Abstract
DNA methyltransferase 1 (DNMT1) is the primary enzyme that maintains DNA methylation. We describe a previously unknown mode of regulation of DNMT1 protein stability through the coordinated action of an array of DNMT1-associated proteins. DNMT1 was destabilized by acetylation by the acetyltransferase Tip60, which triggered ubiquitination by the E3 ligase UHRF1, thereby targeting DNMT1 for proteasomal degradation. In contrast, DNMT1 was stabilized by histone deacetylase 1 (HDAC1) and the deubiquitinase HAUSP (herpes virus–associated ubiquitin-specific protease). Analysis of the abundance of DNMT1 and Tip60, as well as the association between HAUSP and DNMT1, suggested that during the cell cycle the initiation of DNMT1 degradation was coordinated with the end of DNA replication and the need for DNMT activity. In human colon cancers, the abundance of DNMT1 correlated with that of HAUSP. HAUSP knockdown rendered colon cancer cells more sensitive to killing by HDAC inhibitors both in tissue culture and in tumor xenograft models. Thus, these studies provide a mechanism-based rationale for the development of HDAC and HAUSP inhibitors for combined use in cancer therapy.
INTRODUCTION
DNA methylation is involved in key biological processes including differentiation, imprinting, and X chromosome inactivation (1). Failure to maintain proper DNA methylation results in developmental disorders as well as cancer (2). DNA methyltransferases (DNMTs) 3a and 3b are required for de novo DNA methylation, whereas DNMT1 is involved in the maintenance of DNA methylation patterns from parental cells to progeny cells (3). During DNA replication, DNMT1 is recruited to replication forks through its interaction with proliferating cell nuclear antigen (PCNA) (4). UHRF1 [ubiquitin-like with plant homeodomain (PHD) and ring finger domains 1; also called nuclear protein of 95 kD (NP95) and inverted CCAAT box binding protein of 90 kD (ICBP90)] recognizes hemimethylated DNA templates and tethers DNMT1 to replication forks to methylate newly synthesized DNA strands (5, 6). UHRF1 also contains a RING domain with ubiquitin E3 ligase activity that mediates ubiquitination of itself and histone H3 (7, 8). In addition to PCNA and UHRF1, DNMT1 interacts with histone deacetylases (HDACs) (9, 10). HDACs are generally believed to be recruited by DNMT1 and UHRF1 to repress gene expression or to form heterochromatin structures (11). However, Zhou et al. recently demonstrated that HDAC inhibitors induce degradation of DNMT1 (12), suggesting that HDACs may be directly involved in regulating DNMT1 stability.
Mounting evidence suggests that tumor development can be suppressed by maintenance of the proper abundance of DNMT1. Mice that undergo transient inactivation of DNMT1 during embryogenesis suffer global loss of imprinting and develop widespread tumors in adulthood (13). Conversely, overexpression of DNMT1 transforms NIH 3T3 cells (14), and DNMT1 abundance is increased in many cancer types (15). Furthermore, earlier studies suggested that the increased DNMT1 abundance seen in cancers is largely due to dysregulation of DNMT1 protein stability rather than higher messenger RNA (mRNA) abundance (16, 17). Thus, these observations support the premise that fine-tuning DNMT1 abundance through posttranslational regulation is important in suppressing tumorigenesis. However, the mechanisms underlying DNMT1 regulation are poorly understood. Here, we provide support for a model of regulation of DNMT1 abundance and report how an array of interacting proteins can coordinately regulate stability or degradation of this critical enzyme by respectively controlling its deubiquitination and its acetylation-driven ubiquitination.
RESULTS
DNMT1 associates with the deubiquitinase HAUSP
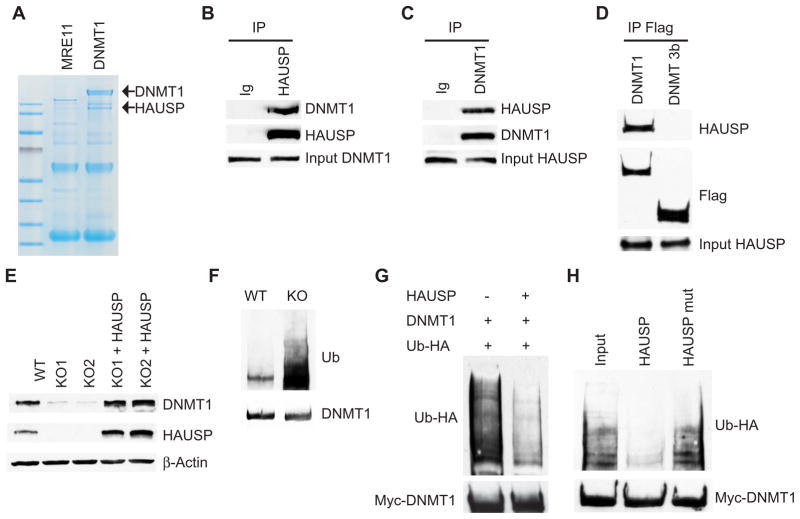
To identify DNMT1-associated proteins, we generated 3xFlag-tagged DNMT1 knock-in cells, using recombinant adeno-associated virus (rAAV)–mediated homologous recombination to place three Flag epitope tag sequences (18) at the C terminus of endogenous DNMT1 protein (fig. S1, A and B). Antibodies directed against Flag immunoprecipitated a protein of ~130 kD from 3xFlag-tagged DNMT1 knock-in cells, but not from knock-in cells harboring endogenously Flag-tagged Mre11 protein (Fig. 1A). Analysis by mass spectrometry (MS) identified the protein as HAUSP (herpes virus–associated ubiquitin-specific protease; also known as USP7) (fig. S2). This interaction was validated through reciprocal immuno-precipitation (Fig. 1, B and C) and by demonstrating nuclear colocalization of DNMT1 and HAUSP (fig. S3A). HAUSP specifically interacts with DNMT1 and does not coimmunoprecipitate with DNMT 3b (Fig. 1D and fig. S1C). Using a series of DNMT1 deletion constructs, we determined that DNMT1 interaction with HAUSP requires a region encompassing the zinc finger and the two BAH (bromo-adjacent homology) domains (fig. S3, B and C).
Fig. 1.
HAUSP regulates DNMT1 protein stability. (A) Copurification of DNMT1 with HAUSP. Lysates from DNMT1-3xFlag knock-in (KI) or Mre11-3xFlag KI cells were immunoprecipitated with antibody-conjugated beads against Flag, resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and stained with GelCode Blue Reagent. (B and C) DNMT1 interacts with HAUSP. RKO cell lysates were immunoprecipitated (IP) with control immunoglobulin G (IgG), antibody against HAUSP or against DNMT1, and then blotted with the indicated antibodies. (D) HAUSP does not interact with DNMT 3b. Cell lysates from DNMT1 and DNMT 3b 3xFlag-tagged KI cells were immunoprecipitated with antibody against Flag and blotted with antibody against HAUSP. (E) DNMT1 abundance is decreased in HAUSP knockout (KO) cells. Cell lysates from DLD1 parental [wild-type (WT)], KO, or KO cells ectopically expressing HAUSP were blotted with the indicated antibodies. KO1 and KO2 are two independently derived HAUSP KO clones. (F) DNMT1 ubiquitination is increased in HAUSP KO cells. HAUSP WT and KO DLD1 cells were treated with 5 μM MG132 for 6 hours. DNMT1 immunoprecipitates were blotted with an antibody against ubiquitin (Ub). (G) HAUSP deubiquitinates DNMT1 in cells. HEK 293 cells were transfected with the indicated plasmids and treated with MG132. DNMT1 immunoprecipitates were blotted with an antibody against HA to detect ubiquitinated DNMT1. (H) HAUSP deubiquitinates DNMT1 in vitro. Ubiquitinated DNMT1 proteins were immunoprecipitated from HEK 293 cells. Equal amounts of recombinant HAUSP or active-site mutant (HAUSP C223S) proteins were added to the immunocomplexes. Western blots were performed to detect either ubiquitinated or total DNMT1 proteins.
HAUSP deubiquitinates DNMT1 and protects it from proteasomal degradation
HAUSP is particularly interesting because it deubiquitinates MDM2, FOXO4, PTEN, and Claspin (19–23). Thus, we proceeded to determine whether HAUSP stabilizes DNMT1 through deubiquitination to protect it from proteasomal degradation. First, we showed that knockout of HAUSP in DLD1 colorectal cancer cells or knockdown of HAUSP in RKO colorectal cancer cells and human embryonic kidney (HEK) 293 cells led to reduced DNMT1 abundance (Fig. 1E and fig. S4, A and B), indicating that HAUSP regulates DNMT1 protein stability in both normal and cancer cells. Furthermore, reconstitution of HAUSP expression in knockout cells restored DNMT1 abundance to an extent comparable to that in parental cells (Fig. 1E), suggesting that loss of HAUSP is the sole cause of DNMT1 instability. Second, DNMT1 ubiquitination was increased in the HAUSP knockout cells (Fig. 1F), whereas overexpression of HAUSP led to deubiquitination of DNMT1 (Fig. 1G). Third, purified HAUSP recombinant proteins directly deubiquitinated DNMT1 in vitro (Fig. 1H and fig. S4E). Experimental controls further showed that (i) a HAUSP C223S (active site) mutant could not remove ubiquitin moieties from DNMT1 (Fig. 1H), and (ii) overexpressed HAUSP failed to deubiquitinate a DNMT1 mutant lacking the HAUSP interaction domain (DNMT1Δ) (fig. S4, C and D). A HAUSP knockout DLD1 cell line, which lacks fully functional p53, was engineered for this study, because the HCT116 HAUSP knockout cells generated by Cummins et al. (19) exhibited increased p53 abundance, which we speculate may be why these cells undergo senescence and stop growing after a few passages.
Acetylation of DNMT1 is directly involved in its proteasomal degradation
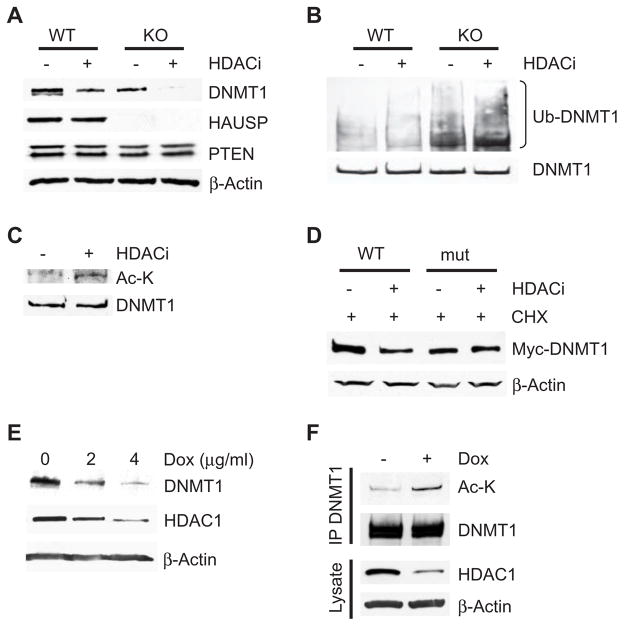
In our search for factors that might oppose the effect of HAUSP on DNMT1 proteasomal degradation, we noted that HDAC inhibitors induce ubiquitination-mediated proteasomal degradation of DNMT1 (12). We subsequently found that the HDAC inhibitor MS-275 directly destabilizes DNMT1 by enhancing its ubiquitination (Fig. 2, A and B) but does not appear to decrease its gene transcription (fig. S5A). Furthermore, loss of HAUSP combined with inhibition of HDAC activity had a synergistic effect on DNMT1 protein stability, because treatment of HAUSP knockout cells with various HDAC inhibitors reduced DNMT1 abundance to nearly undetectable amounts (Fig. 2A and fig. S5B). Moreover, inhibition of HDAC activity induced more DNMT1 ubiquitination in HAUSP knockout cells than in comparably treated wild-type cells (Fig. 2B).
Fig. 2.
Inhibition of HDAC1-mediated DNMT1 deacetylation promotes DNMT1 proteasomal degradation. (A) Knockout of HAUSP potentiates HDAC inhibitor (HDACi)–induced DNMT1 degradation. Parental or HAUSP KO DLD1 cells were treated or not with 5 μM HDACi MS-275 for 72 hours and cell lysates were blotted with the indicated antibodies. (B) HDAC inhibition induces DNMT1 ubiquitination. HAUSP WT or KO cells were treated with or without HDACi for 24 hours and MG132 for 12 hours before being harvested to make cell lysates. DNMT1 immunoprecipitates were blotted with an antibody against ubiquitin. Because the abundance of DNMT1 in the HAUSP KO cells is lower than in WT cells, more KO cells were used than WT cells to obtain equal amounts of precipitated DNMT1 proteins. (C) DNMT1 is acetylated after HDACi treatment. DNMT1 immunoprecipitates from cells treated with HDACi were blotted with an antibody against acetylated lysine (Ac-K). (D) A DNMT1 acetylation site mutant is resistant to HDACi-induced degradation. HEK 293 cells were transfected with WT DNMT1 or a DNMT1 mutant lacking four known acetylation sites (K173R, K1113R, K115R, and K117R) and treated with MS-275 for 48 hours and with CHX for 24 hours. Cell lysates were blotted with the indicated antibodies. (E) Knockdown of HDAC1 decreases the abundance of DNMT1. RKO cells were treated with the indicated concentration of doxycycline (Dox) for 48 hours to induce expression of an shRNA directed against HDAC1. Western blots were performed with the indicated antibodies. (F) Knockdown of HDAC1 leads to increased acetylation of DNMT1. RKO cells expressing an inducible HDAC1 shRNA were treated with or without Dox (4 μg/ml) for 36 hours and then with MG132 for 12 hours. DNMT1 immunoprecipitates were blotted with an antibody against Ac-K. Cell lysates were also blotted with antibodies against HDAC1 and β-actin. See fig. S11 for bar graphs and statistical analyses.
We then showed that acetylation of DNMT1 plays a direct role in this process. Inhibition of HDAC activity increased DNMT1 acetylation (Fig. 2C). The functional importance of DNMT1 acetylation was then demonstrated by showing that inhibition of HDAC activity failed to induce degradation of a DNMT1 mutant in which the four previously identified acetylated lysines (24, 25) were altered to arginines (Fig. 2D).
HDAC1 deacetylates DNMT1 and protects it from proteasomal degradation
Because MS-275 is an HDAC1-specific inhibitor and because HDAC1 associates with DNMT1 (9), we set out to determine whether HDAC1-mediated deacetylation protects DNMT1 from degradation. First, we demonstrated that inducible knockdown of HDAC1 in RKO colorectal cancer cells led to reduced DNMT1 abundance, consistent with a similar observation made in a breast cancer cell line (Fig. 2E) (12). Knockdown of HDAC1 led to increased DNMT1 acetylation (Fig. 2F). These data suggest that HDAC1 deacetylates DNMT1, thus protecting it from proteasomal degradation.
Tip60 acetylates DNMT1 and promotes its degradation
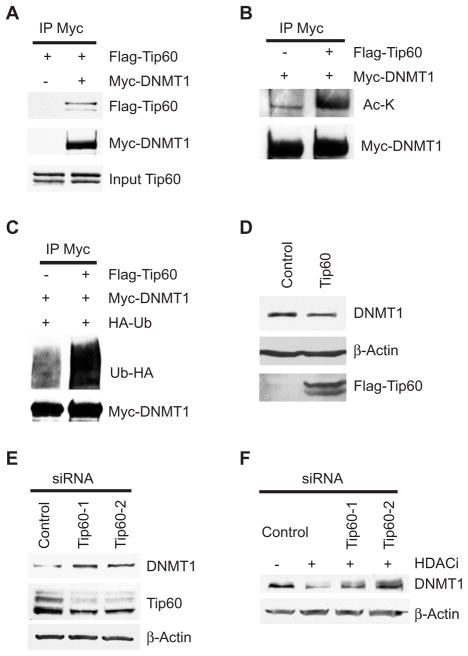
Upon examination of potential acetyltransferases that could acetylate DNMT1, our attention was drawn to Tip60 because of its ability to form complexes with UHRF1 (also known as NP95), which associates with DNMT1 (26). We demonstrated that Tip60 bound not only to UHRF1 but also to DNMT1 (Fig. 3A) and that overexpression of Tip60 led to increased DNMT1 acetylation and ubiquitination (Fig. 3, B and C). In support of the premise that Tip60-mediated acetylation regulates DNMT1 stability, overexpression of Tip60 also led to reduced abundance of endogenous DNMT1 (Fig. 3D). Conversely, knockdown of Tip60 by two different small interfering RNAs (siRNAs) resulted in increased DNMT1 abundance (Fig. 3E). Because commercially available antibodies directed against Tip60 were not optimal for Western blot analysis, we generated a 3xFlag-tagged Tip60 knock-in HCT116 cell line (fig. S1D).
Fig. 3.
Acetylation of DNMT1 by Tip60 promotes its degradation. (A) DNMT1 interacts with Tip60. (B) Tip60 acetylates DNMT1. (C) Overexpression of Tip60 leads to increased ubiquitination of DNMT1. Antibody against Myc immunoprecipitates from HEK 293 cells transfected with the indicated plasmids were immunoblotted with antibody against Flag to detect Tip60 (A), antibody against acetylated lysine to detect DNMT1 acetylation (B), or antibody against HA to detect ubiquitin (C). (D) Over-expression of Tip60 reduced the abundance of endogenous DNMT1. HEK 293 cells were transfected with either control plasmid or Tip60. Cell lysates were blotted with antibody against DNMT1 or antibody against Flag to detect Tip60. β-Actin was used as the loading control. (E) Knockdown of Tip60 leads to increased abundance of endogenous DNMT1. Tip60 Flag-tagged knock-in HCT116 cells were transfected with control siRNA or two independent siRNAs against Tip60. Western blots were used to quantify DNMT1 and Tip60 with antibodies against DNMT1 and Flag, respectively. (F) Knockdown of Tip60 blocks HDACi-induced DNMT1 degradation. Cells were transfected with control siRNA or siRNAs against Tip60 and treated with or without MS-275. Western blots were performed with the indicated antibodies. See fig. S11 for bar graphs and statistical analyses.
Furthermore, knockdown of Tip60 blocked DNMT1 degradation induced by inhibition of HDAC activity (Fig. 3F). Therefore, Tip60 enables HDAC inhibitor–induced degradation of DNMT1 by mediating DNMT1 acetylation.
UHRF1 mediates ubiquitination of DNMT1
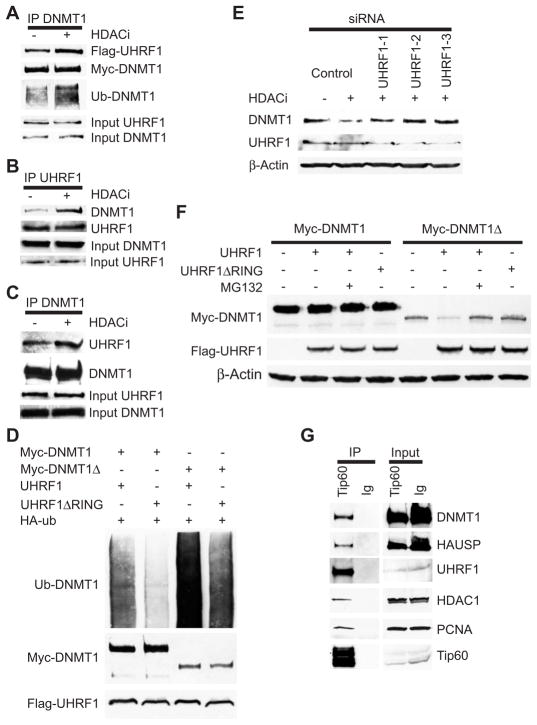
We postulated that acetylation of DNMT1 might promote its ubiquitination by recruiting or enhancing its interaction with an E3 ligase. As noted above, the RING domain–containing E3 ligase UHRF1 associates with DNMT1 (5, 6, 27); therefore, we tested whether acetylation of DNMT1 enhances its interaction with UHRF1. HDAC inhibition increased the interaction between endogenous and transfected DNMT1 and UHRF1 (Fig. 4, A to C). Furthermore, overexpression of UHRF1 led to increased ubiquitination of DNMT1 (Fig. 4D). However, knockdown of UHRF1 did not affect DNMT1 abundance (fig. S6A), but UHRF1-mediated ubiquitination was required for HDAC inhibitor–induced degradation of DNMT1. In addition, knockdown of UHRF1 by three different siRNAs blocked HDAC inhibitor–induced degradation of DNMT1 (Fig. 4E). Conversely, overexpression of UHRF1 reduced the abundance of a DNMT1 mutant lacking the HAUSP interaction domain (DNMT1Δ) (Fig. 4F). Furthermore, overexpression of UHRF1 ΔRING did not decrease the abundance of DNMT1Δ (Fig. 4F), showing that this regulation requires the E3 ligase activity of UHRF1. Overexpression of UHRF1 had no effect on the abundance of full-length DNMT1 (Fig. 4F). This observation is consistent with a requirement of HAUSP to interact with full-length DNMT1 to antagonize UHRF1-mediated ubiquitination of DNMT1.
Fig. 4.
The E3 ligase UHRF1 ubiquitinates DNMT1. (A) HDAC inhibition enhances DNMT1 interaction with UHRF1. HEK 293 cells were transfected with plasmids expressing Myc-DNMT1 and Flag-UHRF1 and treated with or without MS-275 for 24 hours. Myc-DNMT1 immunoprecipitates were blotted with the indicated antibodies. (B and C) HDAC inhibition enhances the interaction of endogenous DNMT1 and UHRF1. Cells were treated with or without MS-275 and UHRF1 (B) or DNMT1 (C) immunoprecipitates were blotted with the indicated antibodies. (D) UHRF1 ubiquitinates DNMT1. HEK 293 cells were transfected with the indicated plasmids. Antibodies against Myc immunoprecipitates were blotted with antibody against HA to detect ubiquitinated DNMT1. Myc-DNMT1Δ, DNMT1 mutant lacking the HAUSP-interacting domain. UHRF1ΔRING, UHRF1 with a RING domain deletion. (E) Knockdown of UHRF1 blocks HDACi-induced DNMT1 degradation. HEK 293 cells were transfected with control siRNA or siRNAs against UHRF1 and treated with or without MS-275. Western blotting was performed with the indicated antibodies. (F) Overexpression of UHRF1 leads to degradation of a DNMT1 mutant lacking the HAUSP-interacting domain (DNMT1Δ). Full-length DNMT1 or DNMT1Δ was cotransfected into HEK 293 cells with the indicated expression vectors. Cell lysates were blotted with the indicated antibodies. (G) DNMT1, HAUSP, UHRF1, HDAC1, and PCNA associate with Tip60. Flag-tagged Tip60 immunoprecipitates were blotted with the indicated antibodies. See fig. S11 for bar graphs and statistical analyses.
DNMT1, HAUSP, UHRF1, Tip60, HDAC1, and PCNA interact with each other
Results to this point demonstrated that DNMT1 abundance can be increased by interaction with HAUSP and decreased by interaction with UHRF1 and Tip60. We wanted to determine whether the proteins that regulate DNMT1 abundance also interacted with PNCA, a factor supporting possessive DNA replication that interacts with proteins at DNA replication forks. Immunoprecipitation of Tip60 not only coprecipitated DNMT1, HAUSP, UHRF1, and HDAC1, but also PCNA (Fig. 4G). In addition, these protein interactions were also validated by immunoprecipitation of both UHRF1 and PCNA (fig. S6, B and C). Moreover, the specificity of HDAC inhibitors for DNMT1 was supported by the observation that they induced DNMT1 degradation without affecting the intracellular abundance of the other components (HAUSP, UHRF1, HDAC1, Tip60, and PCNA) in the complex (fig. S5C).
Tip60 and HAUSP appear to regulate DNMT1 protein stability during the cell cycle
Although we had shown that a complex of proteins regulated DNMT1 stability, we had not shown that these interactions tracked with any cellular function. Therefore, we analyzed the temporal profile of DNMT1 abundance throughout the cell cycle to determine whether the interactions and specific abundance of the proteins regulating DNMT1 abundance changed during cell replication processes. Cells were synchronized at early S phase by sequential thymidine and aphidicolin blocks and then released. The abundance of DNMT1 started to decrease at either late S phase or G2 (5 hours after release) and the reduced abundance of DNMT1 persisted into the G1 phase of the next cell cycle (fig. S7, A and B). Furthermore, the abundance of Tip60 increased as the cell cycle progressed, and the abundance of DNMT1 and Tip60 appeared to be inversely correlated during the cell cycle. These data provide another line of evidence suggesting that Tip60-mediated DNMT1 acetylation triggers DNMT1 degradation. Furthermore, the abundance of UHRF1 was decreased between 11 and 13 hours but was restored by 24 hours (fig. S7, A and B). In contrast, the abundance of HAUSP, HDAC1, and PCNA remained constant throughout the cell cycle (fig. S7A). Given that HAUSP protects DNMT1 from degradation and that physical interaction between the two proteins is required for HAUSP to stabilize DNMT1, we tested whether HAUSP dissociates from DNMT1 during S phase before DNMT1 abundance is decreased by degradation. Less DNMT1 coprecipitated with HAUSP at both 3 and 5 hours after release relative to time point 0 (fig. S7, C to F), suggesting that HAUSP is not associated with DNMT1 during mid or late S phase. Furthermore, the amount of UHRF1 associated with DNMT1 increased with cell cycle progression (fig. S7, C and D), suggesting that increased interaction between UHRF1 and DNMT1 during late S phase facilitates DNMT1 degradation.
The abundance of DNMT1 and HAUSP correlates in human colon cancer
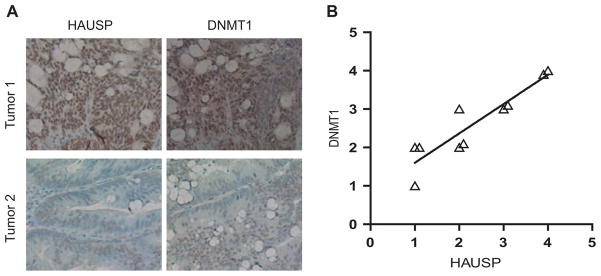
Because of the role of DNMT1 in maintaining epigenetic methylation required to silence genes involved in development and cancer, modulation of its stability and degradation may be potential therapeutic targets. To determine the clinical relevance of regulation of DNMT1 stability by HAUSP, we examined the abundance of DNMT1 and HAUSP in human colon cancer tissues with immunohisto-chemical (IHC) approaches. Ten colon tumor and normal pairs were examined. Five of these tumors showed higher abundance of DNMT1 relative to matched normal colon tissues, and these tumors also stained strongly with the antibody against HAUSP (Fig. 5, A and B). Overall, the abundance of HAUSP correlated with the abundance of DNMT1 in these tumors (Fig. 5B; R2 = 0.84). In normal colon, both DNMT1 and HAUSP were predominantly found in the proliferative crypt epithelial cells (fig. S8A). Furthermore, IHC staining of colon cancer cells could be completely blocked by competition with either DNMT1 peptides or recombinant HAUSP proteins (fig. S8B), indicating that the IHC staining was specific to the target proteins. These results suggest that increased abundance of HAUSP may be a mechanism through which the abundance of DNMT1 is increased in human cancers and that HAUSP could be therapeutically targeted in such tumors.
Fig. 5.
The abundance of DNMT1 correlates with that of HAUSP in colon cancer cells. (A) DNMT1 and HAUSP IHC staining of two representative tumors. Formalin-fixed paraffin-embedded sections of colon adenocarcinomas were stained with antibodies against HAUSP and DNMT1. (B) Correlation of IHC intensity between DNMT1 and HAUSP staining in 10 colon carcinomas (R2 = 0.84). Ten low-grade, moderately differentiated adenocarcinomas were examined and the intensity of DNMT1 staining was plotted against the intensity of HAUSP staining for each tumor.
HAUSP knockout cells are more sensitive to HDAC inhibitors
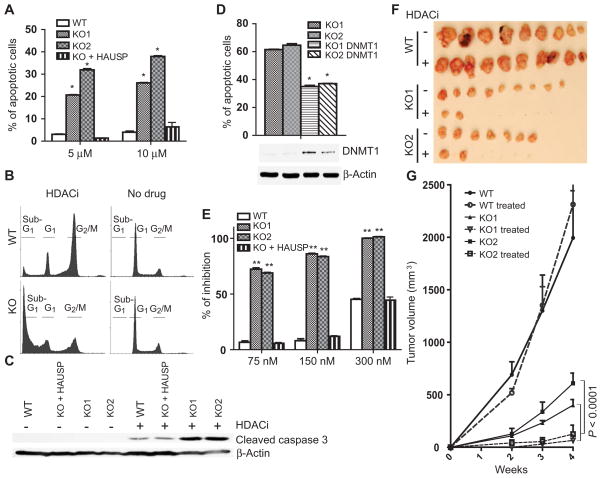
To explore whether loss of HAUSP can be exploited as a cancer therapy strategy, we treated wild-type and HAUSP knockout cells with HDAC inhibitors, which induced 5 to 10 times as many apoptotic cells in HAUSP knockout cells relative to wild-type cells (Fig. 6A and fig. S9A) (P < 0.001, t test). Tested HDAC inhibitors included MS-275, LBH589, SAHA (suberoylanilide hydroxamic acid), and TSA (trichostatin A) (Fig. 6A and fig. S9A). Furthermore, HDAC inhibition increased the number of apoptotic cells (sub-G1 cells) (Fig. 6B and fig. S9B) and increased the abundance of apoptotic cell markers including cleaved caspases 3, 6, and 9, and poly(adenosine diphosphate–ribose) polymerase (PARP) (Fig. 6C and fig. S9C). In contrast, HDAC inhibition predominantly induced G2-M arrest in wild-type cells (Fig. 6B and fig. S9B), and reconstituting the HAUSP knockout cells with ectopically expressed HAUSP suppressed apoptosis induced by HDAC inhibitors (Fig. 6A and fig. S9A). Furthermore, degradation of DNMT1 was the major cause of cell death, because ectopic overexpression of DNMT1 in the HAUSP knockout cells partially rescued HDAC inhibitor–induced apoptosis (Fig. 6D). Moreover, HAUSP knockout cells were more sensitive to growth arrest caused by MS-275 (Fig. 6E) when grown in cell culture (P < 0.001, t test).
Fig. 6.
HAUSP KO cells are more sensitive to HDACi-induced apoptosis. (A) HDAC inhibition induces apoptosisinHAUSPKOcells. HAUSP WT or KO cells were treated with or without MS-275 at the indicated concentration for 72 hours, then fixed and stained with propidium iodide. Flow cytometric analyses were used to profile sub-G1, G1, and G2-M cell populations. Apoptotic cells were quantified after the indicated clones were treated with either 5 or 10 μM MS-275. The means and SDs of three independent experiments were plotted (*P < 0.001, t test). (B) HDAC inhibition induces apoptosis in HAUSP KO cells but leads to G2-M arrest in WT cells. Cell cycle profiles of HAUSP WT or KO cells that were treated or not with 5 μM MS-275. (C) HDAC inhibition increases the abundance of apoptotic cell markers. The indicated cells were treated with or without MS-275 for 72 hours. Cell lysates were blotted with antibodies against cleaved caspase 3 and β-actin. (D) Ectopic over-expression of DNMT1 in HAUSP KO cells suppresses apoptosis. HAUSP KO clones or HAUSP KO cells inducibly overexpressing DNMT1 were treated with 10 μMMS-275. Apoptotic cell populations were quantified by fluorescence-activated cell sorting (FACS) analyses (*P < 0.001, t test). Cell lysates from these cells were blotted with the indicated antibodies. (E) HDAC inhibition arrests the growth of HAUSP KO cells. DLD1, HAUSP KO, and KO cells ectopically expressing HAUSP were treated with the indicated concentration of MS-275 for 4 days. Cell numbers were determined and data from eight replicates were plotted (**P < 0.001, t test). (F and G) HDACi inhibits tumor xenograft formation of HAUSPKO cells. Athymic nude mice (five in each group) were injected subcutaneously and bilaterally with cells of the indicated genotypes. Mice were treated with or without MS-275 at 15 mg/kg for 4 weeks. Tumors were harvested and photographed (F). Tumor sizes of the indicated groups were measured weekly and the average volumes at each time point were plotted (G). MANOVA analyses were performed to determine whether there was an overall difference of the tumor sizes, as well as whether there was a difference in development over time of tumor sizes between the two groups (P < 0.0001).
Finally, in a tumor xenograft model, treating HAUSP knockout cells with the HDAC inhibitor MS-275 led to almost complete suppression of xenograft tumor formation (Fig. 6, F and G), even though HAUSP knockout cells grew slower as xenograft tumors than the wild-type cells. In contrast, HDAC inhibition did not affect wild-type tumor growth. Together, these results demonstrated that HAUSP-deficient colorectal cancer cells are more sensitive to HDAC inhibitors and provide a compelling rationale for targeting both HAUSP and HDAC1 in combined cancer therapy strategies.
DNA methylation in the imprinted H19 locus is impaired in the HAUSP knockout cells
Given that DNMT1 is the maintenance enzyme for DNA methylation and that its abundance is reduced in the HAUSP knockout cells, we set out to determine whether DNA methylation status was altered in knockout cells. We first used pyrosequencing to quantify methylated CpG amounts in the p16 promoter region and in LINE-1 elements. No significant difference was observed among wild-type and knockout clones at these two regions (fig. S10A). Global DNA methylation also was not altered in the HAUSP knockout clones as measured by liquid chromatography–tandem MS (LC-MS/MS) analyses (fig. S10B). However, the knockout clones had reduced CpG methylation at the imprinted H19 locus relative to wild-type cells (fig. S10C). These data suggest that relatively high amounts of DNMT1 are required to maintain DNA methylation at the imprinted regions, but that maintenance of global DNA methylation requires relatively lower amounts of DNMT1.
DISCUSSION
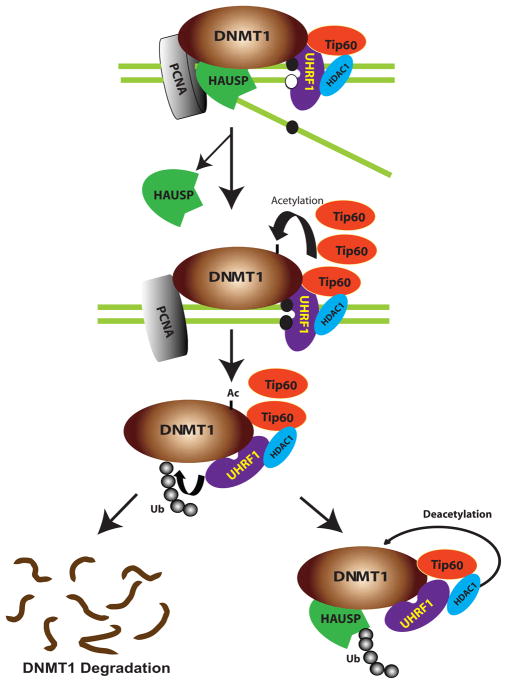
Our study has illuminated a previously unknown pathway that coordinately regulates DNMT1 protein stability through control of its ubiquitination. A schematic of the proposed mechanism is shown in Fig. 7. Our model suggests that DNMT1, HAUSP, Tip60, UHRF1, HDAC1, and PCNA form a macromolecular protein complex that binds at the DNA replication fork to coordinately regulate the functional availability of DNMT1. Interactions within the complex are proposed to proceed as follows. First, Tip60 acetylates DNMT1, which serves as a trigger for UHRF1 to ubiquitinate DNMT1, thereby leading to proteasomal degradation. Second, HDAC1 and HAUSP protect DNMT1 from degradation by deacetylation and deubiquitination, respectively. Third, the protectors (HAUSP and HDAC1) and functional destroyers (Tip60 and UHRF1) form a multifunctional protein complex with DNMT1, and components of this macromolecular complex localize to the replication forks through their association with PCNA. The concerted actions of the DNMT1 protectors and destroyers in this complex maintain the proper abundance of DNMT1 at different stages during the cell cycle. Fourth, at the end of S phase or the beginning of G2, when DNA replication and methylation are completed and hemimethylated, DNA is no longer available as a substrate; excess amounts of DNMT1 protein are destroyed through ubiquitination-mediated proteasomal degradation. Fifth, two critical events trigger DNMT1 degradation: (i) increased abundance of Tip60, which impedes HDAC1-mediated deacetylation and results in increased acetylation of DNMT1, and (ii) dissociation of HAUSP from DNMT1, which enables UHRF1 to ubiquitinate DNMT1. This model provides a series of sequential posttranslational events at the DNA replication fork that control the abundance of DNMT1 and the consequent regulation of its stability during the cell cycle.
Fig. 7.
A model of posttranslational regulation of DNMT1 stability. We propose that DNMT1 physically interacts with HAUSP, Tip60, UHRF1, HDAC1, and PCNA on chromatin. After the completion of DNA methylation in S phase, HAUSP dissociates from DNMT1 to enable DNMT1 degradation. Moreover, increased abundance of Tip60 results in increased acetylation of DNMT1, which in turn triggers the ubiquitination of DNMT1 by UHRF1. This sequence of events results in proteasomal degradation of DNMT1. In contrast, HAUSP and HDAC1 protect DNMT1 from degradation through deubiquitination and deacetylation, respectively.
The abundance of DNMT1 tracks with DNA synthesis during the cell cycle: It is increased in S phase, decreased after S phase, and is lowest in G1. This observation is consistent with previous reports that the abundance of Dnmt1 transcript reaches its peak in S phase and lowest abundance in G1 during the cell cycle (28, 29). Robertson et al. found that the abundance of Dnmt1 transcript remains high in G2 and M phases (28). However, we observed that the abundance of DNMT1 is decreased after the S phase, suggesting that Tip60- and UHRF1-mediated proteasomal degradation is the major mechanism through which DNMT1 abundance is decreased after DNA replication. When compared to G1, there is still a considerable amount of DNMT1 remaining in the G2 and M phases. We speculate that these remaining DNMT1 proteins might perform some DNA methylation–independent functions.
It appears that UHRF1 has dual roles in regulating the activities of DNMT1. UHRF1 is required for loading DNMT1 onto hemimethylated DNA templates (5, 6). Here, we demonstrated that UHRF1 ubiquitinates DNMT1 and triggers its proteasomal degradation (Fig. 4D). Consistent with this premise, UHRF1 possesses ubiquitin ligase activity and mediates ubiquitination of itself and histone H3 (7, 8). However, UHRF1-mediated degradation of DNMT1 is tightly and temporally correlated during the cell cycle. Our results suggest that in S phase, the ubiquitin ligase activity of UHRF1 must be inhibited when DNMT1 is required for methylation of newly synthesized DNA strands, and HAUSP helps to keep the ubiquitin ligase activity of UHRF1 in check. First, HAUSP counteracts UHRF1 enzymatic activity by directly deubiquitinating DNMT1. Second, we hypothesize that HAUSP directly inhibits the E3 ubiquitin ligase activity of UHRF1. In support of this notion, overexpression of UHRF1 led to greater ubiquitination of a DNMT1 construct lacking the HAUSP interaction domain compared to full-length DNMT1 (Fig. 4D). Therefore, it appears that HAUSP must be removed from the DNMT1 complexes before degradation of DNMT1. Indeed, our observations suggest that HAUSP dissociates from DNMT1 as S phase progresses (fig. S7C). As part of this mechanism, acetylation of DNMT1 also apparently acts to trigger the E3 ligase activity of UHRF1. HDAC inhibitor treatment enhances UHRF1-DNMT1 interaction as well as DNMT1 ubiquitination (Fig. 3, A to C). Consequently, it appears that increased abundance of UHRF1 is associated with DNMT1 in late rather than in early S phase (fig. S7C). Our proposed model also provides a probable explanation for the observations that knockdown of UHRF1 protects DNMT1 from HDAC inhibitor–induced degradation, but knockdown or knockout of UHRF1 does not result in increased DNMT1 abundance in unchallenged cells (fig. S6A) (5).
Moreover, our observations may have important therapeutic implications. The HDAC inhibitor SAHA and depsipeptide have been clinically used to treat cutaneous T cell lymphoma (30). However, HDAC inhibitors have failed to show efficacy in single-agent treatment of solid tumors (31). Our study demonstrates that loss of HAUSP in colorectal cancer cells potentiates cell death by apoptosis after exposure to various HDAC inhibitors, thus suggesting the potential of a combination of HDAC1 and HAUSP inhibitors as an effective cancer therapy.
MATERIALS AND METHODS
Cell culture
DLD1, HCT116, RKO, and HEK 293T cells were obtained from the American Type Culture Collection. DLD1, HCT116, and RKO colorectal cancer cells were maintained in McCoy 5A medium plus 10% fetal bovine serum (FBS). HEK 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% FBS. Cell proliferation was assayed with the Cell Counting Kit-8, which quantifies viable cells (Dojindo Molecular Technologies).
Chemicals and antibodies
MS-275, SAHA, and LBH589 were purchased from Selleck Chemicals. Trichostatin A (TSA) was purchased from Sigma. The following antibodies were used: antibodies against β-actin and Flag M2 and antibody-conjugated beads (Sigma); antibodies against Myc, hemagglutinin (HA), and HAUSP (Santa Cruz Biotechnology); antibody against DNMT1 (Abcam); antibody against ubiquitin (BIOMOL); acetylated lysine antibodies (ImmuneChem) and antibodies against UHRF1 (Abnova), cleaved caspase 3, caspase 6, caspase 9, and PARP (Cell Signaling Technology).
Somatic cell gene targeting
The approach for generating targeted cells with AAV was performed as described (18). The targeting vectors were constructed by polymerase chain reaction (PCR) with genomic DNA as the template for the homologous arms. The targeting AAV viruses were packaged in 293T cells by transfecting equal amounts of the targeting vector, pHelper, and pRC plasmids. Viruses were harvested 72 hours after transfection. RKO or DLD1 cells were infected with the targeting viruses and selected with geneticin for 20 days. The geneticin-resistant clones were then screened for homologous recombination by genomic PCR with primers derived from the neomycin resistance gene (5′-GTTGTGCCCAGTCATAGCCG-3′) and the upstream region of the left homologous arm. After the first allele was targeted, the neomycin resistance gene was excised by Cre recombinase. In the case of HAUSP knockout, two independently targeted clones were retargeted to obtain homozygous knockout clones.
In-gel digestion
Each gel piece was cut into 0.5-mm cubes and destained by repeated addition of NH4CO3/acetonitrile (1:1, v/v) solution until most of the dye was extracted from the gel cubes. The gel cubes were dried with 100% acetonitrile followed by vacuum centrifugation. The dried gel cubes were then reduced by 20 mM dithiothreitol (DTT) and alkylated by 55 mM iodoacetamide. After removal of excess DTT and iodoacetamide by 100 mM NH4CO3 and 100 mM NH4CO3/acetonitrile (1:1, v/v) solution, the gel cubes were dried with 100% acetonitrile followed by vacuum centrifugation. For one gel piece, 10 μl of the trypsin solution (0.2 μg/μl; Promega, PR-V5111) was added to rehydrate the gel plugs and incubated at 37°C overnight. The digestion solution was collected and combined with more extraction buffer (60% acetonitrile/5% formic acid) containing extracted peptides after the vortex and sonication steps. The final volume was reduced to 10 μl by vacuum centrifugation and addition of 0.1% formic acid.
Mass spectrometric analyses
Separation of peptides by capillary liquid chromatography was performed with a Dionex Ultimate 3000 capillary high-performance liquid chromatography (HPLC) system. MS analyses of samples were performed with a Thermo-Finnegan LTQ Orbitrap mass spectrometer with an octopole collision cell.
Mass spectrometric data analysis
Raw LC-MS/MS data were processed by Mascot version 2.2.0 (Matrix Science). The raw data were searched against the human International Protein Index database (released in 2009 and containing 74,017 protein sequences) with fixed modification carbamidomethyl (C) and variable modification oxidation (M). Peptides were filtered at a significance threshold of P < 0.05 (Mascot).
Generation of stable cell lines conditionally inducible for expression of DNMT1 in HAUSP knockout cells
The pcDNA6/TR plasmid (Invitrogen) was transfected into DLD1 HAUSP knockout cells. After selection with blasticidin, several TR stable colonies were transfected with pcDNA4/TO/Myc-His-DNMT1 plasmid. Blasticidin- and zeocin-resistant colonies were induced with tetracycline, and the abundance of DNMT1 was determined with antibodies directed against Myc.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer with complete protease inhibitor mixture [50 mM tris-HCl (pH 8.0), 0.5% Triton X-100, 0.25% sodium deoxycholate, 150 mM sodium chloride, 1 mM EDTA, 1 mM sodium orthovanadate, 50 mM NaF, 80 μM β-glycerophosphate, and 20 mM sodium pyrophosphate]. Western blots were performed essentially as described (32), and multiple replicates of each blot were performed. For Fig. 2D, protein translation was blocked with cycloheximide (CHX) for 24 hours before cells were harvested, because unlike the endogenous DNMT1 promoter, which is not affected by HDAC inhibitor treatment, HDAC inhibition increased DNMT1 mRNA expression driven by the cytomegalovirus promoter in the construct.
Immunofluorescence staining
DNMT1 3xFlag-tagged knock-in RKO cells were fixed with 4% para-formaldehyde for 30 min at room temperature, permeabilized with 0.2% Triton X-100 at room temperature for 5 min, and then blocked with Image-iT FX signal enhancer (Invitrogen) at room temperature for 30 min. Immunofluorescence staining was performed with mouse antibody against Flag, rabbit antibody against HAUSP, and Alexa Fluor 488–conjugated antibody against mouse and Alexa Fluor 594–conjugated secondary antibody against rabbit (Invitrogen). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml) at room temperature for 20 min. Images were captured with an inverted Leica DM IRE2 microscope and a Leica TCS SP2 AOBS filter-free UV/spectral confocal laser scanner.
In vitro deubiquitination assay
Hexahistidine-tagged wild-type HAUSP and HAUSP-C223S mutant proteins were expressed in Escherichia coli BL21 cells and purified with Ni-NTA beads. Ubiquitinated DNMT1 was incubated with 100 ng of either wild-type or mutant HAUSP protein in a deubiquitination buffer [50 mM tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 10 mM dithiothreitol (DTT), and 5% glycerol] for 2 hours at 37°C.
Immunohistochemistry
Paraffin-embedded human tissues were deparaffinized in xylene twice for 7 min. Antigen retrieval was performed by boiling the sample in a steamer for 20 min. Samples were blocked with Dako Serum-Free Protein Block for 20 min and then incubated with primary antibodies at 4°C overnight. The sections were stained with secondary antibody for 30 min at room temperature and then stained with an EnVision-HRP kit (Dako). The immunohistochemical staining was reviewed blindly and independently by two individuals. Stained sections were classified according to the intensity of staining and the percentage of cells showing HAUSP or DNMT1 staining. This was assessed in a semiquantitative manner with assignment of staining range graded from 1+ to 4+.
Flow cytometry
Cells were collected by trypsinization, fixed in 70% ethanol overnight at 4°C, and stained with propidium iodide. Fluorescence was measured with an EPICS-XL MCL flow cytometer.
Reverse transcription PCR
Total RNAs were isolated from 1 million cells with the Qiagen RNeasy Mini Kit according to the manufacturer’s instructions, and complementary DNAs (cDNAs) were synthesized with the SuperScript III First-Strand Synthesis System (Invitrogen). Primers (5′-gtgggggactgtgtctctgt-3′ and 5′-accaactcggtacaggatgc-3′) were used to PCR-amplify DNMT1.
Cell cycle synchronization
HCT116 cells were treated with 2.5 mM thymidine for 18 hours, washed with phosphate-buffered saline (PBS) twice, and then grown in complete medium for 8 hours. Cells were then treated with aphidicolin (2 μg/ml) for 15 hours. At time 0, the cells were washed with PBS to release the block and grown in complete medium.
siRNA knockdown of HAUSP, Tip60, and UHRF1
Cells were transfected with 150 pmol of each of the siRNAs below with the Lipofectamine RNAiMAX transfection reagent (Invitrogen). An ON-TARGETplus SMARTpool siRNA against HAUSP was purchased from Dharmacon (target sequence: 5′-AAGCGUCCCUUUAGCAUUA-3; 5′-GCAUAGUGAUAAACCUGUA-3′; 5′-UAAGGACCCUG-CAAAUUAU-3; 5′-GUAAAGAAGUAGACUAUCG-3′).
Two siRNA duplexes for Tip60 were synthesized by Dharmacon: 5′-ACGGAAGGUGGAGGUGGUUdTdT-3′/5′-AACCACCUCCAC-CUUCCGUdTdT-3′; and 5′-GUACGGCCGUAGUCUCAAGdTdT-3′/5′-CUUGAGACUACGGCCGUACdTdT-3′. Three siRNA duplexes against UHRF1 were purchased from Invitrogen: 5′-UUGUAGUUGAGCAU-GACCACCUGGC-3′/5′-GCCAGGUGGUCAUGCUCAACUACAA-3′; 5′-UAAAUGACGUCCUCCUCCAGCGCCG-3′/5′-CGGCGCUGGAG-GAGGACGUCAUUUA-3′; and 5′-UUCAUCUGGACCACGCCGUU-CUCCG-3′/5′-CGGAGAACGGCGUGGUCCAGAUGAA-3′.
Generation of RKO cells inducibly expressing a short hairpin RNA against HDAC1
To conditionally induce RNA interference with HDAC1 expression, we transduced an RKO cell line to harbor a conditional short hairpin RNA (shRNA)–expressing lentivirus construct. This cell line was constructed following the methods described in (33). The doxycycline-inducible system carried an shRNA construct derived from a sequence provided by Dharmacon RNA Technologies (mature siRNA sense sequence: 5′-GAAAGUCUGUUACUACUACUU-3′). For these experiments, HEK 293T cells were cotransduced with pLV-tTRKRAB-red vector and pLVTHsiGFP, and then RKO cells were multiply transduced (three times) with the transducing replication-defective virus stock carrying the conditional HDAC1-targeted knockdown vector system. Experiments were carried out at 2 days after induction of HDAC1 shRNA with doxycycline (4 μg/ml) treatment.
Xenograft and HDAC inhibitor treatment
Five million cells were injected subcutaneously and bilaterally into 4- to 6-week-old female nude mice (five nude mice in each group). The mice were randomized to vehicle or MS-275 (15 mg/kg) treatment intra-peritoneally daily. Tumor formation and size were assessed by weekly caliper measurements. After 28 days, the mice were killed and tumors were harvested.
Pyrosequencing to quantify LINE-1 and p16 methylation
Genomic DNA was prepared with the QIAamp DNA Blood Mini Kit. Bisulfite treatment of the genomic DNA samples was carried out with the Qiagen EpiTect kit. The methylation status of LINE-1 and p16 was determined with the pyrosequencing assay (PyroMark MD, Qiagen) according to the manufacturer’s instructions. Briefly, 150 ng of bisulfate-converted DNA from HCT116 wild type, HCT116 HAUSP−/−, DLD1 wild type, DLD1 HAUSP−/−, and DLD1 HAUSP−/− with HAUSP reconstitution was PCR-amplified in triplicate in a 50-μl reaction volume with prede-signed biotinylated LINE-1–specific primers that span part of the LINE-1 element, and with p16-specific primers that span a region of +148 to +161 within the exon 1 of the p16 gene (Qiagen). The amplification products were then captured with streptavidin Sepharose beads (GE Healthcare BioSciences) and pyrosequenced with the respective pre-designed sequencing primers per the manufacturer’s instructions (Qiagen). Next, the individual methylation frequencies of four CpG sites located in both LINE-1 and p16 amplification products were determined with the Pyro Q-CpG software (Qiagen). The average methylation frequency of the four CpG sites was then calculated, and the mean frequency of the triplicate reactions was considered as the overall methylation rate for each sample.
Quantification of m5dC by LC-MS/MS
About 1 μg of genomic DNA and 10 μl of the internal standard working mixture solution of cytosine-2,4-13C2-15N3 (10.0 μg/ml) and 5-methyl-2′-deoxycytidine (m5dC) (500 ng/ml) were transferred to a 4-ml glass vial and dried with nitrogen gas. The residues were dissolved in 30 μl of formic acid. The glass vials were sealed with a polytetrafluoroethylene-faced silicone septum with an aluminum crimp cap and heated at 150°C for 3 hours in a dry-bath incubator. The glass vials were then cooled, and the formic acid solution was dried with nitrogen gas at room temperature. The residue was reconstituted in 100 μl of 5 mM perfluoropentanoic acid aqueous solution before analysis. The Shimadzu HPLC system has two LC-20AD pumps and one SIL-20AC autosampler, and a Waters Xterra C18 analytical column (2.1 × 50 mm, 3.5 μm) with a Waters Xterra guard column (2.1 × 10 mm). Mobile phase A (MPA) was 5 mM perfluoropentanoic acid in deionized water, and mobile phase B (MPB) was 5 mM perfluoropentanoic acid in acetonitrile. The analytes were eluted by a linear gradient with 5 to 75% MPB from 0 to 7 min, 75 to 5% MPB from 7 to 7.5 min, and 5% MPB from 7.5 to 13 min. The flow rate was 0.2 ml/min and the injection amount was 10 μl.
The API 3200 MS/MS mass spectrometer was operated under a positive turbospray ionization condition. Quantification was carried out by multiple reaction monitoring (MRM) mode with the following settings: mass transitions: mass/charge ratio (m/z) 112.1 → 95.1 for cytosine, 126.1 → 109.1 for 5-methylcytosine, 117.1 → 99.1 for cytosine-2,4-13C2-15N3 (internal standard), and 130.1 → 113.2 for m5dC (internal standard); dwell time, 50 ms; curtain gas (CUR), 40; collision gas, 5; ion spray voltage, 5500 V; source temperature (TEM), 650°C; ion source gas 1 (GS1), 45; ion source gas 2 (GS2), 50; declustering potential (DP), 50; entrance potential (EP), 8; collision cell entrance potential (CEP), 12; collision energy (CE), 25; collision cell exit potential (CXP), 2.0; and unit resolution for both Q1 and Q2. Data acquisition and peak integration were done with Analyst software (version 1.5) with the IntelliQuan-MQII algorithm. The peak area ratios of cytosine (m/z 112.1 → 95.1) to cytosine-2,4-13C2-15N3 (internal standard, m/z 117.1 → 99.1) were plotted against cytosine concentrations in standard samples for a linear regression equation with a weighting factor of 1/x2. The cytosine and 5-methyl cytosine concentrations of the unknown samples were determined automatically by the Analyst software from the linear regression equation after obtaining the peak area ratios from their mass chromatograms.
Bisulfate sequencing of H19 CpG islands
Bisulfite treatment of the genomic DNA samples was carried out with the Qiagen EpiTect kit according to the manufacturer’s instructions, followed by PCR amplification with specific primers for H19 promoter region (forward: 5′-ATGTAAGATTTTGGTGGAATAT-3′; reverse: 5′-ACAAACT-CACACATCACAACC-3′). The PCR products were gel-purified, inserted into TA cloning vectors (Invitrogen), and sequenced.
Statistical analyses
We applied the t test to compare the means between two groups assuming unequal variances. For xenograft growth, we performed multivariate analysis of variance (MANOVA) for repeated measurements to determine whether there was an overall difference of the tumor sizes, as well as whether there was a difference in development over time of tumor sizes between the two groups. The Wilcoxon rank sum test was used to analyze the normalized Western blot data.
Supplementary Material
Fig. S1. Engineering of DNMT1, DNMT 3b, and Tip60 3xFlag-tagged knock-in cells.
Fig. S2. Identification of HAUSP by LC-MS/MS.
Fig. S3. Specific interaction between DNMT1 and HAUSP.
Fig. S4. Physical interaction is required for HAUSP to deubiquitinate DNMT1.
Fig. S5. HDAC inhibitors induce DNMT1 degradation.
Fig. S6. DNMT1-interacting proteins associate with each other.
Fig. S7. The abundance of DNMT1 changes during the cell cycle.
Fig. S8. IHC staining of HAUSP and DNMT1 in normal colon tissues.
Fig. S9. HDAC inhibitors induce apoptosis in HAUSP knockout cells.
Fig. S10. DNA methylation status of wild-type and HAUSP knockout cells.
Fig. S11. Quantification and statistical analysis of Western blots.
Acknowledgments
We thank B. Vogelstein for providing an HCT116 HAUSP knockout clone; M. Miyagi, C.-Y. Bao, and A. Polinkovsky for technical assistance; Y. Zhang, H. Lou, A. Ting, and M. Veigl for helpful discussions and critical reading of the manuscript; and X. Zhu for advice on statistical analyses.
Funding: This research was supported by the NIH (grants R01-CA127590, R01-HG004722, and HG004722-02S1) and the V foundation (to Z.W.); R01-CA101983 (to D.S.); DK078965 and HL093269 (to H.-Y.K.); and the core facilities of the Case Comprehensive Cancer Center (P30 CA43703).
Footnotes
Author contributions: Z.W., Z.D., R.M.E., Y.X., D.S., S.D.M., and H.-Y.K. designed the experiments. Z.D., J.S., Y.W., Y.Z., K.G., and S.Y. performed the experiments. D.S. and J.W. provided essential reagents. Z.D., Z.W., R.M.E., J.S., and Y.X. analyzed the data. Z.W., D.S., Z.D., S.D.M., and R.M.E. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests. A materials transfer agreement (MTA) is required by Case Western Reserve University for DNMT1, DNMT3B, Tip60 3xFlag knock-in cell lines, the HAUSP knockout cell line, the HDAC1-inducible knockdown cells, and the plasmids generated for this study.
REFERENCES AND NOTES
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA–(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 5.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 6.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 7.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 10.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 11.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, III, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Issa JP, Herman J, Bassett DE, Jr, Nelkin BD, Baylin SB. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome—components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 16.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 17.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng Z, Scacheri PC, Wang Z. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: Disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. following 486. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 21.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 22.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP–PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFβTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184:13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 26.Achour M, Fuhrmann G, Alhosin M, Rondé P, Chataigneau T, Mousli M, Schini-Kerth VB, Bronner C. UHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activity. Biochem Biophys Res Commun. 2009;390:523–528. doi: 10.1016/j.bbrc.2009.09.131. [DOI] [PubMed] [Google Scholar]
- 27.Achour M, Jacq X, Rondé P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, Schini-Kerth VB, Bronner C. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 28.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G0/G1 to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991;266:10027–10030. [PubMed] [Google Scholar]
- 30.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–1031. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 31.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15:3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, Iwakura Y, Asano M, Wei L, Yang Z, Zheng W, Dawson D, Willis J, Markowitz SD, Satake M, Wang Z. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci USA. 2010;107:2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Engineering of DNMT1, DNMT 3b, and Tip60 3xFlag-tagged knock-in cells.
Fig. S2. Identification of HAUSP by LC-MS/MS.
Fig. S3. Specific interaction between DNMT1 and HAUSP.
Fig. S4. Physical interaction is required for HAUSP to deubiquitinate DNMT1.
Fig. S5. HDAC inhibitors induce DNMT1 degradation.
Fig. S6. DNMT1-interacting proteins associate with each other.
Fig. S7. The abundance of DNMT1 changes during the cell cycle.
Fig. S8. IHC staining of HAUSP and DNMT1 in normal colon tissues.
Fig. S9. HDAC inhibitors induce apoptosis in HAUSP knockout cells.
Fig. S10. DNA methylation status of wild-type and HAUSP knockout cells.
Fig. S11. Quantification and statistical analysis of Western blots.