Abstract
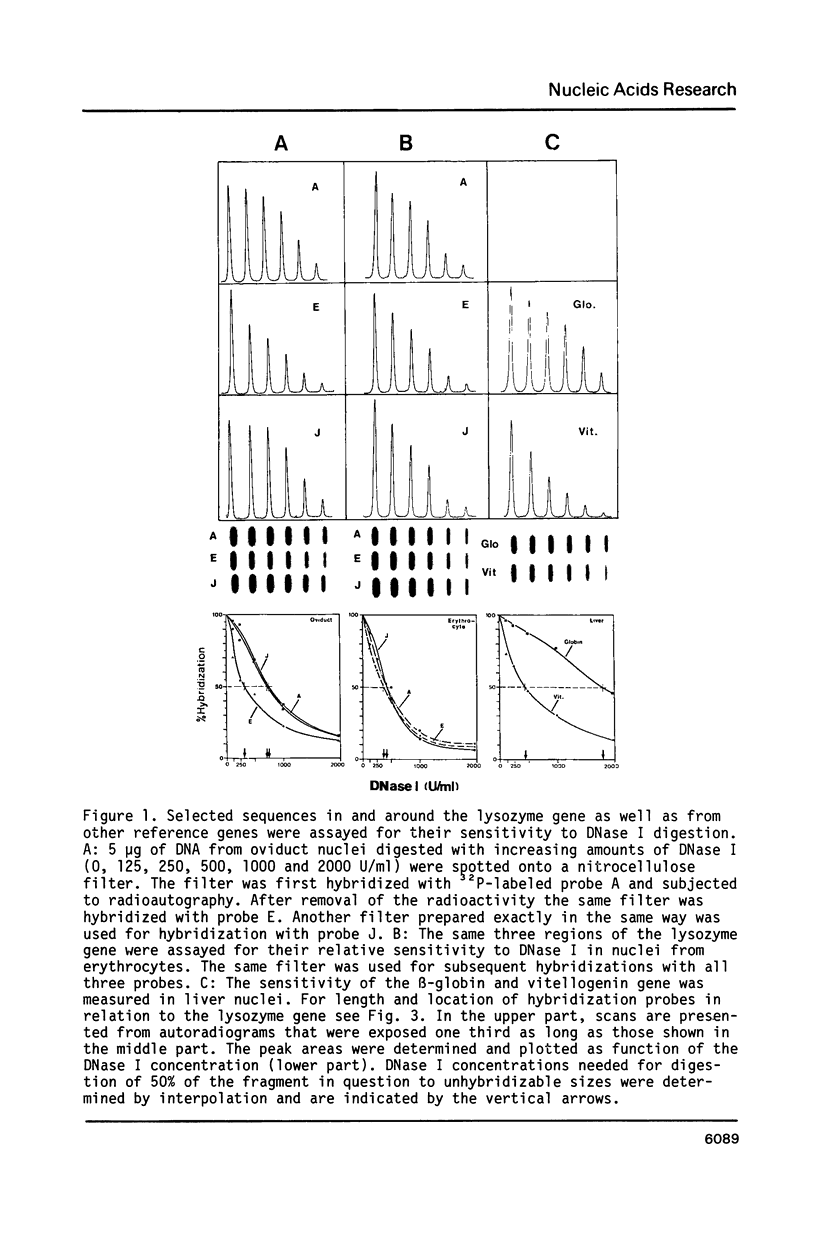
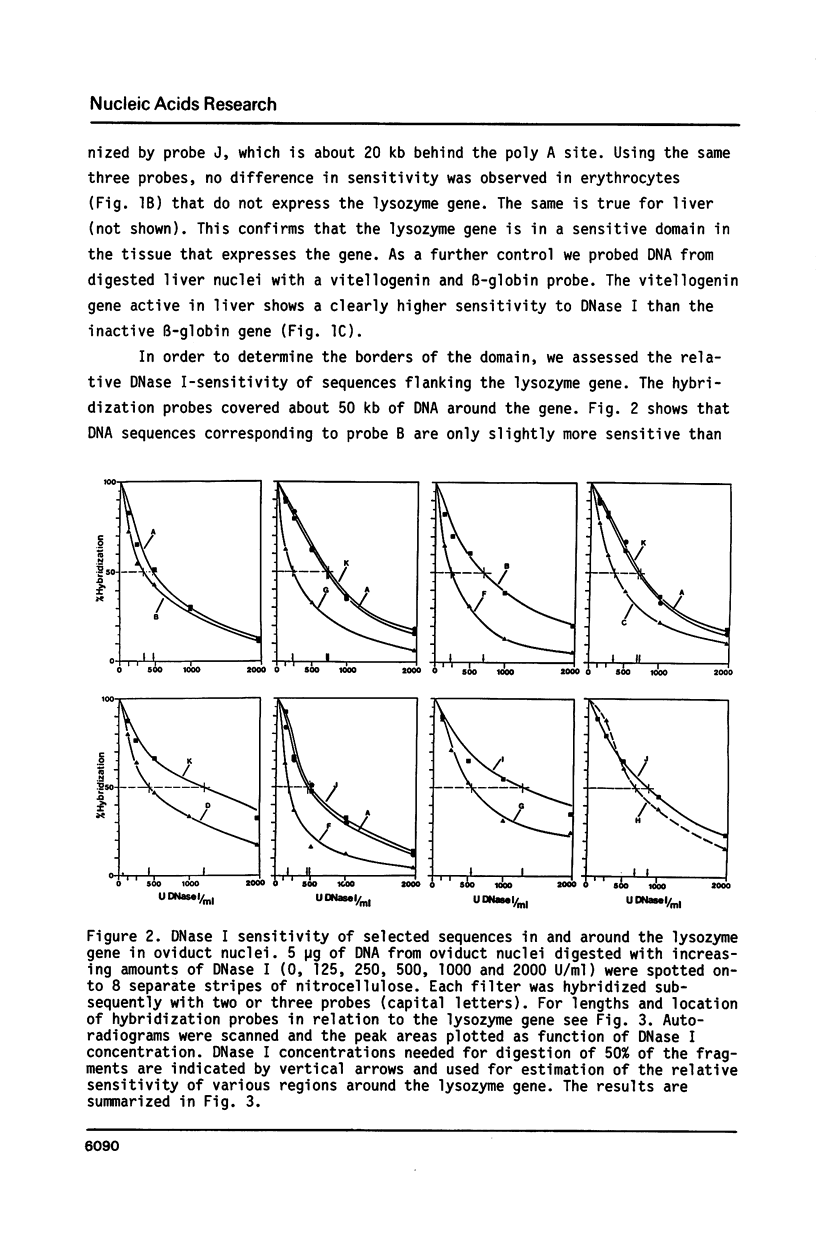
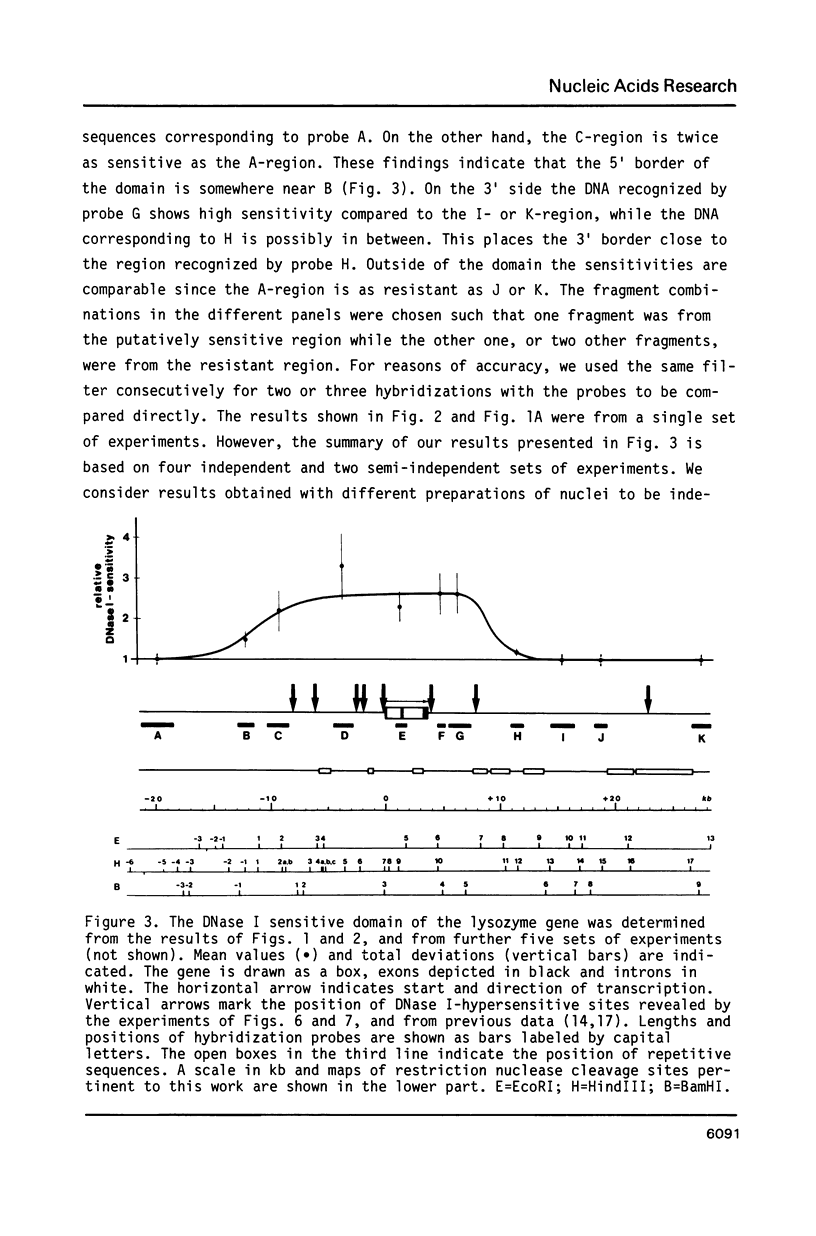
We have determined the DNase I sensitive chromatin domain of the lysozyme gene in the hen oviduct. When nuclei were digested with DNase I, about 14 kb of upstream and 6 kb of downstream sequences in addition to the 4 kb long transcribed region were preferentially degraded. The transcription start site is located near the center of the approximately 24 kb long sensitive domain. At the 3' boundary there is a rather abrupt transition from the DNase I sensitive to the resistant chromatin configuration whereas at the 5' border this transition occurs in a gradual fashion over 6-7 kb of DNA. No obvious correlation between the boundaries of the domain and repetitive sequences could be established. DNase I-hypersensitive sites are clustered within the boundaries of the sensitive domain which seems to represent a functional unit of the gene.
Full text
PDF




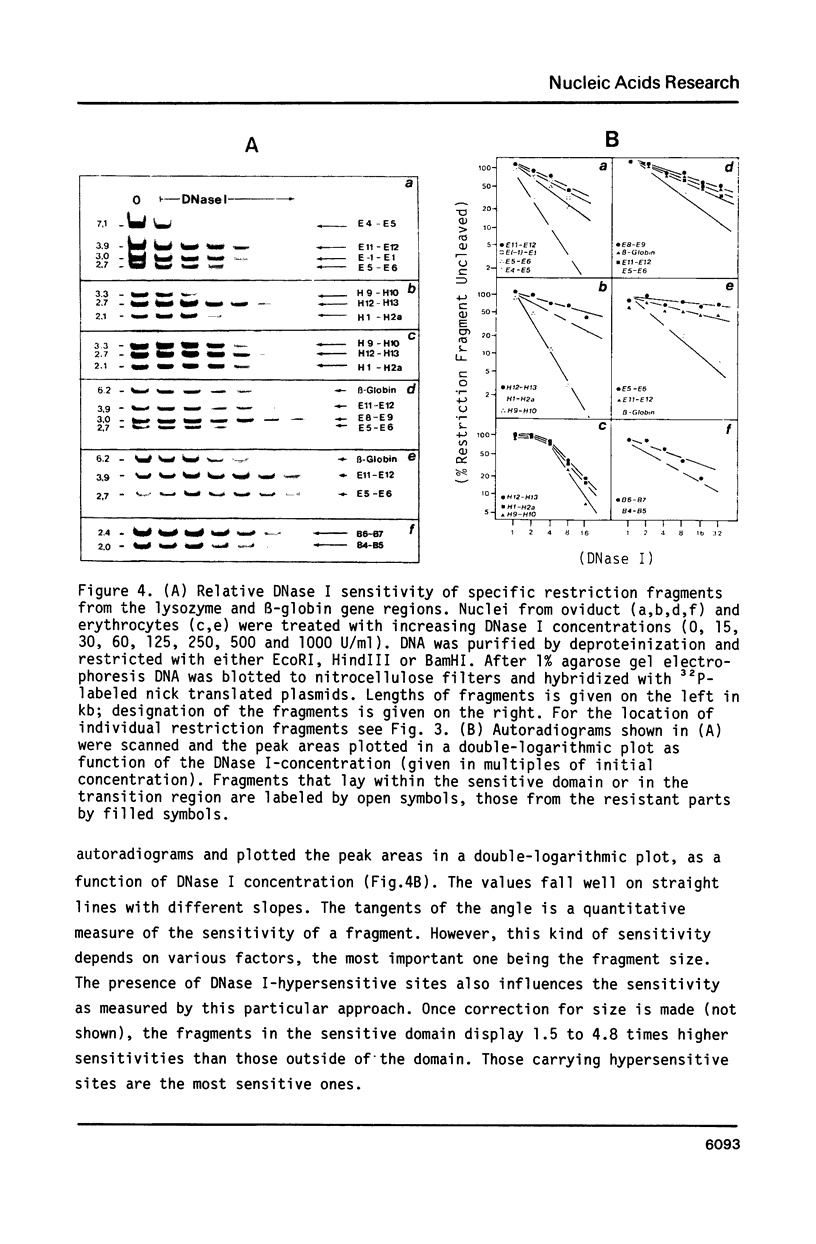
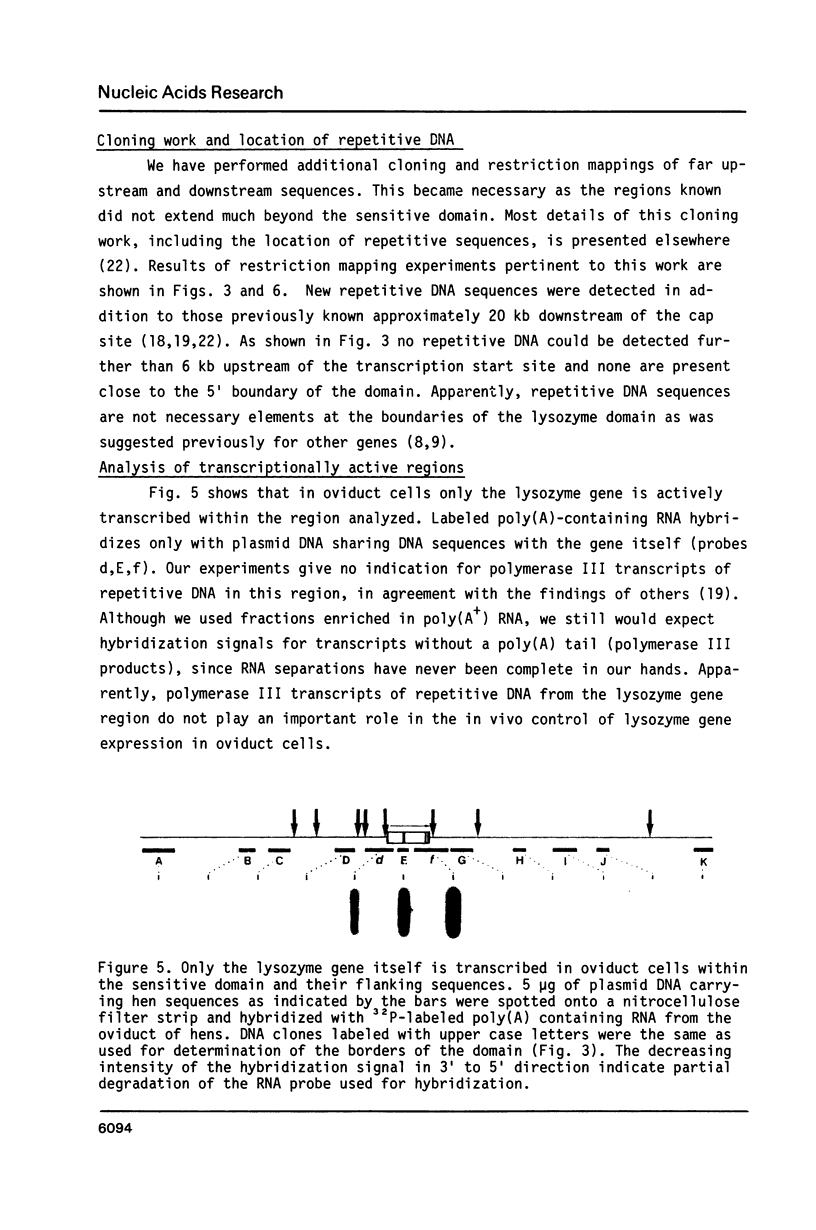





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alevy M. C., Tsai M. J., O'Malley B. W. DNase I sensitive domain of the gene coding for the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1984 May 8;23(10):2309–2314. doi: 10.1021/bi00305a034. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Baldacci P., Royal A., Brégégère F., Abastado J. P., Cami B., Daniel F., Kourilsky P. DNA organisation in the chicken lysozyme gene region. Nucleic Acids Res. 1981 Aug 11;9(15):3575–3588. doi: 10.1093/nar/9.15.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Strommer J., Engel J. D. Isolation of the chicken beta-globin gene and a linked embryonic beta-like globin gene from a chicken DNA recombinant library. Cell. 1979 Aug;17(4):879–887. doi: 10.1016/0092-8674(79)90328-3. [DOI] [PubMed] [Google Scholar]
- Dolan M., Dodgson J. B., Engel J. D. Analysis of the adult chicken beta-globin gene. Nucleotide sequence of the locus, microheterogeneity at the 5'-end of beta-globin mRNA, and aberrant nuclear RNA species. J Biol Chem. 1983 Mar 25;258(6):3983–3990. [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Forrester W. C., Thompson C., Elder J. T., Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H. J., Domdey H., Lossow C., Jank P., Raba M., Alberty H., Sänger H. L. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978 May 18;273(5659):203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Lawson G. M., Knoll B. J., March C. J., Woo S. L., Tsai M. J., O'Malley B. W. Definition of 5' and 3' structural boundaries of the chromatin domain containing the ovalbumin multigene family. J Biol Chem. 1982 Feb 10;257(3):1501–1507. [PubMed] [Google Scholar]
- Leegwater P. A., van der Vliet P. C., Rupp R. A., Nowock J., Sippel A. E. Functional homology between the sequence-specific DNA-binding proteins nuclear factor I from HeLa cells and the TGGCA protein from chicken liver. EMBO J. 1986 Feb;5(2):381–386. doi: 10.1002/j.1460-2075.1986.tb04223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmaier W., Nguyen-Huu M. C., Lurz R., Stratmann M., Blin N., Wurtz T., Hauser H. J., Sippel A. E., Schütz G. Arrangement of coding and intervening sequences of chicken lysozyme gene. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6196–6200. doi: 10.1073/pnas.76.12.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Nicolas R. H., Wright C. A., Cockerill P. N., Wyke J. A., Goodwin G. H. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983 Feb 11;11(3):753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek M., Ziegenmeyer J., Heckman J., Rajbhandary U. L. Absence of the sequence G-T-psi-C-G(A)- in several eukaryotic cytoplasmic initiator transfer RNAs. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1041–1045. doi: 10.1073/pnas.70.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D., Yu J., Annunziato A., Seale R. L. beta-Globin gene family in murine erythroleukemia cells resides within two chromatin domains differing in higher order structure. Biochemistry. 1984 Jun 19;23(13):2970–2976. doi: 10.1021/bi00308a019. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanev R. Transcriptionally active chromatin. Mol Biol Rep. 1983 May;9(1-2):9–17. doi: 10.1007/BF00777468. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]