Abstract
NY-ESO-1 is a tumor-specific shared antigen with distinctive immunogenicity. Both CD8+ T cells and class-switched Ab responses have been detected from patients with cancer. In this study, a CD4+ T cell line was generated from peripheral blood mononuclear cells of a melanoma patient and was shown to recognize NY-ESO-1 peptides presented by HLA-DP4, a dominant MHC class II allele expressed in 43–70% of Caucasians. The ESO p157–170 peptide containing the core region of DP4-restricted T cell epitope was present in a number of tumor cell lines tested and found to be recognized by both CD4+ T cells as well as HLA-A2-restricted CD8+ T cells. Thus, the ESO p157–170 epitope represents a potential candidate for cancer vaccines aimed at generating both CD4+ and CD8+ T cell responses. More importantly, 16 of 17 melanoma patients who developed Ab against NY-ESO-1 were found to be HLA-DP4-positive. CD4+ T cells specific for the NY-ESO-1 epitopes were generated from 5 of 6 melanoma patients with NY-ESO-1 Ab. In contrast, no specific DP4-restricted T cells were generated from two patients without detectable NY-ESO-1 Ab. These results suggested that NY-ESO-1-specific DP4-restricted CD4+ T cells were closely associated with NY-ESO-1 Ab observed in melanoma patients and might play an important role in providing help for activating B cells for NY-ESO-1-specific Ab production.
A number of studies suggest that tumor-reactive T cells play an important role in mediating tumor regression. The molecular basis of T cell-mediated antitumor immunity has been elucidated by the identification of a number of tumor antigens recognized by CD8+ T cells (1, 2). These MHC class I-restricted tumor antigens can be divided into several categories. The tissue-specific differentiation antigens, which include MART-1 (3), TRP-1/gp75 (4), TRP-2 (5), and gp100 (6) are expressed in melanoma as well as normal melanocytes. Tumor-specific shared antigens such as MAGE-1 (7) and NY-ESO-1 (8, 9) are expressed in a wide variety of tumors such as breast cancer and lung cancer. The expression of these products is limited in cancer cells with the exception of normal testis. Tumor-specific unique or mutated antigens such as β-catenin (10) also have been described. Among these antigens, NY-ESO-1 is of particular interest because both cytotoxic T lymphocyte and Ab responses have been shown to react with this antigen (9, 11). NY-ESO-1 encodes two gene products recognized by CD8+ T cell clones (9). In addition, high titers of Ab were detected from about 50% of patients with NY-ESO-1-positive tumor (12). Because of its strict tumor-specific expression pattern (8), NY-ESO-1 is potentially an important immune target for the development of immunotherapy for a variety of cancer types.
Increasing evidence from both human and animal studies has indicated that CD4+ T cells play a central role in initiating and maintaining host immune responses against cancer (2, 13). The observation that high titers of NY-ESO-1 antibodies were present in a high proportion of patients suggested that CD4+ T cell responses also might be found in these patients. Recently MHC class II-restricted T cell epitopes from NY-ESO-1 presented by DRB1*0401 (14) and DRB4*0101 (15) have been identified. Nevertheless, the observation that the majority of patients with NY-ESO-1 Ab did not express the above-mentioned MHC class II alleles (14, 15) suggested that CD4+ T cells from most patients might recognize epitopes in the context of additional MHC class II alleles.
In this study, a CD4+ T cell line was generated and shown to recognize NY-ESO-1 peptides presented by HLA DP4, a prevalent MHC class II allele present in approximately 43–70% of Caucasians (16). More importantly, 16 of 17 (94%) of the melanoma patients who produced high titer Ab against NY-ESO-1 are DP4-positive. The results of in vitro stimulation demonstrated that the HLA DP4-restricted T cells could be generated from 5 of 6 patients with NY-ESO-1 Ab. These results suggest that recognition of NY-ESO-1 by CD4+ T cells in the context of DP4 is closely associated with the ability of these patients to mount an Ab response against this antigen.
Materials and Methods
Cell Lines, Tissue Culture Reagents, and Abs Used in the Study.
293CIITA is a cell line generated by transduction of 293 cells with a retrovirus encoding the MHC class II transactivator (17). All melanoma lines and Epstein–Barr virus transformed B lymphocytes (EBVB) lines were generated and maintained in RPMI 1640 (Life Technologies, Rockville, MD) supplemented with 10% FCS (Biofluids, Gaithersburg, MD). Culture medium for lymphocytes was RPMI 1640 with 0.05 mM β-mercaptoethanol, 50 cetus units/ml IL-2 plus 10% human male AB serum (Valley Biochemicals, Winchester, VA). W6/32 (HLA class I), L243 (HLA DR), IVA12 (HLA class II), B7/21 (HLA DP), Genox 3.53, and IVD12 (both HLA DQ) were purchased from Becton Dickinson Immunocytometry Systems.
Construction of Plasmids.
The pESO plasmid was an expression vector containing the NY-ESO-1 cDNA driven by a cytomegalovirus promoter as described (9). The pIi-ESO plasmid was constructed by inserting an NheI and NotI-digested PCR product of the whole NY-ESO-1 cDNA into the pTi80 vector digested with the same enzymes (18). The NY-ESO-1 cDNA was fused in-frame with the first 80 amino acid residues of invariant chain (Ii) leader sequence at its N terminus. PCR primers used to amplify NY-ESO-1 were as follows: forward primer, 5′-cattgctagcATG CAG GCC GAA GGC CGG GGC A-3′ containing an NheI site, and the reverse primer, 5′-aaggctacattGC GGC CGC TTA GCG CCT CTG CCC TGA G-3′ containing an NotI site.
Peptides and Generation of CD4+ T Cells.
Synthetic peptides used in this study were made by using a solid-phase method on a peptide synthesizer (Gilson) at the Surgery Branch, National Cancer Institute. The in vitro sensitization procedure was carried out as described (14). Briefly, approximately 2.5 × 105 peripheral blood mononuclear cells (PBMCs) were plated in a 96-well flat-bottom plate in the presence of 20 μg/ml peptide. On days 7 and 14, 1 × 105 nonirradiated PBMCs were pulsed with 20 μg/ml peptide, washed twice, and added to each well, and IL-2 at 120 units/ml was added on days 8, 11, 15, and 18. On day 21, cells were harvested and incubated with target cells overnight before the supernatants were taken for cytokine release assays.
T cells from those wells with specific activities were pooled and expanded by using a rapid expansion method (19). After expansion, CD8+ T cells were depleted from cultures by using magnetic beads selection (Dynal), and the cell lines subsequently were analyzed for CD4+ and CD8+ expression by flow cytometry.
Cytokine Release Assays.
To prepare protein or peptide pulsed targets, peptides were used at a final concentration of 20 μg/ml, and proteins were used at a final concentration of 5 μg/ml. Cells were washed in serum-free RPMI medium and pulsed at 37οC in the absence of serum for 4 h, followed by two time washes. Unless specified, approximately 3 × 104 target cells were incubated with the same number of T cells for at least 16 h before a cytokine release assay was carried out. Granulocyte–macrophage colony-stimulating factor (GM-CSF) ELISA kit (R&D Systems) and other cytokine kits were obtained from R&D Systems and Endogen (Woburn, MA).
Molecular Typing of HLA DP Molecules.
The following primer pairs were used for PCR amplification of HLA-DPA and -DPB. DPA forward primer was 5′-ATG CGC CCT GAA GAC AGA ATG T-3′; DPA reverse primer was 5′-TCA CAG GGT CCC CTG GGC CCG GGG GA-3′; DPB forward primer was 5′-ATG ATG GTT CTG CAG GTT TCT G-3′, and DPB reverse primer was 5′-TTA TGC AGA TCC TCG TTG AAC TTT C-3′. The PCR product was subsequently purified and sequenced by using the identical primers that were used to carry out the PCR. Alternatively, PCR product was first cloned into a pCR4 vector (Invitrogen) and sequenced by using 5′ and 3′ primers complementary to the vector sequence. The final sequence was searched against the IMGT-HLA database to confirm the HLA DP identity (http://www3.ebi.ac.uk/Services/imgt/hla).
Results
Generation of a CD4+ T Cell Line TE4–2 Against NY-ESO-1.
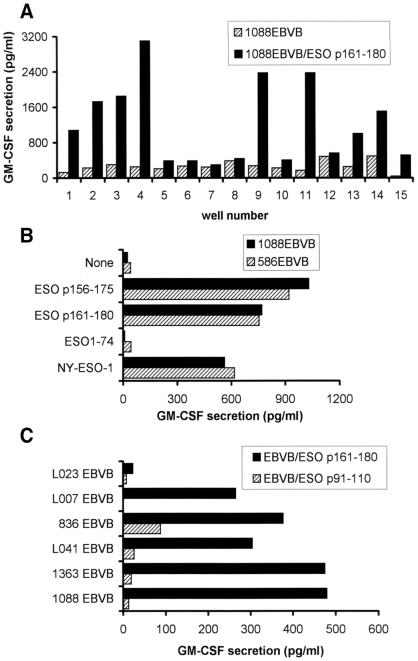
Initial studies were carried out to identify NY-ESO-1 epitopes restricted by the HLA DR4 alleles. Eight 20-mer peptides that contained predicted 9-mer DR4 binding motifs were examined for recognition by lymphocytes from HLA-DR4-IE transgenic mice immunized with the NY-ESO-1 recombinant protein and stimulated in vitro (14). Three 20-mer peptides were found to be positive in these experiments. One of them was characterized as a promiscuous epitope of both DRB1*0401 and DRB1*0101 (14). To further characterize two other peptides, ESO p161–180 and ESO p141–160, we used them to stimulate PBMCs from a DRB1*0401-positive patient (TE) who had high titer antibodies. After three rounds of weekly stimulation, 15 of 24 wells showed marked growth from PBMCs that were stimulated with ESO p161–180. Nine of the 15 growth-positive wells tested showed specific cytokine release against peptide-pulsed DRB1*0401-expressing 1088EBVB cells (Fig. 1A). Specific CD4+ T cells also were generated from the PBMCs stimulated with ESO p141–160 (data not shown). T cells from cultures that specifically responded to the ESO p161–180 peptide stimulation then were combined and expanded. After the depletion of CD8+ T cells, this CD4+ T cell line was designated TE4–2. Analysis of the cytokine secretion profile of TE4–2 demonstrated that this T cell line secreted IFN-γ, tumor necrosis factor α, IL-4, and GM-CSF, but not transforming growth factor type β in response to peptide pulsed targets (data not shown), suggesting that both Th1 and Th2 types of CD4+ T cells may be present in this cell line.
Figure 1.
Generation of CD4+ T cells from PBMCs after in vitro stimulation with synthetic peptides. (A) Specific peptide reactivity was detected in multiple wells after three in vitro stimulations. Fifteen of 24 wells each containing 2.5 × 105 PBMCs in a 96-well plate showed marked growth and tested for specific activity against 1088EBVB cells alone and 1088EBVB cells pulsed with the ESO p161–180 peptide. GM-CSF release was measured from supernatants. (B) TE4–2 T cells specifically reacted with NY-ESO-1 peptides and protein. Overlapping peptides ESO p161–180 and ESO p156–175 were pulsed onto 1088EBVB (DR4+) and 586EBVB (DR1+) cells at 20 μg/ml for 90 min. ESO p91–110 was used as an irrelevant peptide for pulsing. Purified NY-ESO-1 and ESO1–75 proteins were pulsed overnight at 5 μg/ml and 2 μg/ml, respectively, to maintain the same molar ratio. After three washes, TE4–2 T cells were added and incubated overnight. GM-CSF release was measured. (C) A panel of EBVB cells pulsed with ESO p161–180 were used as targets for TE4–2 CD4+ T cells. These EBVB lines were known to express different HLA DR and DQ alleles. Their HLA DP alleles were molecularly typed in this study (Table 1).
Recognition of NY-ESO-1 by TE4–2 in the Context of HLA DPB1*0401–0402.
DR4-positive target cells pulsed with ESO p161–180, an overlapping peptide (ESO p156–175), as well as the full-length NY-ESO-1 protein were further tested for T cell recognition. An irrelevant peptide, ESO p91–110, and a purified truncated recombinant protein, ESO1–74, comprising amino acids 1–74 (14) were used as controls. TE4–2 T cells specifically recognized DR4+ 1088EBVB cells pulsed with the full-length NY-ESO-1 protein, but not the truncated ESO1–74 protein (Fig. 1B). Both the ESO p161–180 and p156–175 were recognized by TE4–2, indicating that the minimal peptide epitope resided between amino acids 161 and 175. Unexpectedly, the TE4–2 T cell line appeared to respond equally well to peptides and proteins pulsed on 586EBVB cells, which expressed DRB1*0101 but not DRB1*0401. One possibility was that peptides were presented by different class II molecules. An alternative explanation was that a common MHC class II restriction element shared by both 1088EBV and 586EBVB cells presented the peptides to TE4–2 T cells. To test these possibilities, a number of other EBVB cells with known HLA DR and DQ types were tested in an attempt to identify the restriction element used by TE4–2 T cells. All but one of the EBVB cell lines tested were able to present the ESO p161–180 peptide to TE4–2 (Fig. 1C).
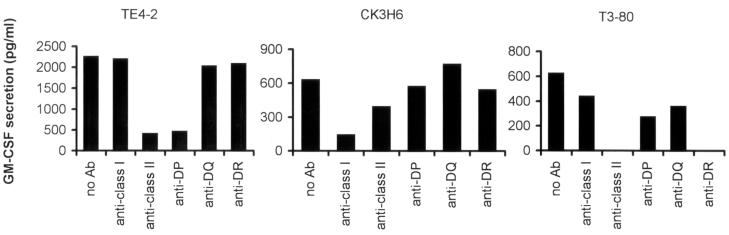
T cell recognition of peptides then was carried out in the presence of specific antibodies that blocked the recognition of peptides restricted by different MHC restriction elements. The results in Fig. 2 demonstrated that anti-MHC class II (IVA12) and anti-HLA DP (B7/21) antibodies abolished the ability of TE4–2 T cells to recognize ESO p161–180. Anti-class I (W6/32), anti-DR (L243), and anti-DQ (Genox 3.53 and IVD12) Ab had little or no effect on the stimulation of TE4–2 T cells. Thus, these results suggested that the TE4–2 T cells recognized ESO p161–180 in the context of a highly prevalent HLA DP allele shared by EBVB cell lines used in this study.
Figure 2.
Blocking of T cell recognition of NY-ESO-1 epitopes by an anti-DP Ab. 1088EBVB cells pulsed with 20 μg/ml ESO p161–180 peptide were used as target cells in the presence of different blocking antibodies. Specificity of Abs used was as follows: anti-MHC class I (HLA A, B, C) Ab (W6/32), anti-MHC class II (HLA DP, DQ, DR) Ab (IVA12), anti-HLA DP Ab (B7/21), anti-HLA DR Ab (L243), and anti-HLA DQ Abs [mixture of Genox 3.53 (anti-DQw1) and IVD12 (anti-DQw3)]. All Abs were used at a final concentration of 20 μg/ml each. CK3H6 T cells specifically recognized gp100 p209–218 peptide in the context of HLA A2 and was used as the specificity control for anti-MHC class I Ab. 1088EBVB (A2+) pulsed with gp100 p209–218 peptide was used as targets. A CD4+ T cell line (T3–80) recognized 1362mel in an HLA-DR1-restricted fashion and was used as the specificity control for anti-MHC class II and anti-DR Abs.
We then molecularly cloned and sequenced HLA-DP alleles for cell lines used in Fig. 1C and showed that 1088EBVB and 586EBVB lines were both homozygous for the HLA DPB1*0401 gene product (Table 1). The L023 EBVB cell line, which did not present the ESO p161–180 peptide to TE4–2, was negative for DPB1*0401 and 0402. The 1363, 1088, 836, and L007 EBVB cell lines all expressed DPB1*0401. Thus, it appeared that both DPB1*0401 and DPB1*0402 were capable of presenting the ESO p161–180 epitope to TE4–2 CD4+ T cells.
Table 1.
HLA (DP, DQ, and DR alleles) typing of patients used in this study
| HLA-DP | HLA-DQ | HLA-DR | |
|---|---|---|---|
| Patients with NY-ESO-1 Abs | |||
| TE | B1*0401, nd | 0302, 06† | B1*0401, 1501; B4*0101, B5*0101 |
| BE | B1*0401 | 0301, 0302 | B1*0401, 1102; B3*0202, B4*01† |
| AC | B1*04 negative | 0603, 0604 | B1*1301, 1302; B3*0202, B3*0301 |
| FJ | B1*0401 | 0502, 0601 | B1*1502, 1601; B5*0102, B5*02† |
| LD | B1*0401 | 0303, 0603 | B1*0901, 1301; B3*0101, B4*01† |
| CJ | B1*0401, nd | 0201, 0301 | B1*0701, 1101; B3*0202, B4*01† |
| BFE | B1*0402, nd | 0303, 0602 | B1*0701, 1501; B4*01†, B5*0101 |
| KF | B1*0401, 0402 | 0301, 0603 | B1*0401, 1301; B3*0101, B4*0101 |
| CT | B1*0401, 0402 | 0301, 0603 | B1*1101, 1502; B3*0202, B5*0102 |
| DA | B1*0401 | 06† | B1*08†, 15†; nd |
| BL | B1*0401, nd | 0201, 0602 | B1*0301, 1501; B3*0101, B3*0202 |
| BD | B1*0401, nd | 02†, 03† | B1*0301, 0404; B3*01†, B4*01† |
| KR | B1*0401, nd | nd | nd |
| PV | B1*0402, nd | 02†, 06† | B1*0901, 13†; B3*02†, B4*01† |
| PJ | B1*0401 | nd | nd |
| MJO | B1*0401 | 03†, 06† | B1*0401, 1501; B4*01†, B5*0100 |
| GL | B1*0401, nd | 02†, 0303 | B1*03†, 0901; B3*01†, B4*01† |
| Patients with NY-ESO-1-expressing tumor but no detectable Ab | |||
| FS | B1*04 negative | 0301, 0501 | B1*0101, 1101; B3*0202 |
| BFJ | B1*04 negative | 0201, 05† | B1*0701, 1601; B3*0101, B3*0202 |
| MJC | B1*04 negative | 0501 | B1*1501; B5*0101 |
| SH | B1*04 negative | nd | nd |
| LM | B1*04 negative | 03† | B1*0404, 0405; B4*01† |
| LB | B1*04 negative | 03†, 04† | B1*04†; B4*01† |
| KD | B1*0401 | 02†, 03† | B1*0301, 0401; B3*01†; B4*01† |
| CE | B1*0401, nd | nd | nd |
| EBVB lines used for antigen presentation | |||
| L007 EBVB | B1*0401 | 0602 | B1*1501 B5*0101 |
| L023 EBVB | B1*04 negative | 0301 | B1*1201 B3*0202 |
| L041 EBVB | B1*0402, nd | 0402 | B1*0822 nd |
| 836 EBVB | B1*0401, nd | 02† | B1*0701; B4*01† |
| 1363 EBVB | B1*0401 | 0501 | B1*0101; nd |
| 1088 EBVB | B1*0401 | 0201, 0301 | B1*0301, 04†; B3*0101, B4*01† |
| 586 EBVB | B1*0401 | 0501, 0201 | B1*0101, 07†; B4*01† |
nd: allele(s) not determined. The detection of the presence of NY-ESO-1 Abs in melanoma patients has been described (14). In this study, 17 of 148 patients were positive for NY-ESO-1 Ab. Fisher's exact test P value was 0.0009 for the association of DPB1*0401 and NY-ESO-1 Ab production.
Subtype unknown.
Recognition of a Naturally Processed NY-ESO-1 Epitope on Tumor Cells by TE4–2.
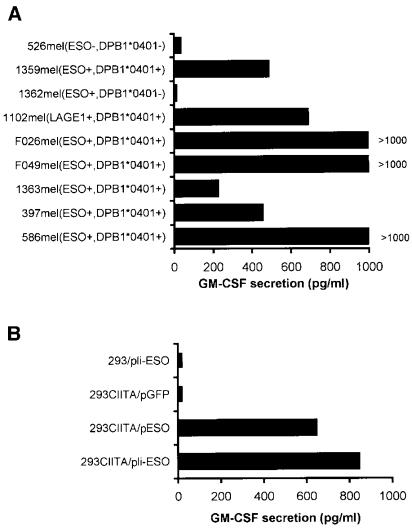
To investigate whether the T cell epitope recognized by TE4–2 was naturally processed and presented on the surface of tumor cells, TE4–2 T cells were tested against a number of tumor cell lines with known expression patterns for both NY-ESO-1 and DP4, and 293CIITA transfected with NY-ESO-1. As shown in Fig. 3A, TE4–2 T cells recognized multiple tumor lines expressing both NY-ESO-1 and DPB1*0401, but failed to recognize tumor lines 1362mel (NY-ESO-1+ DP4−) and 526mel (NY-ESO-1− DP4−). Interestingly, 1102mel (NY-ESO-1− DP4+) was recognized by TE4–2 T cells, but reverse transcriptase–PCR analysis demonstrated that 1102mel expressed the LAGE-1 gene, a cancer/testis antigen possessing approximately 90% amino acid similarity to NY-ESO-1 (20). A sequence identical to ESO p161–175 was also present in the LAGE-1 protein. In addition, TE4–2 T cells also recognized 293CIITA (HLA DPB1*0401 positive) transfected with either NY-ESO-1 or Ii-fused NY-ESO-1, but not with 293 (DPB1*0401 negative) transfected with the NY-ESO-1 gene or 293CIITA transfected with control plasmid pEGFP (Fig. 3B). These results suggested that epitopes derived from NY-ESO-1 and LAGE-1 were present on the surface of tumor cells for T cell recognition. Taken together, these results demonstrated that TE4–2 T cells recognized a naturally processed NY-ESO-1 epitope on the cell surface.
Figure 3.
Recognition of tumor cells and 293CIITA/NY-ESO-1 by TE4–2 CD4+ T cells. (A) TE4–2 recognized melanoma lines expressing both NY-ESO-1 and DP4. Melanoma lines with known NY-ESO-1 expression (by reverse transcriptase–PCR) and HLA DP types (determined by reverse transcriptase–PCR and sequencing) were used as targets. IFN-γ treatment (500 units/ml) was conducted for F026mel, 526mel, and 397mel to up-regulate their MHC class II expression in this experiment. TE4–2 T cells were cocultured with tumor cells overnight before cytokine release was measured. (B) TE4–2 CD4+ T cell line recognized 293CIITA transfected with NY-ESO-1 with or without the Ii targeting sequence. Parental 293 cells and 293CIITA cells were transfected with plasmid encoding NY-ESO-1 (pESO), Ii-NY-ESO-1 (pIi-ESO), or green fluorescent protein (pGFP), respectively. TE4–2 T cells were cocultured with the transfectants overnight before cytokine release was assayed.
Overlapping of DP4-Restricted Epitopes with an HLA-A2 Restricted Epitope.
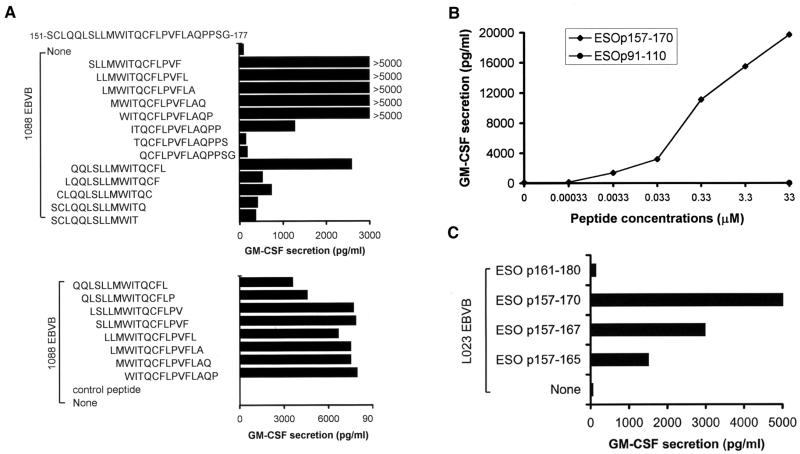
In an attempt to identify the anchor residues present between amino acids 161 and 175, a series of overlapping 13-mer peptides were used to pulse 1088EBVB cells and tested for their abilities to stimulate TE4–2 T cells. As shown in Fig. 4A, a partial loss of activity was observed when the W residue at position 161 was removed; and a complete loss of activity was observed when the I residue at position 162 was deleted. The deletion of a C-terminal L residue at position 167 also abolished the recognition of the peptide by TE4–2 T cells. Moreover, the residue V at position 169 appeared to be important, as deletion of this residue resulted in a 2-fold decrease in the peptide's stimulatory activity. These results indicated that the W residue at position 161 may be a P1 anchor, and the L residue at position 167 represented the P7 anchor. The V residue at position 169 also appeared to contribute to the stimulatory capacity of the peptide epitope, indicating that it may represent the P9 anchor residue. These putative anchor residues closely matched the previously described consensus HLA DPB1*0401 binding motif (21).
Figure 4.
Characterization of T cell epitopes recognized by TE4–2. (A) Determination of the anchor residues and minimal length of the NY-ESO-1 epitope for T cell recognition. Synthetic peptides with amino acid deletions at the N or C terminus were used to pulse 1088EBVB cells at 40 μg/ml. EBVB cells were then thoroughly washed and used as target cells to stimulate TE4–2 T cells. Two separate experiments (Upper and Lower) were conducted. (B) Determination of the minimal peptide concentration required for T cell recognition. The ESO p157–170 peptide was used to pulse 1088EBVB cells at various concentrations. The peptide-pulsed cells then were washed and used as targets to stimulate the TE4–2 line. A control peptide ESO p91–110 was used only at the highest concentration, 33 μM. (C) Recognition of the DPB1*0401-restricted CD4+ T cell epitope by the NY-ESO-1-specific CD8+ T cells. The TE8–1 cell line was generated from PBMCs of patient TE by in vitro stimulation with the ESO p157–167 peptide. L023 EBVB cells (HLA-A2+, DP4−) were pulsed with peptides covering the DPB1*0401 epitope region in a serum-free medium, washed, and used to stimulate TE8–1 T cells.
The ESO p157–170 peptide, which contained all three anchored residues, was used in the titration experiment to determine the minimal stimulatory concentration for the peptide. The results demonstrated that ESO p157–170 was able to stimulate significant cytokine release from TE4–2 T cells at a minimum concentration between 3 and 33 nM (Fig. 4B). These results indicated that TE4–2 T cells recognized ESO p157–170 with a relatively high affinity compared with several known MHC class II binding epitopes from nonmutated peptides, such as those from gp100 (22), tyrosinase (26), and CDC-27 (18).
Interestingly, a previously identified HLA-A2 epitope, ESO p157–167 (11), was contained within the DPB1*0401–0402 epitope, ESO p157–170. To assess whether the HLA-DP epitope may be presented by HLA-A2 and cross-react with CD8+ T cells, ESO p157–170 was tested for recognition by TE8–1, a CD8+ T cell line specifically recognizing the HLA-A2-restricted ESO p157–167. ESO p157–170 was able to stimulate significant cytokine releases from TE8–1 T cells when pulsed onto L023 EBVB cells (HLA-A2+ DP4−) (Fig. 4C), indicating that the ESO p157–170 epitope had dual specificity for stimulating both CD4+ and CD8+ T cells. Therefore, ESO p157–170 might be an attractive candidate for cancer vaccines aimed at eliciting both tumor-specific CD4+ and CD8+ T cell immune responses.
Association of the NY-ESO-1 Ab Production with HLA DP4.
HLA DP4 is a dominant MHC class II allele present in a large portion of Caucasians, ranging between 43% and 70% in population studies involving different ethnic groups (16). A previous study (12) showed that about 10% of patients with advanced disease developed NY-ESO-1-specific Ab. In our study, we found that 11 of 88 (13%) melanoma patients tested were found to have high titers of NY-ESO-1 Ab (14). Because NY-ESO-1 is expressed in 20% of melanoma and HLA-DR4 expressed in 15% of the population, the previously identified DR4-restricted CD4+ T cell peptides only account for 3% of patients who have the potential to develop NY-ESO-1-specific Ab. Evidently, DR4-restricted CD4+ T cells cannot account for the Ab production in 10–13% of patients observed. To further investigate whether NY-ESO-1-specific DP4-restricted CD4+ T cells were associated with the production of NY-ESO-1-specific Ab in these melanoma patients, we first analyzed their HLA DP subtypes. Sixteen of seventeen (94%) patients with NY-ESO-1 Ab were found to be positive for DP4, whereas no predominant DQ or DR restriction elements could be identified in this group of patients (Table 1). Six of eight patients with known NY-ESO-1-expressing tumors but with no detectable Ab were negative for DP4. A P value of 0.0009 was obtained from a Fisher's exact test, indicating the significance of the association between Ab responses and the HLA-DP4 expression. Based on the expression frequency for NY-ESO-1 (20%) and DP4 (43–70%), the percentage of patients expressing both NY-ESO-1 and DP4 and with the potential to develop Ab responses is in the range of 8.6% to 14%. This prediction is very close to the observed 10–13% frequency of patients with NY-ESO-1 Ab.
To obtain additional evidence as to the association between NY-ESO-1 Ab response and the DP4 expression, PBMCs from six patients with NY-ESO-1 Ab were used for in vitro stimulation with the ESO p161–180 peptide, and PBMCs from two DP4-negative patients with no detectable NY-ESO-1 Ab were used as controls. T cells were examined for their response to 293CIITA cells pulsed with the ESO p161–180 peptide after two or three rounds of in vitro stimulation. CD4+ T cells from five of six patients with NY-ESO-1 Ab showed a specific recognition of the ESO p161–180 epitope presented by 293CIITA cells (DP4+ and HLA-A2−) (Table 2). Two of them also showed significant tumor recognition of DP4+ and NY-ESO-1+ melanoma lines without further enrichment of the CD4+ T cells (data not shown). In contrast, no NY-ESO-1-reactive T cells were generated by using PBMCs from two patients (WC and EW) with no detectable NY-ESO-1 Ab after three stimulations. It should be noted that although in this particular experiment T cells from patients TE and FJ exhibited relatively low activities, high T cell activity was originally generated from the same patient TE as well as in other experiments. On the other hand, we were not able to generate any T cell activity from patients without NY-ESO-1 Ab. These results suggested a strong correlation between the frequency of CD4+ T cells precursor reactive with the DP4-restricted NY-ESO-1 epitope and the production of anti-NY-ESO-1 Ab in melanoma patients. Taken together, these results support the notion that NY-ESO-1-specific DP4-restricted CD4+ T cells may contribute to the development of Ab responses against the NY-ESO-1 tumor antigen.
Table 2.
Recognition of DPB1*0401-restricted ESO p161-180 by CD4+ T cells generated from patients with and without specific Ab responses
| Patients | T cell reactivity, pg/ml IFN-γ
secretion
|
Ab responses | DPB1*0401 | |
|---|---|---|---|---|
| Irrelevant peptide† | ESO p161–180 | |||
| BE | 0 | 0 | + | + |
| FJ | 0 | 160 | + | + |
| CJ | 150 | 475 | + | + |
| CT | 150 | 2,350 | + | + |
| BL | 180 | 1,089 | + | + |
| TE | 90 | 207 | + | + |
| WC‡ | 0 | 0 | − | + |
| EW‡ | 0 | 0 | − | + |
293CIITA cells (DPB1*0401 positive and HLA-A2 negative) were pulsed with the indicated peptides and used as targets. Cultures showing more than 100 pg/ml IFN-γ production in response to ESO p161–180 pulsed targets and at least 2-fold above the background were defined as positive. Values of cytokine secretion were from representative positive wells.
Tumor sample of WC was not available for the determination of the expression of NY-ESO-1. Expression of NY-ESO-1 in tumors from patient EW was negative.
Discussion
NY-ESO-1 is an important immune target because it gives rise to both humoral and cellular immune responses (9, 11). Of particular interest, high titered NY-ESO-1 reactive Abs were frequently detected in patients with cancer (12, 14) whereas a very low percentage of patients developed high titers of Abs against the MAGE antigens or differentiation antigens such as tyrosinase, gp100, TRP-1, and TRP-2 (12). The use of recombinant protein expressed in bacteria may not detect Ab responses against glycosylated epitopes as well as native conformational epitopes, but a similar conclusion has been made based on the use of protein translated in an in vitro eukaryotic expression system (23). These studies strongly suggest that NY-ESO-1-reactive CD4+ T cells may be involved in Ab production and cytotoxic T lymphocyte proliferation. In this end, HLA-DR4-restricted T cell epitopes derived from NY-ESO-1 were shown to be presented by HLA-DR4 (DRB1*0401) and (DRB4*0101–0103) molecules to CD4+ T cells (14, 15). However, HLA-DR4 is only present in about 15% of the population, suggesting that other MHC class II molecules may play an important role in presenting NY-ESO-1 epitopes to CD4+ T cells. The studies described above provide several lines of evidence that DP4-restricted CD4+ T cells may play a role in antitumor immunity.
First, in animal studies, antitumor immunity and autoimmunity mediated by gp75/TRP-1 appeared to require CD4+ T cells and Abs (24). Immunization of mice with hTRP-2, but not mTRP-2, broke tolerance to the self-antigen and the antitumor immunity required the participation of both CD4+ and CD8+ T cells (25). These studies suggested that antitumor immunity could be mediated by either Abs or CD8+ T cells, but both require the critical help of CD4+ T cells (25). DP4-restricted CD4 T cells may provide help for both CD8+ T cell proliferation as well as B cell activation. Second, CD4+ T cell responses are restricted by the highly polymorphic MHC class II molecules. A few MHC class II-restricted tumor antigens recently identified by using tumor-reactive CD4+ T cells are presented by HLA-DR molecules with a frequency of less than 20% of the population. The majority of these antigens are mutated antigens. Because of this and the polymorphic property of DR alleles, these antigens are not very useful in clinical application. The T cell epitopes identified in this study are nonmutated peptides presented by a dominated DP4 allele with a frequency of 43–70% of the population. Hence, these CD4+ T cell epitopes can be used in up to 70% of patients with cancer. Third, 16 of 17 (94%) patients with high titered NY-ESO-1-specific Ab are DP4-positive, whereas six of eight (75%) patients with NY-ESO-1 expressing tumor and no detectable NY-ESO-1 Ab are negative for DP4 (Table 1). More importantly, DP4-restricted CD4+ T cells were generated from five of six DP4-positive patients with high titered Ab (Table 2). These results suggest a strong association between Ab production and DP4-restricted CD4+ T cells. Finally, the DP4-restricted NY-ESO-1 peptides identified in this study contain both MHC class I and II-restricted T cell epitopes and may be useful in immunotherapy aimed at inducing both CD4+ and CD8+ T cell responses. These findings will provide an opportunity for developing more effective cancer vaccines for the treatment of patients with cancer.
Acknowledgments
We thank Mona El-Gamil and Yong F. Li for technical assistance, Dr. John Wunderlich and the T cell lab for clinical samples, Dr. Maria Parkhurst and John Riley for peptide synthesis, and Arnold Mixon and Shawn Farid for FACS analysis. We also thank Drs. C. E. Touloukian, K. Hanada, N. P. Restifo, and S. L. Topalian for helpful discussions.
Abbreviations
- EBVB
Epstein–Barr virus transformed B lymphocytes
- Ii
invariant chain
- PBMC
peripheral blood mononuclear cell
- GM-CSF
granulocyte–macrophage colony-stimulating factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rosenberg S A. Immunity. 1998;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 2.Wang R F, Rosenberg S A. Immunol Rev. 1999;170:85–100. doi: 10.1111/j.1600-065x.1999.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 3.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Rivoltini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R F, Robbins P F, Kawakami Y, Kang X, Rosenberg S A. J Exp Med. 1995;181:799–804. doi: 10.1084/jem.181.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R F, Appella E, Kawakami Y, Kang X, Rosenberg S A. J Exp Med. 1996;184:2207–2216. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Sakaguchi K, Appella E, Yannelli J R, Adema G J, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y T, Scanlan M J, Sahin U, Tureci O, Gure A O, Tsang B, Williamson E, Stockert E, Pfreundschuh M, Old L J. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R F, Johnston S L, Zeng G, Topalian S L, Schwartzentruber D J, Rosenberg S A. J Immunol. 1998;161:3596–3606. [PubMed] [Google Scholar]
- 10.Robbins P F, El-Gamil M, Li Y F, Kawakami Y, Loftus D, Appella E, Rosenberg S A. J Exp Med. 1995;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jager E, Chen Y T, Drijfhout J W, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, et al. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockert E, Jager E, Chen Y T, Scanlan M J, Gout I, Karbach J, Arand M, Knuth A, Old L J. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardoll D M, Topalian S L. Curr Opin Immunol. 1998;10:588–594. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- 14.Zeng G, Touloukian C E, Wang X, Restifo N P, Rosenberg S A, Wang R F. J Immunol. 2000;165:1153–1159. doi: 10.4049/jimmunol.165.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager E, Jager D, Karbach J, Chen Y T, Ritter G, Nagata Y, Gnjatic S, Stockert E, Arand M, Old L J, Knuth A. J Exp Med. 2000;191:625–630. doi: 10.1084/jem.191.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gjertson D W, Geer L, Lee S-H, Kawata J, Sutrisno R. In: HLA 1997. Terasaki P, Gjertson D W, editors. Los Angeles: UCLA Tissue Typing Laboratory; 1997. pp. 174–427. [Google Scholar]
- 17.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang R F, Wang X, Atwood A C, Topalian S L, Rosenberg S A. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 19.Walter E A, Greenberg P D, Gilbert M J, Finch R J, Watanabe K S, Thomas E D, Riddell S R. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 20.Lethe B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. Int J Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Rammensee H G, Friede T, Stevanoviic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 22.Touloukian C, Leitner W W, Topalian S L, Li Y F, Robbins P F, Rosenberg S A, Restifo N P. J Immunol. 2000;164:3535–3542. doi: 10.4049/jimmunol.164.7.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp E H, Gawkrodger D J, Watson P F, Weetman A P. Clin Exp Immunol. 1997;109:495–500. doi: 10.1046/j.1365-2249.1997.4781381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overwijk W W, Lee D S, Surman D R, Irvine K R, Touloukian C E, Chan C C, Carroll M W, Moss B, Rosenberg S A, Restifo N P. Proc Natl Acad Sci USA. 1999;96:2982–2987. doi: 10.1073/pnas.96.6.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowne W B, Srinivasan R, Wolchok J D, Hawkins W G, Blachere N E, Dyall R, Lewis J J, Houghton A N. J Exp Med. 1999;190:1717–1722. doi: 10.1084/jem.190.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topalian S L, Gonzales M L, Parkhurst M, Li Y F, Southwood S, Sette A, Rosenberg S A, Robbins P F. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]