulated individuals, and hence applied these markers to identify the ancestry of attacked trees. Here we show for the first time successful MPB attack in natural jack pine stands at the leading edge of the epidemic. This once unsuitable habitat is now a novel environment for MPB to exploit, a potential risk which could be exacerbated by further climate change. The consequences of host-range expansion for the vast boreal ecosystem could be significant.
Keywords: host-range expansion, hybrid, jack pine, lodgepole pine, mountain pine beetle
Introduction
Increasing global temperatures have lead to elevational and/or latitudinal shifts in species ranges (Parmesan 1996; Bale et al. 2002; Walther et al. 2002). This is especially true for insects because they are poikilothermic organisms and therefore quickly respond to changes in their thermal environment. For forest insect pests this is expected to result in negative economic and ecological impacts (Ayres & Lombardero 2000; Logan et al. 2003; Battisti et al. 2006). Range shifts can expose novel habitats, where naïve hosts may not have evolved appropriate defences to ward off attack (Cudmore et al. 2010). Changes in climate suitability may go beyond the range of the host species, which could mean a dead end for the insect. However hybrid zones between forest species can provide a phenotypic stepping-stone which can help mediate range expansion into a new host (Floate & Whitham 1993; Pilson 1999).
The mountain pine beetle (MPB; Dendroctonus ponderosae Hopkins) is a bark beetle indigenous to western North America that primarily feeds on lodgepole pine (Pinus contorta Dougl. ex Loud. var. latifolia), but also feeds on sugar pine (P. lambertiana Dougl.), western white pine (P. monticola Dougl. Ed. D. Don) and ponderosa pine (P. ponderosa, P. Laws. Ex C. Laws; Safranyik & Carroll 2006). Beetle populations typically infest damaged trees or trees with compromised defence capacity; however, given the right conditions they can erupt into large-scale outbreaks and cause significant losses of mature healthy stands (Safranyik & Carroll 2006). The most recent outbreak has affected over 14 million hectares of forest land in western Canada (Nealis & Peter 2008) with considerable losses recorded in the USA (Hicke et al. 2006). This is the largest outbreak that has been documented since record taking began approximately 125 years ago (Taylor & Carroll 2004; Raffa et al. 2008) and brings negative economic impacts, inefficient nutrient cycling and carbon sequestration, and reduced biodiversity (Ayres & Lombardero 2000; Kurz et al. 2008).
Until recently, the range of MPB in Canada has been primarily restricted to the British Columbia interior due to physiological restrictions for MPB (Bentz et al. 2010). Colder temperatures at higher elevations and increasing latitudes can affect their synchrony and over-winter survival, suppressing population growth at their range limit (Logan & Powell 2001). However, a suite of studies examining future climate change scenarios have predicted a geographic range shift into previously marginal habitat (Logan & Bentz 1999; Logan & Powell 2001; Carroll et al. 2003; Fauria & Johnson 2009). As predicted, the beetle has traversed the Rocky Mountains and has reached the eastern edge of the range of lodgepole pine in north-central Alberta (Robertson et al. 2009; Bentz et al. 2010) where forest composition shifts to jack pine (P. banksiana, Lamb) through a hybrid zone (Zavarin et al. 1969; Pollack & Dancik 1985). This hybrid zone could conceivably help mediate MPB host-range expansion into jack pine. Jack pine is a boreal species whose range extends from Alberta to Nova Scotia. There has been no record of MPB infection in natural hybrid or jack pine stands (Bentz et al. 2010; Safranyik et al. 2010). However, there is considerable evidence to suggest hybrids and jack pine would be suitable hosts for MPB reproduction (reviewed in Safranyik et al. 2010). Given the close evolutionary relationship between lodgepole and jack pine (Wheeler et al. 1983), together with the ability of MPB to attack more distantly related pine (Bentz et al. 2010) and instances of MPB attack on jack pine in nursery settings (Furniss & Schenk 1969; Safranyik et al. 2010), hybrids and jack pine are likely to be compatible hosts for MPB. Consequently, the arrival of MPB in north-central Alberta has raised major concerns regarding the potential for further expansion to the vast boreal forest.
Evidence for host-range expansion by MPB to jack pine and hybrids has been difficult since it has not been possible to reliably distinguish lodgepole and jack pine from their hybrids (Zavarin et al. 1969; Pollack & Dancik 1985; Rweyongeza et al. 2007). Seed and cone morphometry are the current criteria for distinguishing species (Wheeler & Guries 1987); however, considerable morphological variation in the hybrid zone makes positive identification difficult (Rweyongeza et al. 2007). Previous efforts to identify hybrids using molecular approaches including chemical profiles, allozymes, organellar DNA, random amplified polymorphism and restriction fragment length polymorphisms (Pollack & Dancik 1985; Wheeler & Guries 1987; Dong & Wagner 1993; Yang et al. 2007) have also been ineffective in resolving hybrids from parents in the hybrid zone. Microsatellites have been effectively utilized for a variety of taxa where hybrids have been difficult to distinguish from parentals using other characters (Thulin et al. 2006; Burgarella et al. 2009; Quintela et al. 2010). For that reason, our first objective was to develop a panel of microsatellite markers that amplify in both species and test their efficacy to distinguish jack pine, lodgepole pine and their hybrids. We verified our ability to accomplish this by analysing simulated datasets containing multiple levels of admixture generated using pure lodgepole pine (central and southern British Columbia) and jack pine (Saskatchewan and Ontario) as benchmarks. Due to the level of differentiation described between these species (FST = 0.108, Wheeler & Guries 1987; GST = 0.247, Ye et al. 2002) we expected accurate resolution of first and second generation hybrids, however, we anticipated diminishing power with advanced generations of backcrossing given these species are closely related (Wheeler et al. 1983). Our second objective was to analyse samples collected from MPB attacked and un-attacked trees from the region of MPB expansion including the leading edge in north-central Alberta where the ranges of lodgepole pine and jack pine overlap (Fig. 1). We classified the hybrid and species status of the trees, focusing on MPB-attacked individuals sampled at the leading edge of the expansion based on the information from our simulations and species assignment analysis. We hypothesized that some of the attacked trees in these regions would be jack pine because of the geographic location and the reported susceptibility of jack pine to MPB in nursery settings (Furniss & Schenk 1969; Safranyik et al. 2010).
Fig. 1.
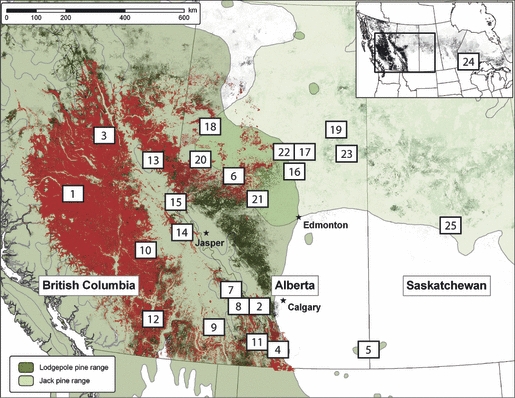
Sampling locations for lodgepole pine, jack pine and hybrids across western Canada analysed at 12 microsatellite loci. MPB attack data from 1958 to 2009 for British Columbia and from 1975 to 2009 for Alberta are indicated in red (Thandi & Taylor unpublished data), and approximate pine volume is shaded in green, where maximum densities are over 500 m3/ha (Yemshanov & McKenney unpublished data). Top-right inset of North America illustrates the location of the Ontario/Minnesota samples. Range distributions for jack pine and lodgepole pine were obtained from USGS (http://esp.cr.usgs.gov/data/atlas/little/, accessed 29 July 2010) and are based on Little (1971).
Methods
Sample collection
Foliage samples were collected from lodgepole pine within 10 locations in British Columbia (n= 160), from 14 jack pine locations in Ontario and Minnesota (n = 70) and one location in Saskatchewan (n = 43), and from lodgepole pine, jack pine and putative hybrids within 12 locations in Alberta (n = 381) (Table 1, Fig. 1). In British Columbia and Alberta both MPB attacked and non-attacked trees were sampled. The majority of the foliage samples were collected during two time periods, from February 2007 to May 2007 and September 2007 to April 2008. Stands for sampling were selected on the basis of aerial and ground survey data of MPB attack provided by Alberta Sustainable Resource Development (ASRD). Aerial surveys were conducted in the late summer and early fall each year by ASRD to identify newly attacked trees by the presence of fading foliage, an indicator of MPB infestation. The majority of aerially identified trees were subsequently ground-truthed by ASRD. For each sampling period, waypoints from the most recently available aerial survey were used to locate candidate trees for sampling. Based on 2009 survey data, additional foliar samples from putative jack pine trees were collected in April and May 2010 in the easternmost extent of detected MPB attack by ASRD (n = 6, ‘Smith’–Table 1, Fig. 1) and by the Canadian Forest Service (n = 18, ‘Wildwood’–Table 1, Fig. 1).
Table 1.
Sample size (N), number of effective alleles (Ne), observed heterozygosity (HO), expected heterozygosity (HE) and fixation index (F) calculated for lodgepole pine (Pl), jack pine (Pj) and their hybrids at each location across our study area, the numbers (‘Pop’) correspond to the locations in Fig. 1. Expected species composition is included based on Little's (1971) distribution maps. All measures were calculated in GenAlEx 6.4 (Peakall & Smouse) and were estimated without locus Pcon54
| Pop | Location | Prov. | Species | N | Na | Ne | Ho | HE | F |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Bulkley | BC | Pl | 3 | 3.63 | 3.23 | 0.667 | 0.770 | −0.072 |
| 2 | Canmore | AB | Pl | 28 | 12.45 | 7.24 | 0.765 | 0.838 | 0.066 |
| 3 | Plateau | BC | Pl | 4 | 4.36 | 3.50 | 0.705 | 0.786 | −0.046 |
| 4 | Crowsnest Pass | AB | Pl | 12 | 8.36 | 5.73 | 0.744 | 0.826 | 0.054 |
| 5 | Cypress Hills | AB/SK | Pl | 12 | 7.72 | 5.27 | 0.757 | 0.805 | 0.000 |
| 6 | Fox Creek | AB | Pl | 16 | 10.00 | 5.98 | 0.746 | 0.809 | 0.040 |
| 7 | Golden | BC | Pl | 38 | 13.18 | 7.69 | 0.725 | 0.828 | 0.109 |
| 8 | Kootenay/Yoho | BC | Pl | 20 | 11.36 | 6.30 | 0.720 | 0.817 | 0.098 |
| 9 | Nelson | BC | Pl | 3 | 4.18 | 3.64 | 0.727 | 0.836 | −0.061 |
| 10 | Prince George | BC | Pl | 5 | 5.36 | 3.96 | 0.595 | 0.791 | 0.178 |
| 11 | Sparwood | BC | Pl | 29 | 12.90 | 6.81 | 0.711 | 0.817 | 0.112 |
| 12 | Okanagan | BC | Pl | 7 | 5.45 | 4.24 | 0.672 | 0.787 | 0.081 |
| 13 | Tumbler Ridge | BC | Pl | 28 | 12.27 | 6.69 | 0.704 | 0.821 | 0.123 |
| 14 | Valemount | BC | Pl | 23 | 11.36 | 6.70 | 0.671 | 0.815 | 0.156 |
| 15 | Willmore-Kakwa | AB | Pl | 21 | 11.18 | 6.63 | 0.711 | 0.834 | 0.110 |
| 16 | Smith | AB | Pj × Pl | 6 | 4.54 | 3.28 | 0.539 | 0.655 | 0.067 |
| 17 | Wildwood | AB | Pj × Pl | 18 | 10.81 | 6.89 | 0.764 | 0.855 | 0.076 |
| 18 | Fairview | AB | Pj × Pl | 27 | 12.18 | 7.61 | 0.753 | 0.847 | 0.091 |
| 19 | FtMcMurray | AB | Pj × Pl | 89 | 12.54 | 4.12 | 0.571 | 0.659 | 0.131 |
| 20 | Grande Prairie | AB | Pj × Pl | 30 | 13.09 | 7.49 | 0.707 | 0.847 | 0.150 |
| 21 | Hinton | AB | Pj × Pl | 8 | 6.90 | 5.01 | 0.727 | 0.822 | 0.048 |
| 22 | Wabasca | AB | Pj × Pl | 34 | 13.63 | 8.36 | 0.761 | 0.856 | 0.097 |
| 23 | Conklin | AB | Pj | 104 | 12.00 | 3.83 | 0.548 | 0.610 | 0.113 |
| 24 | Ontario/Minnesota | ON/MN | Pj | 70 | 7.45 | 3.56 | 0.536 | 0.596 | 0.103 |
| 25 | Saskatchewan | SK | Pj | 43 | 8.36 | 3.71 | 0.537 | 0.614 | 0.096 |
| Average | 9.415 | 5.503 | 0.683 | 0.782 | 0.077 |
Prior to sampling, each tree was first confirmed for MPB attack and colonization by identification of diagnostic entrance holes in the bark, followed by bark removal around the area of these entrance holes to confirm the presence of MPB larval galleries, as opposed to galleries created by other species such as Ips pini Say, which co-occur in this region. For trees sampled in 2010, the existence of pupal chambers was specifically noted as proof that the host is suitable for completing development of all feeding larval stages. Further development of these pupae had likely not occurred because of the time of year that the samples had been taken. In all cases, foliage was collected from the crown using pole pruners or a shotgun. For samples collected in 2007 and 2008, MPB (typically as larvae) from attacked trees were sampled simultaneously for genetic analysis (Samerasekera et al., unpublished). All sampled trees were geo-referenced using Garmin GPS units (Garmin International, Olathe, KS, USA). Samples were stored in coolers until they could be brought to the laboratory, where they were processed and stored at –20 °C or –80 °C until DNA extraction could be performed.
DNA extraction and genotyping
Genomic DNA was isolated from ground needle tissue using a CTAB (hexadecyl trimethyl ammonium bromide) protocol modified from Chang et al. (1993), according to Roe et al. (2010). Additional changes include 5 μL of RNase A (70 units/mg protein, Sigma) being added to each sample along with the CTAB buffer. As well, incubation time at 65 °C was increased to 2 h and all centrifugation steps were performed at 5800 g with the duration of the final two centrifugation steps being increased to 5 min. Pellets were resuspended in 125 μL Milli-Q water.
We screened twenty-three microsatellite loci that were previously tested in lodgepole pine (Auckland et al. 2002) and found 10 to be polymorphic in a panel of eight individuals of each species (Appendix I). Additional loci were isolated from a microsatellite enriched (GTn/CTn) library constructed from a single lodgepole pine individual using the methods of Glenn & Schable (2005). We selected 368 clones and obtained bi-directional sequences using T3 and T7 primers using BigDye v3.1 sequencing chemistry (Applied Biosystems, Carlsbad, CA, USA) and resolved on a 3730 DNA Analyzer (Applied Biosystems). Sequences were aligned in SeqMan (LaserGene; DNASTAR, Madison, WI, USA) resulting in 222 contigs. Twenty-five contigs were selected that contained long uninterrupted (11–33) repeats with sufficient flanking sequence for primer design. Amplification primers were designed using Primer3 with the default parameters except the optimal Tm was set to 56 °C and the maximum Tm difference between pairs of primers was restricted to 1 °C (Rozen & Skaletsky 2000). Two loci were retained (GENBANK accession numbers: HQ404301– 2) that were polymorphic in both species and had clean amplification products (Appendix I).
Genotyping was completed for all individuals at 12 microsatellite loci. These loci were amplified in two single, and five multiplex 15 μL reactions (A–G; Appendix I) containing: ∼400 ng DNA (∼200 ng DNA for the single reactions), 1X PCR Buffer, 160 μM each dNTP, 1% dimethyl sulfoxide (volume to volume), 1U Taq DNA polymerase (AB), and optimized MgCl2 and primer amounts (Appendix I). Amplifications were completed using an Eppendorf Mastercycler using the following cycling parameters: 94 °C for 5 min, 33 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 15 s, and a final extension at 72 °C for 30 min. These reactions were co-loaded into three injections (1–3; Appendix I) on an ABI 3730 DNA Analyzer and genotyped using GeneMapper software (Applied Biosystems) with allele sizes being determined relative to GeneScan-500LIZ (Applied Biosystems). Genotyping error rate was quantified by running duplicate genotypes for 46 samples.
Diversity measures
We assessed our microsatellite scoring for stutter errors, large-allele drop-out and null alleles using microchecker (Oosterhout et al. 2004); any loci with scoring issues were removed from further analyses. Hardy–Weinberg equilibrium and linkage disequilibrium was assessed in genepop 4.0 (Raymond & Rousset 1995; web version, http://genepop.curtin.edu.au/) within locations (where n ≥ 20) for each species (hybrids excluded) as defined by analysis of outputs from Newhybrids and Structure (see Results section). Significance was assessed using Bonferroni corrected alpha values for multiple comparisons (α = 0.05, Rice 1989). Standard measures of allelic diversity including number of alleles, effective number of alleles (defined as the number of alleles with equal frequency that would achieve the observed level of diversity, Hedrick 2000) and observed and expected heterozygosities (HO and HE, respectively) were calculated in GenAlEx 6.0 (Peakall & Smouse 2006). Allelic richness and private allelic richness corrected for sample size differences were calculated in hp-rare 1.0 (Kalinowski 2005). All measures were estimated for the entire data-set, for each species (hybrids excluded) once identified, and basic diversity measures were calculated for each location. In addition, we calculated the level of differentiation (FST) at several hierarchical levels: among locations in the entire dataset, between species and among locations within species, using GenAlEx.
Species identification
The detection of hybrid classes will depend on the degree of differentiation between the parental species and the loci used (Anderson & Thompson 2002; Vähä & Primmer 2006). Previous studies that have assessed hybrid zones and the resolving capacity of microsatellites have included only first (F1) and second generation hybrids (F2, F1 backcrosses) in their simulations (Thulin et al. 2006; Vähä & Primmer 2006; Burgarella et al. 2009; Quintela et al. 2010). We also included a third generation of hybrids (F2 backcrosses and F1 double backcrosses) to assess our ability to resolve advanced introgression. We simulated five datasets using Hybridlab ver. 1.0 (Nielsen et al. 2006). Hybridlab was developed to create artificial parental and hybrid genotypes to evaluate the power to correctly identify hybrids. As input, we used genetic profiles from 100 jack pine from Ontario and Saskatchewan and 100 lodgepole pine from British Columbia (we selected samples far removed from the hybrid zone) to represent the microsatellite allele frequency variation for each species. For each dataset we simulated profiles for 300 jack pine and 300 lodgepole pine; we chose 300 as we felt this would simulate our dataset closely. Using these 600 simulated genotypes we generated 100 F1 hybrids. With these hybrid profiles we were able to simulate F2 hybrids, F1 × jack pine (F1Pj), F1 × lodgepole pine (F1Pl), F2 × jack pine (F2Pj), F2 × lodgepole pine (F2Pl), F1-jackpine × jack pine (F1Pj-Pj), and F1-lodgepole pine × lodgepole pine (F1Pl-Pl) (Fig. 3). Each dataset for hybrid analysis was comprised of simulated genotypes for 300 jack pine and 300 lodgepole pine and 10 individuals from each hybrid class: F1, F2, F1Pj, F1Pl, F2Pj, F2Pl, F1Pj-Pj and F1Pl-Pl. We then analysed these datasets with two different Bayesian methods (Structure and Newhybrids) to establish a threshold (QT) for assigning parental and hybrid status. We used both Bayesian methods in our approach to maximize the accuracy of assigning individuals (Thulin et al. 2006; Vähä & Primmer 2006; Burgarella et al. 2009).
Fig. 3.
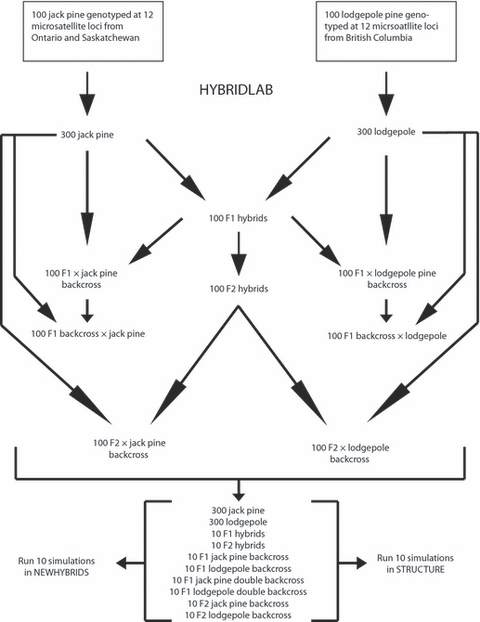
Work-flow for generating genotypes for simulations to assess the capability of 12 microsatellite loci to resolve the species identify of jack pine, lodgepole pine and their hybrids.
In Newhybrids v. 1.0 (Anderson & Thompson 2002), we implemented the model with uninformative priors for both the allele frequency and admixture distributions. For each individual Newhybrids calculates the probability of belonging to the parental categories and each specific hybrid classes (e.g. F1, F2, F1 backcross). However, assignment to a specific hybrid class may be uncertain in which case the probability is often split among the hybrid classes (Burgarella et al. 2009). Therefore, to assign hybrids we used two different approaches; if the assignment to a hybrid class was of high probability (i.e. an assignment to the F1 hybrid class with over 0.9 probability) we took the assignment. However, if we found the probability was divided among the hybrid classes, we summed the probability over classes, and if the sum was ≥ 0.9 we considered this a hybrid with class undefined. To assign a QT for the parental categories we looked at the range and average assignment across all individuals. We ran each data-set through ten simulations with a burn-in of 50 000 and 500 000 Markov-chain Monte Carlo sweeps for data collection and we did not use prior population information as species designations were not included for the collected samples.
In Structure 2.3.1 (Pritchard et al. 2000; Falush et al. 2003, 2007), we set K = 2, and ran the admixture model with uncorrelated allele frequencies, inferring lambda for each population 10 times. The algorithm was run for 550 000 Markov-chain Monte Carlo sweeps with a burn-in of 50 000 and 500 000 for data collection. We used Clumpp (Jakobsson & Rosenberg 2007) to summarize the ten iterations for each of the five simulated data-sets then looked at the Q values for each cluster to determine the most appropriate QT. Again, we looked at the range and average of Q values for each class as we did with Newhybrids. We assessed the standard deviation among individuals across the 10 iterations for each of the five simulated datasets to determine the margin of error for both programs. We also compared the outputs from both programs for each simulated dataset to assess the level of agreement between the methods.
We analysed our 678 genotyped samples in Newhybrids and Structure using the same parameters and summary methods as our simulations. Our choice of the most appropriate QT for assigning species class was based on the simulations (see Results section). The results from the two programs were compared and a final species class was assigned based on a combination of the two. If the assignment between the two programs did not agree we made a decision based on the Q values from the simulations (see Results section).
Results
Sample collection
Six-hundred and seventy-eight individuals were used for genotyping (Dryad entry doi:10.5061/dryad.8677), 154 of these represented individuals that had been successfully attacked by MPB. In the putative jack pine sampled in 2010, signs of successful reproduction were observed where pupal chambers were present (Fig. 2), which indicates the eggs hatched and the insects completed all larval stages.
Fig. 2.

Images from putative jack pine attacked by MPB in north-central Alberta. Left panel shows the red needles indicating tree die-off typical of trees one-year after MPB infection. Right panel shows a set of well-developed larval galleries, including a pupal chamber, indicating successful completion of larval development. Adult beetles from the parental generation (bottom left) occupy a different gallery. Images courtesy of James D. Weber, Canadian Forest Services.
Diversity measures
Individuals were genotyped at 12 microsatellite loci with less than 2% missing data (555 complete profiles, 101 missing one locus and 22 missing two loci). Our genotyping error rate was very low at 0.8%. Some loci were not in Hardy–Weinberg equilibrium at the location level (Appendix II) with a homozygote excess, however only the locus Pcon54 was consistently and extremely out of Hardy–Weinberg equilibrium, therefore we removed this locus from analyses. After Bonferroni correction we did not find any pairs of loci in linkage disequilibrium in lodgepole pine, and only two comparisons for jack pine. Genetic diversity was high among locations, average HO = 0.683 (Table 1). Genetic diversity measured for each species across loci by HO, HE, allelic richness and private allelic richness was higher in lodgepole pine (Appendix III). Across the entire dataset, differentiation among locations (FST = 0.125) and between species (FST = 0.133) was high. Within species however, differentiation among locations was very low (FSTlodgepole = 0.033, FSTjack = 0.016). However, there were some significant pair-wise comparisons; most notably for jack pine, where all locations were differentiated from Ontario/Minnesota.
Species identification
Across the five simulated datasets the assignment of individuals to their correct class was never less than 96% using either assignment method (Fig. 4, Table 2). The calculated proportion of ancestry for both programs were highly consistent, the standard deviation among individuals across the ten iterations for each simulated dataset were extremely low (Structure = 0.00014, Newhybrids = 0.00003). As well, the consistency in assignment across the programs was also very high with few discrepancies (Table 3). For both methods detection of 1st generation hybrids was 100%, for 2nd generation hybrids this decreased to 87% and 81% and for 3rd generation hybrids this decreased further to 68% and 63%, in Newhybrids and Structure, respectively (Table 2). Based on the results from Newhybrids the most accurate method to assign hybrids was to sum the estimated proportions across the hybrid categories. When we used a QT of 0.9 for the parental and hybrid classes in newhybrids, similar to other studies (Thulin et al. 2006; Vähä & Primmer 2006; Burgarella et al. 2009; Quintela et al. 2010), we found a large number of individuals that did not assign to any category. However, over 98% of these individuals with QT < 0.90 were hybrids, therefore we used a QT ≥ 0.9 to assign pure species, and all other individuals were assigned hybrid status.
Fig. 4.
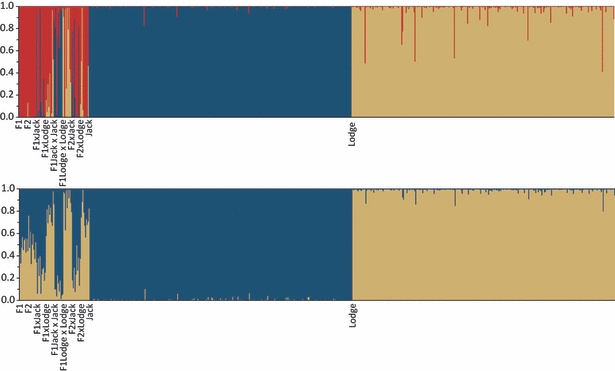
Ancestry plots from simulated lodgepole pine (tan), jack pine (blue), and various hybrid crosses (red for Newhybrids) generated in Newhybrids (top) and Structure (bottom).
Table 2.
Accuracy of assignment among five simulated datasets of lodgepole pine, jack pine and their hybrids using Newhybrids 1.0 (NH, Anderson & Thompson 2002) and Structure 2.3.1 (STR, Pritchard et al. 2000; Falush et al. 2003, 2007). Hybrid categories are as follows: 1st Gen – F1, 2nd Gen – F2 and F1 backcrosses, and 3rd Gen – F2 backcrosses and F1 double backcrosses
| Sim 1 | Sim 2 | Sim 3 | Sim 4 | Sim 5 | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | NH | STR | NH | STR | NH | STR | NH | STR | NH | STR | NH | STR |
| 1st Gen | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2nd Gen | 0.90 | 0.90 | 0.90 | 0.87 | 0.83 | 0.63 | 0.80 | 0.79 | 0.93 | 0.90 | 0.87 | 0.81 |
| 3rd Gen | 0.73 | 0.65 | 0.63 | 0.60 | 0.73 | 0.70 | 0.65 | 0.55 | 0.65 | 0.63 | 0.68 | 0.63 |
| Hybrid Avg | 0.83 | 0.79 | 0.78 | 0.75 | 0.80 | 0.71 | 0.75 | 0.69 | 0.80 | 0.78 | 0.79 | 0.74 |
| Jack | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Lodgepole | 0.96 | 0.99 | 0.98 | 1.00 | 0.98 | 0.99 | 0.97 | 0.98 | 0.96 | 0.99 | 0.97 | 0.99 |
Table 3.
Classification of simulated data sets (Sim01–Sim05) and trees sampled in Alberta, British Columbia, Ontario and Saskatchewan (Data) based on outputs from Newhybrids (NH) and Structure (ST) to jack pine, lodgepole pine and hybrids. There was a high level of agreement between the two methods (8–16 discrepancies)
| Sim01 | Sim02 | Sim03 | Sim04 | Sim05 | Data | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NHST | Jack | Lodge | Hybrid | Jack | Lodge | Hybrid | Jack | Lodge | Hybrid | Jack | Lodge | Hybrid | Jack | Lodge | Hybrid | Jack | Lodge | Hybrid |
| Jack | 306 | 0 | 4 | 302 | 0 | 6 | 310 | 0 | 0 | 303 | 0 | 2 | 307 | 0 | 4 | 294 | 0 | 1 |
| Lodge | 0 | 295 | 9 | 0 | 304 | 6 | 0 | 301 | 8 | 0 | 307 | 8 | 0 | 298 | 12 | 0 | 287 | 2 |
| Hybrid | 0 | 0 | 66 | 0 | 0 | 62 | 0 | 0 | 61 | 0 | 0 | 60 | 0 | 0 | 59 | 6 | 1 | 87 |
Using Structure, we found that QT ≥ 0.9 gave the most accurate pure assignment for simulated data, but cases of advanced introgression were difficult to detect (Fig. 4). The average disagreement between Structure and Newhybrids across the five simulations was 1.4%. All of these discrepancies were either hybrids correctly assigned by Newhybrids, or pure individuals correctly assigned by Structure. Based on these discrepancies we developed a set of rules for assigning individuals when the methods had conflicting results: (i) if the probability of being a hybrid in Newhybrids is ≥ 0.9 and the probability of being a parental in Structure is > 0.9 but < 0.95 the individual was assigned hybrid status. (ii) If the probability is most likely to be a pure parental in Newhybrids but < 0.9 and in Structure is > 0.95, then the individual is assigned to the parental category.
We were able to clearly delineate the two species and our power to detect hybrid individuals was 0.74 averaged across three generations of hybrids in the simulated data (Table 2). We therefore used a QT ≥ 0.9 in both Structure and Newhybrids for the assignment of pure lodgepole pine and jack pine, and considered the unassigned individuals as hybrids. The two assignment methods agreed for 668 of the 678 samples analysed (Table 3). The 10 disagreements were resolved using the rules that we developed based on the simulations (see above). The final breakdown of assignment for our sample data was 87 hybrids, 301 jack pine and 290 lodgepole pine (Figs 5 and 6). Ancestry of the hybrid trees was predominately lodgepole pine (Fig. 5). Of the trees sampled that were designated as attacked there were eight jack pine (five from the samples from Smith, and three from Wildwood), 127 lodgepole pine, and 19 hybrids. The eight trees assigned as jack pine had Q values > 0.99.
Fig. 5.
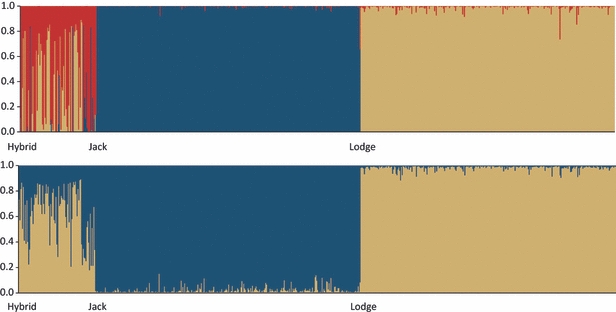
Ancestry plots generated in Newhybrids (top) and Structure (bottom) for 678 trees sampled across Alberta, British Columbia, Ontario, Minnesota, and Saskatchewan illustrating lodgepole pine (tan), jack pine (blue) and hybrid (red for Newhybrids) ancestry.
Fig. 6.
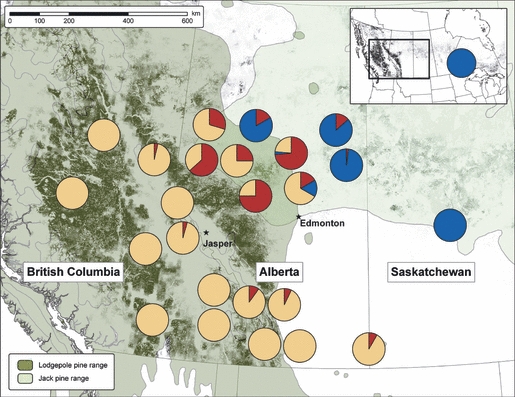
Proportion of lodgepole pine (tan), jack pine (blue), and hybrids (red) at 24 locations in western Canada. Assignment to species categories was based on results from Newhybrids and Structure. Range distributions for jack pine and lodgepole pine were obtained from USGS (http://esp.cr.usgs.gov/data/atlas/little/, accessed 29 July 2010) and are based on Little (1971).
Discussion
Using simulated microsatellite genotypes and Bayesian cluster analyses, we were able to test the power of a set of microsatellites to distinguish lodgepole pine, jack pine and variable levels of hybrid ancestry. We found high agreement between the two Bayesian algorithms and low assignment error in the analysis of simulated data. Where discrepancies arose between the two methods, we generally found that Newhybrids had better power to detect hybrids while Structure performed better with pure individuals, highlighting the complementarily of the methods. While our microsatellite panel had excellent resolving power for parentals and recent hybrids, resolution declined with increasing generations of hybrid back-crossing. We expected decreased resolution based on the close evolutionary relationship between lodgepole pine and jack pine (Wheeler et al. 1983), however this may not be a major issue for our dataset. Given the colonization times for lodgepole pine (7000 YBP; Yeatman 1967; MacDonald & Cwynar 1985; MacDonald et al. 1998) and jack pine (6000 YBP; Ritchie & Yarranton 1978; McLeod & MacDonald 1997) in central Alberta, the long generation times for pine (Critchfield 1985; Muir 1993) and low hybrid vigour (Pollack 1980), the potential for geographically spread, advanced introgression is not likely (Pollack & Dancik 1985).
Our ability to distinguish species classes enabled us to address the second objective of our study, namely to determine whether jack pine have been successfully attacked by MPB. We identified eight pure jack pine trees collected from MPB attacked stands at the edge of MPB range expansion in Alberta (Figs 1 and 2). In addition, we also identified 19 hybrid trees that have also been attacked confirming their susceptibility within the hybrid zone.
The discovery that MPB has expanded its host repertoire to include jack pine has prompted us to consider whether MPB will be able to sustain eruptive populations locally and thus spread further eastward into the boreal forest. Host and beetle interaction is influenced by physiology, population dynamics and environment (Safranyik & Carroll 2006; Raffa et al. 2008) making this a complex system where many biotic and abiotic factors need to be considered concomitantly. This study demonstrates that both hybrids and pure jack pine are susceptible to MPB within the hybrid zone. It is currently unknown whether hybrids and jack pine have different susceptibilities to MPB attack relative to lodgepole pine. Differential susceptibility of jack pine and hybrids to MPB is plausible. For example these two species have different susceptibilities to western gall rust fungus (Yang et al. 1999). Also, lodgepole pine and MPB appear to share a long co-evolutionary history that has presumably allowed this tree species to adapt at some level to MPB (Raffa et al. 2008). In fact, Cudmore et al. (2010) recently showed that naïve lodgepole pine stands had higher MPB reproductive output than stands that experienced epidemic outbreaks. This suggests that MPB may have higher reproductive success in hybrid and jack pine trees which would further sustain the epidemic and support its eastward expansion. There is a potential for introgression of lodgepole pine genes – including genes that condition defence – into the jack pine genome through historical hybridization. However, widespread hybridization and introgression of defence genes seems unlikely based on our findings, therefore any potential benefits of introgression would likely be limited to the hybrid zone. As well, those trees identified as hybrids in our study exhibited higher lodgepole than jack pine ancestry (Fig. 5) suggesting a higher percentage of lodgepole pine backcrosses and limited introgression of lodgepole genes into the jack pine range. Further, chemical defences produced by lodgepole pine differ in their composition from jack pine and hybrids (Zavarin et al. 1969; Pollack & Dancik 1985). For example, Clark et al. (2008) found α-pinene, a chemical that may facilitate a successful mass attack by MPB, to be at considerably higher concentrations in jack pine than in lodgepole pine. Safranyik & Linton (1982) and Cerezke (1995) found that measures of beetle performance were similar between lodgepole pine and jack pine, suggesting the potential of jack pine to sustain populations in a manner similar to lodgepole pine. As well, there are MPB fungal associates that are part of the beetle invasion process, and it has been shown that hybrids and jack pine are susceptible (Rice et al. 2007a,b; Rice & Langor 2009). It should be noted that many of the published experiments were carried out using cut bolts and artificially infested rather than naturally infested live trees, limiting extrapolation to the natural boreal forest.
While tree-level defences that are partly under genetic control will conceivably contribute to the probability of MPB eruptive population dynamics in jack pine, there also needs to be a sufficient density of available hosts. The severity of the recent MPB outbreak has been partly attributed to the present-day distribution of lodgepole pine, where the continuity of even-aged stands resulting from forest management practices has been ideal for maintaining beetle populations (Taylor et al. 2006). In contrast, jack pine is not uniformly distributed in the boreal forest, typically occurring in a patchy distribution (Nealis & Peter 2008). Nealis & Peter (2008) and Safranyik et al. (2010) have analysed the potential for MPB spread in the boreal forest region and have suggested that Alberta populations are susceptible; however, jack pine occurrence may be too fragmented east of Alberta to sustain the type of epidemic that British Columbia has experienced.
Climate has influenced the severity of the recent MPB outbreak (Hicke et al. 2006; Régnière & Bentz 2007; Powell & Bentz 2009), and will likely play an important role in determining whether this epidemic will be maintained in the boreal forest. Temperature has received the most attention and from this there are two considerations: (i) in the past, the summer climate in northern Alberta has not been suitable to sustain synchronous beetle populations which are necessary to maintain eruptive populations (Carroll et al. 2003) and (ii) beetles are not completely cold tolerant and can incur high mortality within the jack pine range from cold exposure (Régnière & Bentz 2007; Cooke 2009). However, changes in global temperatures have improved the climatic suitability for both summer (Logan & Powell 2001) and winter (Carroll et al. 2003) seasons promoting conditions for eruptive behaviour.
The discovery of successful MPB attack and evidence of completed larval development in jack pine is a critical first step in assessing future impacts of this destructive forest pest on the boreal forest and how climate change may affect the system. We have considered whether MPB populations can be maintained in the boreal forest and there are many factors that need to be met for continued MPB expansion and population growth. If jack pine can sustain endemic populations and thus maintain this host-range expansion it is critical that forest management incorporates these considerations in their future planning. MPB is not endemic to the boreal forest and therefore should be considered an invasive species and managed as such. Forest ecosystems in North America have already been challenged with numerous pest invasions that represent a considerable threat (Liebhold et al. 1995). When we factor in climate change, the vulnerability of ecosystems such as the boreal forest to disturbance is further increased putting an extremely important ecosystem in jeopardy.
Acknowledgments
The authors would like to thank Alberta Sustainable Resources and Development – particularly Daniel Lux, Sunil Ranasinghe and Tom Hutchinson – for logistical support and sample collection, Jim Weber (Canadian Forest Service, Natural Resources Canada) for sample collection and images; Brad Jones, Darryl Edwards, Ed Hunt, and Stephane Bourassa (University of Alberta) for sample collection; Michael Carlson (Government of British Columbia) for sample material, Rory McIntosh and Rob Moore (Saskatchewan Ministry of the Environment) for sample collection; Gurp Thandi and Steve Taylor (Canadian Forest Service, Pacific Forestry Centre) for access to MPB attack data; Denys Yemshanov and Daniel McKenney (Canadian Forest Service, Great Lakes Forestry Centre) for access to pine volume data; and Matthew Bryman (University of Alberta) for logistical support. We acknowledge funding for this research from the Government of Alberta (AAET/AFRI-859-G07), as well as grants from Genome Canada, the Government of Alberta through Genome Alberta, and Genome British Columbia in support of the Tria I and Tria II projects (http://www.thetriaproject.ca) of which J.E.K. Cooke, B.J. Cooke and D.W. Coltman are principle investigators.
Appendix I
Primer sequences and PCR conditions for 14 microsatellite loci used to type lodgepole and jack pine from British Columbia, Alberta, Saskatchewan, Ontario and Minnesota. VIC, NED, PET and 6-FAM are fluorescent dyes (Applied Biosystems)
| Locus | Forward primer | Reverse primer | *Multiplex PCR and co-loading | Primer (μmol) | MgCl2 (mm) |
|---|---|---|---|---|---|
| PtTx2123† | VIC-GAAGAACCCACAAACACAAG | GGGCAAGAATTCAATGATAA | A1 | 0.32 | 1 |
| PtTx2146 + 16bp† | VIC-TCCCCTTAAGCCTGGGGATTTGGATTGGGTATTTG | GTTTCTATATTTTCCTTGCCCCTTCCA | D2 | 0.96 | 1 |
| PtTx3011† | 6-FAM-AATTTGGGTGTATTTTTCTTAGA | AAAAGTTGAAGGAGTTGGTGATC | B1 | 0.64 | 1 |
| PtTx3025† | NED-CACGCTGTATAATAACAATCTA | TTCTATATTCGCTTTTAGTTT | B1 | 0.96 | 1 |
| PtTx3030† | 6-FAM-AATGAAAGGCAAGTGTCG | GAGATGCAAGATAAAGGAAGTT | G2 | 0.64 | 1 |
| PtTx3034† | NED-TCAAAATGCAAAAGACG | ATTAGGACTGGGGATGAT | C2 | 0.96 | 1 |
| PtTx3049† | VIC-GAAGTGATAATGGCATAGCAAAAT | CAGACCCGTGAAAGTAATAAACAT | B1 | 0.96 | 1 |
| PtTx3127† | PET-ACCCTTACTTTCAGAAGAGGATA | AATTGGGGTTCAACTATTCTATTA | A1 | 0.64 | 1 |
| PtTx4054† | NED-TGCATTCACCTTGGAGTT | TAGGAGATAATATAAAATGTT | F3 | 0.64 | 3 |
| PtTx4139† | 6-FAM-TGGCATGCTAGGAAGAAGA | TTGTATGTTGCCTGTGGAGA | E3 | 0.96 | 1 |
| Pcon3‡ | 6-FAM-CGACGAATATGTGATTGGATA | TGCTCCTAAATTTTTCAACCT | C2 | 0.32 | 1 |
| Pcon54‡ | VIC-CAGATGATGGTGTACCTTTGA | TCCAAATCTTCATTGTGTGTC | E3 | 0.96 | 1 |
Loci were amplified in seven reactions (A–G) and detected in three co-loaded sets (1–3).
In house.
Appendix II
Hardy–Weinberg calculations for sample locations where n ≥ 20. Included are the standard error and FIS (calculated using Weir & Cockerham 1984), all values were calculated in Genepop. P-values in bold are significant folowing Bonferroni correction
| Lodgepole pine | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Canmore | Golden | Sparwood | Tumbler Ridge | Valemount | Willmore-Kakwa | |||||||||||||
| Locus | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS |
| PtTx2123 | 0.354 | 0.004 | −0.068 | 0.376 | 0.004 | 0.113 | 0.637 | 0.003 | 0.111 | 0.225 | 0.004 | −0.076 | 0.360 | 0.003 | 0.172 | 0.101 | 0.002 | −0.406 |
| PtTx3030 | 0.735 | 0.005 | 0.040 | 0.000 | 0.000 | 0.470 | 0.000 | 0.000 | 0.493 | 0.000 | 0.000 | 0.549 | 0.000 | 0.000 | 0.553 | 0.094 | 0.003 | 0.165 |
| PtTx3127 | 0.019 | 0.002 | 0.275 | 0.253 | 0.005 | 0.020 | 0.859 | 0.002 | −0.015 | 0.468 | 0.006 | 0.111 | 0.098 | 0.003 | 0.169 | 0.451 | 0.004 | 0.088 |
| PtTx3011 | 0.000 | 0.000 | 0.223 | 0.034 | 0.005 | 0.150 | 0.075 | 0.008 | 0.135 | 0.138 | 0.010 | 0.127 | 0.060 | 0.006 | 0.157 | 0.001 | 0.000 | 0.246 |
| PtTx3049 | 0.002 | 0.001 | 0.198 | 0.023 | 0.003 | 0.134 | 0.401 | 0.010 | 0.124 | 0.000 | 0.000 | 0.359 | 0.018 | 0.002 | 0.167 | 0.000 | 0.000 | 0.463 |
| PtTx3025 | 0.776 | 0.007 | −0.089 | 0.990 | 0.001 | −0.171 | 0.534 | 0.008 | −0.086 | 0.151 | 0.005 | 0.066 | 0.408 | 0.010 | 0.029 | 0.401 | 0.008 | −0.171 |
| Pcon3 | 0.026 | 0.003 | 0.300 | 0.037 | 0.005 | 0.167 | 0.034 | 0.004 | 0.102 | 0.033 | 0.004 | 0.104 | 0.045 | 0.004 | 0.097 | 0.003 | 0.001 | 0.238 |
| PtTx3034 | 0.875 | 0.004 | 0.077 | 0.010 | 0.001 | 0.212 | 0.000 | 0.000 | 0.393 | 0.880 | 0.004 | 0.029 | 0.061 | 0.003 | 0.276 | 0.001 | 0.000 | 0.482 |
| PtTx2146 | 0.491 | 0.011 | 0.034 | 0.442 | 0.010 | −0.050 | 0.502 | 0.010 | −0.080 | 0.041 | 0.004 | 0.139 | 0.483 | 0.009 | 0.024 | 0.661 | 0.010 | −0.004 |
| PtTx4139 | 0.784 | 0.008 | 0.047 | 0.000 | 0.000 | 0.296 | 0.036 | 0.004 | 0.224 | 0.344 | 0.009 | 0.095 | 0.293 | 0.009 | 0.131 | 0.000 | 0.000 | 0.227 |
| PtTx4054 | 0.871 | 0.006 | 0.029 | 0.528 | 0.009 | 0.000 | 0.085 | 0.006 | 0.018 | 0.321 | 0.009 | 0.078 | 0.007 | 0.002 | 0.188 | 0.118 | 0.007 | 0.133 |
| Jack pine | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conklin | FtMcMurray | Ontario/Minnesota | Saskatchewan | |||||||||||||||
| Locus | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS | P-val | SE | FIS | ||||||
| PtTx2123 | 0.767 | 0.003 | 0.052 | 0.858 | 0.002 | −0.039 | 0.333 | 0.002 | 0.068 | 0.612 | 0.003 | 0.099 | ||||||
| PtTx3030 | 0.002 | 0.001 | 0.286 | 0.000 | 0.000 | 0.337 | 0.001 | 0.001 | 0.320 | 0.056 | 0.004 | 0.265 | ||||||
| PtTx3127 | 1.000 | 0.000 | −0.015 | 0.001 | 0.000 | 0.432 | 1.000 | 0.000 | −0.031 | 1.000 | 0.000 | −0.017 | ||||||
| PtTx3011 | 0.002 | 0.001 | 0.153 | 0.014 | 0.003 | 0.126 | 0.329 | 0.010 | 0.075 | 0.020 | 0.003 | 0.171 | ||||||
| PtTx3049 | 0.074 | 0.003 | 0.083 | 0.014 | 0.002 | 0.232 | 0.107 | 0.003 | 0.147 | 0.732 | 0.004 | 0.003 | ||||||
| PtTx3025 | 0.288 | 0.010 | 0.074 | 0.207 | 0.007 | −0.062 | 0.036 | 0.001 | 0.190 | 0.406 | 0.005 | 0.156 | ||||||
| Pcon3 | 0.877 | 0.005 | 0.004 | 0.235 | 0.009 | 0.124 | 0.916 | 0.004 | 0.016 | 0.358 | 0.008 | 0.115 | ||||||
| PtTx3034 | 0.000 | 0.000 | 0.412 | 0.000 | 0.000 | 0.465 | 0.002 | 0.000 | 0.286 | 0.000 | 0.000 | 0.455 | ||||||
| PtTx2146 | 0.024 | 0.003 | −0.050 | 0.261 | 0.008 | −0.068 | 0.578 | 0.006 | 0.024 | 0.131 | 0.005 | −0.071 | ||||||
| PtTx4139 | 0.616 | 0.012 | 0.028 | 0.170 | 0.009 | −0.017 | 0.844 | 0.006 | −0.050 | 0.006 | 0.002 | 0.106 | ||||||
| PtTx4054 | 0.459 | 0.012 | −0.014 | 0.727 | 0.010 | −0.084 | 0.029 | 0.002 | 0.162 | 1.000 | 0.000 | −0.099 | ||||||
Appendix III
Allelic diversity measures for all data and for only samples that were assigned to jack pine and lodgepole pine. Number of alleles (N), allelic richness (Na), effective number of alleles (Ne), observed heterozygosity (HO), expected heterozygosity (HE) and the fixation index (F) were calculated in GenAlEx 6 (Peakall & Smouse 2006). Allelic richness (Richness) and private allelic richness (Private) were calculated using rarefaction in hp-rare 1.0 (Kalinowski 2005)
| Pop | FIS | Locus | N | Na | Ne | Ho | He | F | Richness | Private |
|---|---|---|---|---|---|---|---|---|---|---|
| All | 0.242 | PtTx2123 | 677 | 6 | 4.038 | 0.600 | 0.752 | 0.203 | 6.0 | |
| PtTx3030 | 667 | 22 | 5.160 | 0.465 | 0.806 | 0.424 | 21.9 | |||
| PtTx3127 | 662 | 12 | 2.397 | 0.384 | 0.583 | 0.342 | 11.9 | |||
| PtTx3011 | 673 | 49 | 26.051 | 0.798 | 0.962 | 0.170 | 48.8 | |||
| PtTx3049 | 649 | 19 | 8.980 | 0.695 | 0.889 | 0.218 | 18.9 | |||
| PtTx3025 | 674 | 22 | 5.061 | 0.706 | 0.802 | 0.120 | 21.8 | |||
| Pcon3 | 651 | 32 | 11.475 | 0.768 | 0.913 | 0.159 | 31.9 | |||
| PtTx3034 | 649 | 17 | 5.506 | 0.522 | 0.818 | 0.362 | 17.0 | |||
| PtTx2146 | 668 | 24 | 8.130 | 0.823 | 0.877 | 0.061 | 24.6 | |||
| PtTx4139 | 673 | 25 | 8.250 | 0.722 | 0.879 | 0.178 | 24.6 | |||
| Pcon54 | 673 | 17 | 4.004 | 0.429 | 0.750 | 0.428 | 16.9 | |||
| PtTx4054 | 675 | 18 | 5.055 | 0.612 | 0.802 | 0.237 | 18.0 | |||
| Jack | 0.172 | PtTx2123 | 299 | 4 | 2.331 | 0.548 | 0.571 | 0.039 | 4.0 | 0.0 |
| PtTx3030 | 294 | 16 | 1.885 | 0.327 | 0.469 | 0.304 | 15.4 | 6.1 | ||
| PtTx3127 | 287 | 5 | 1.096 | 0.070 | 0.088 | 0.207 | 4.9 | 0.0 | ||
| PtTx3011 | 299 | 31 | 12.353 | 0.793 | 0.919 | 0.138 | 30.2 | 8.1 | ||
| PtTx3049 | 281 | 12 | 5.135 | 0.683 | 0.805 | 0.151 | 11.9 | 0.0 | ||
| PtTx3025 | 299 | 14 | 2.919 | 0.599 | 0.657 | 0.089 | 13.5 | 1.1 | ||
| Pcon3 | 295 | 16 | 5.939 | 0.786 | 0.832 | 0.054 | 15.4 | 0.0 | ||
| PtTx3034 | 285 | 12 | 2.952 | 0.400 | 0.661 | 0.395 | 11.7 | 1.0 | ||
| PtTx2146 | 299 | 14 | 4.330 | 0.796 | 0.769 | −0.035 | 13.8 | 1.0 | ||
| PtTx4139 | 298 | 17 | 3.468 | 0.688 | 0.712 | 0.033 | 16.8 | 0.8 | ||
| Pcon54 | 298 | 11 | 2.308 | 0.168 | 0.567 | 0.704 | 10.5 | 1.0 | ||
| PtTx4054 | 299 | 14 | 1.451 | 0.314 | 0.311 | −0.012 | 13.7 | 0.1 | ||
| Lodgepole | 0.122 | PtTx2123 | 279 | 6 | 2.553 | 0.624 | 0.608 | −0.025 | 6.0 | 2.0 |
| PtTx3030 | 274 | 15 | 4.732 | 0.544 | 0.789 | 0.310 | 14.6 | 5.4 | ||
| PtTx3127 | 277 | 12 | 3.196 | 0.635 | 0.687 | 0.075 | 11.8 | 6.9 | ||
| PtTx3011 | 276 | 41 | 19.515 | 0.804 | 0.949 | 0.152 | 40.5 | 18.4 | ||
| PtTx3049 | 275 | 19 | 10.611 | 0.695 | 0.906 | 0.233 | 18.8 | 6.9 | ||
| PtTx3025 | 277 | 21 | 4.081 | 0.791 | 0.755 | −0.047 | 20.7 | 8.3 | ||
| Pcon3 | 261 | 30 | 13.396 | 0.747 | 0.925 | 0.193 | 29.9 | 14.5 | ||
| PtTx3034 | 268 | 15 | 5.730 | 0.631 | 0.825 | 0.236 | 14.9 | 4.1 | ||
| PtTx2146 | 272 | 23 | 6.257 | 0.824 | 0.840 | 0.020 | 22.4 | 9.7 | ||
| PtTx4139 | 277 | 22 | 11.065 | 0.755 | 0.910 | 0.171 | 21.8 | 5.9 | ||
| Pcon54 | 277 | 16 | 3.884 | 0.675 | 0.743 | 0.091 | 15.9 | 6.4 | ||
| PtTx4054 | 278 | 18 | 9.759 | 0.849 | 0.898 | 0.054 | 17.9 | 4.3 |
Footnotes
C.I.C. is a postdoctoral fellow whose research highlights the use of molecular methods to investigate issues of wildlife management. Currently she has decided to work on tree genetics because they tend to stay in the same place. J.E.K.C. is an assistant professor in the Department of Biological Sciences at the University of Alberta. She is a tree biologist whose research is mainly focused on understanding how environmental cues affect growth and development of forest trees. S.D. is a molecular technician and enjoys optimizing genetic loci in pine. C.S.D. has a keen interest in marker development and applying new molecular techniques to studies of molecular ecology. B.J.C. is a research scientist with the Canadian Forest Service interested in the population dynamics of eruptive boreal forest insect pests. D.W.C. is a Professor of wildlife genetics and a Sasquatch DNA expert at the University of Alberta.
References
- Anderson E, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland LD, Bui T, Zhou Y, Shepard M, Williams CG. Conifer Microsatellite Handbook. Raleigh, NC: Corporate Press; 2002. [Google Scholar]
- Ayres MP, Lombardero MJ. Assessing the consequences of global change for forest disturbance from herbivores and pathogens. Science of the Total Environment. 2000;262:263–286. doi: 10.1016/s0048-9697(00)00528-3. [DOI] [PubMed] [Google Scholar]
- Bale JS, Masters GJ, Hodkinson ID, et al. Herbivory in global climate change research: direct effects of rising temperature on insect herbivores. Global Change Biology. 2002;8:1–16. [Google Scholar]
- Battisti A, Stastny M, Buffo E, Larsson S. A rapid altitudinal range expansion in the pine processionary moth produced by the 2003 climate anomaly. Global Change Biology. 2006;12:662–671. [Google Scholar]
- Bentz BJ, Régnière J, Fettig CJ, et al. Climate change and bark beetles of the western United States and Canada: direct and indirect effects. BioScience. 2010;60:602–613. [Google Scholar]
- Burgarella C, Lorenzo Z, Jabbour-Zahad R, et al. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex) Heredity. 2009;102:442–452. doi: 10.1038/hdy.2009.8. [DOI] [PubMed] [Google Scholar]
- Carroll AL, Taylor SW, Régnière J, Safranyik L. Effects of climate change on range expansion by the mountain pine beetle in British Columbia. In: Shore TL, Brooks JE, Stone JE, editors. Mountain Pine Beetle Symposium: Challenges and Solutions. Victoria, BC: Report BC-X-399, Canadian Forest Service, Pacific Forestry Centre; 2003. pp. 223–232. [Google Scholar]
- Cerezke HF. Egg gallery, brood production, and adult characteristics of mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Scolytidae), in three pine hosts. Canadian Entomologist. 1995;127:955–965. [Google Scholar]
- Chang S, Puryear J, Cairney JW. A simple method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Clark E, Huber D, Carroll A. Induced terpene defense response of lodgepole and jack pine. Mountain Pine Beetle: From Lessons Learned to Community-based Solutions Conference Proceedings, June 10–11, 2008 BC Journal of Ecosystems and Management. 2008;9 [Google Scholar]
- Cooke BJ. Natural Resources Canada, Canadian Forest Service. Victoria, BC: Pacific Forestry Centre; 2009. Forecasting mountain pine beetle overwintering mortality in a variable environment. Mountain Pine Beetle Working Paper 2009-03; pp. 1–25. [Google Scholar]
- Critchfield WB. The late quaternary history of lodgepole and jack pines. Canadian Journal of Forest Research. 1985;15:749–772. [Google Scholar]
- Cudmore TJ, Bjorklund N, Carroll AL, Lindgren BS. Climate change and range expansion of an aggressive bark beetle: evidence of higher beetle reproduction in naïve host tree populations. Journal of Applied Ecology. 2010;47:1036–1043. [Google Scholar]
- Dong J, Wagner DB. Taxonomic and population differentiation of mitochondrial diversity in Pinus banksiana and Pinus contorta. Theoretical and Applied Genetics. 1993;86:573–578. doi: 10.1007/BF00838711. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauria MM, Johnson EA. Large-scale climatic patterns and area affected by mountain pine beetle in British Columbia, Canada. Journal of Geophysical Research. 2009;114:G01012. [Google Scholar]
- Floate KD, Whitham TG. The ‘hybrid bridge’ hypothesis: host shifting via plant hybrid swarms. American Naturalist. 1993;141:651–662. doi: 10.1086/285497. [DOI] [PubMed] [Google Scholar]
- Furniss MM, Schenk JA. Sustained natural infestations by the mountain pine beetle in seven new Pinus and Picea hosts. Journal of Economic Entomology. 1969;62:518–519. [Google Scholar]
- Glenn TC, Schable NA. Isolating microsatellite DNA loci. Methods in Enzymology. 2005;395:202–222. doi: 10.1016/S0076-6879(05)95013-1. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Genetics of Populations. 2nd edn. Sudbury, MA: Jones and Barlettt; 2000. [Google Scholar]
- Hicke JA, Logan JA, Powell J, Ojima DS. Changing temperatures influence suitability for modeled mountain pine beetle (Dendroctonus ponderosae) outbreaks in the western United States. Journal of Geophysical Research. 2006;111:G02019. [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes. 2005;5:187–189. [Google Scholar]
- Kurz WA, Dymond CC, Stinson G, et al. Mountain pine beetle and forest carbon feedback to climate change. Nature. 2008;452:987–990. doi: 10.1038/nature06777. [DOI] [PubMed] [Google Scholar]
- Liebhold AM, MacDonald WL, Bergdahl D, Mastro VC. Invasion by exotic forest pests: a threat to forest ecosystems. Forest Science Monograph. 1995;41(2):1–49. [Google Scholar]
- Little EL., Jr . Atlas of the United States Trees, Conifers and Important Hardwoods, Volume 1. Washington, D.C: US Department of Agriculture Miscellaneous Publication No. 1146; 1971. [Google Scholar]
- Logan JA, Bentz BJ. Model analysis of mountain pine beetle (Coleoptera: Scolytidae) seasonality. Environmental Entomology. 1999;28:924–934. [Google Scholar]
- Logan JA, Powell JA. Ghost forests, global warming, and the mountain pine beetle (Coleoptera: Scolytidae) American Entomologist. 2001;47:160–172. [Google Scholar]
- Logan JA, Régnière J, Powell JA. Assessing the impacts of global warming on forest pest dynamics. Frontiers in Ecology and the Environment. 2003;1:130–137. [Google Scholar]
- MacDonald GM, Cwynar LC. A fossil pollen based reconstruction of the late quaternary history of lodgepole pine (Pinus contorta ssp. latifolia) in the western interior of Canada. Canadian Journal of Forest Research. 1985;15:1039–1044. [Google Scholar]
- MacDonald GM, Cwynar LC, Whitlock C. The late Quarternary dynamics of pines in northern North America. In: Richardson DM, editor. Ecology and Biogeography of Pinus. Cambridge: Cambridge University Press; 1998. pp. 122–136. [Google Scholar]
- McLeod TK, MacDonald GM. Postglacial range expansion and population growth of Picea marina, Picea glauca, and Pinus banksiana in the western interior of Canada. Journal of Biogeography. 1997;24:865–881. [Google Scholar]
- Muir PS. Disturbance effects on structure and tree species composition of Pinus contorta forests of western Montana. Canadian Journal of Forest Research. 1993;23:1617–1625. [Google Scholar]
- Nealis V, Peter B. Risk assessment of the threat of mountain pine beetle to Canada's boreal and eastern pine forests. Victoria, British Columbia: Report BC-X-417, Canadian Forest Service, Pacific Forestry Centre; 2008. [Google Scholar]
- Nielsen EEG, Arvebach L, Kotlicki P. Hybridlab (version 1.0): a program for generating simulated hybrids from population samples. Molecular Ecology Notes. 2006;6:971–973. [Google Scholar]
- Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Parmesan C. Climate and species’ range. Nature. 1996;382:765–766. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilson D. Plant hybrid zones and insect host range expansion. Ecology. 1999;80:407–415. [Google Scholar]
- Pollack JC. A chemical and morphological investigation of the jack pine – lodgepole pine complex in Alberta. Edmonton, Alberta: University of Alberta; 1980. MSc Thesis. [Google Scholar]
- Pollack JC, Dancik BP. Monoterpene and morphological variation and hybridization of Pinus contorta and P. banksiana in Alberta. Canadian Journal of Botany. 1985;63:201–210. [Google Scholar]
- Powell JA, Bentz BJ. Connecting phenological predictions with population growth rates for mountain pine beetle, an outbreak insect. Landscape Ecology. 2009;24:657–672. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintela M, Thulin C-G, Höglund J. Detecting hybridization between willow grouse (Lagopus lagopus) and rock ptarmigan (L. muta) in central Sweden through Bayesian admixture analyses and mtDNA screening. Conservation Genetics. 2010;11:557–569. [Google Scholar]
- Raffa KF, Aukema BH, Berntz J, et al. Cross-scale divers of natural disturbances prone to anthropogenic amplification: the dynamics of bark beetle eruptions. BioScience. 2008;58:501–517. [Google Scholar]
- Raymond M, Rousset F. Genepop (Version-1.2) – population-genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Régnière J, Bentz B. Modeling cold tolerance in the mountain pine beetle, Dendroctonus ponderosae. Journal of Insect Physiology. 2007;53:559–572. doi: 10.1016/j.jinsphys.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rice AV, Langor DW. Mountain pine beetle-associated blue-stain fungi in lodgepole × jack pine hybrids near Grande Prairie, Alberta (Canada) Forest Pathology. 2009;39:323–334. [Google Scholar]
- Rice AV, Thormann MN, Langor DW. Mountain pine beetle-associated blue-stain fungi cause lesions on jack pine, lodgepole pine, and lodgepole-jack pine hybrids in Alberta. Canadian Journal of Botany. 2007a;85:307–315. [Google Scholar]
- Rice AV, Thormann MN, Langor DW. Virulence of and interactions among mountain pine beetle-associated blue-stain fungi on two pine species and their hybrids in Alberta. Canadian Journal of Botany. 2007b;85:316–323. [Google Scholar]
- Ritchie JC, Yarranton GA. The late-quaternary history of the boreal forest of central Canada, based on standard pollen stratigraphy and principal components analysis. Journal of Ecology. 1978;66:199–212. [Google Scholar]
- Robertson C, Nelson TA, Jelinski DE, Wulder MA, Boots B. Spatial-temporal analysis of species range expansion: the case of the mountain pine beetle, Dendroctonus ponderosae. Journal of Biogeography. 2009;36:1446–1458. [Google Scholar]
- Roe AD, Rice AV, Bromilow SE, Cooke JEK, Sperling FAH. Multilocus species identification and fungal DNA barcoding: insights from blue stain fungal symbionts of the mountain pine beetle. Molecular Ecology Resources. 2010;10:946–959. doi: 10.1111/j.1755-0998.2010.02844.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Rweyongeza DM, Dhir NK, Barnhardt LK, Hansen C, Yang R-C. Population differentiation of lodgepole pine (Pinus contorta) and jack pine (Pinus banksiana) complex in Alberta: growth, survival, and responses to climate. Canadian Journal of Botany. 2007;85:545–556. [Google Scholar]
- Safranyik L, Carroll A. The biology and epidemiology of the mountain pine beetle in lodgepole pine forests. In: Safranyik L, Wilson W, editors. The Mountain Pine Beetle: A Synthesis of Biology, Management and Impacts on Lodgepole Pine. Victoria, BC: Pacific Forestry Centre; 2006. pp. 3–66. Natural Resources Canada, Canadian Forest Service. [Google Scholar]
- Safranyik L, Linton D. Survival and development of mountain pine beetle broods in jack pine bolts from Ontario. Canadian Forest Service Research Notes. 1982;2:17–18. [Google Scholar]
- Safranyik L, Carroll AL, Règniére J, et al. Potential for range expansion of mountain pine beetle into the boreal forest of North America. Canadian Entomologist. 2010;142:415–4442. [Google Scholar]
- Taylor SW, Carroll AL. Disturbance, forest age, and mountain pine beetle outbreak dynamics in BC: a historical perspective. In: Shore TL, Brooks JE, Stone JE, editors. Mountain Pine Beetle Symposium: Challenges and Solutions. Victoria, BC: Report BC-X-399, Canadian Forest Service, Pacific Forestry Centre; 2004. pp. 67–94. [Google Scholar]
- Taylor SW, Carroll AL, Alfaro RI, Safranyik L. Forest climate and mountain pine beetle outbreak dynamics in western Canada. In: Shore TL, Brooks JE, Stone JE, editors. Mountain Pine Beetle Symposium: Challenges and Solutions. Victoria, BC: Report BC-X-399, Canadian Forest Service, Pacific Forestry Centre; 2006. pp. 67–94. [Google Scholar]
- Thulin C-G, Stone J, Tegelström H, Walker CW. Species assignment and hybrid identification among Scandinavian hares Lepus europaeus and L. timidus. Wildlife Biology. 2006;12:29–38. [Google Scholar]
- Vähä J-P, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Walther G-R, Post E, Convey P, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wheeler NC, Guries RP. A quantitative measure of introgression between lodgepole and jack pines. Canadian Journal of Botany. 1987;65:1876–1885. [Google Scholar]
- Wheeler NC, Guries RP, O'Malley DM. Biosystematics of the genus Pinus, subsection Contortae. Biochemical Systematics and Ecology. 1983;11:333–340. [Google Scholar]
- Yang R-C, Ye Z, Hiratsuka Y. Susceptibility of Pinus contorta–Pinus banksiana complex to Endocronartium harknessii: host-pathogen interactions. Canadian Journal of Botany. 1999;77:1035–1043. [Google Scholar]
- Yang R-C, Yeh FC, Ye TZ. Multilocus structure in the Pinus contorta–Pinus banksiana complex. Canadian Journal of Botany. 2007;85:774–784. [Google Scholar]
- Ye TZ, Yang R-C, Yeh FC. Population structure of a lodgepole pine (Pinus contorta) and jack pine (P. banksiana) complex as revealed by random amplified polymorphic DNA. Genome. 2002;45:530–540. doi: 10.1139/g02-016. [DOI] [PubMed] [Google Scholar]
- Yeatman CW. Biogeography of jack pine. Canadian Journal of Botany. 1967;45:2201–2211. [Google Scholar]
- Zavarin E, Critchfield WB, Snajberk K. Turpene composition of Pinus contorta × Pinus banksiana hybrids and hybrid derivatives. Canadian Journal of Botany. 1969;47:1443–1453. [Google Scholar]


