Although there are similarities between each segment of the gastrointestinal (GI) tract that distinguish it from other organs, each segment has its own specific functions that are based on unique gene expression patterns that direct responses to the unique environments and stresses of each segment.1 Consequently, modeling these diseases in the proper GI segment with similar pathologies has led to a variety of genetically engineered mouse (GEM) models. The vast majority of tumors carry multiple genetic alterations that accumulate from initiation through progression. Altered tumor susceptibilities can result also from gene polymorphisms. Animal modeling of the major components of this genetic complexity is possible in GEMs, although the full range of such variation is probably not possible. However, information obtained from xenograft models in which this variation a priori exists, will inform GEM modelers. Oncogenes can be introduced and tumor suppressor genes can be ablated. Hereditary cancer can be modeled through germ-line mutations, and nonhereditary cancer can be introduced in tissue-specific and inducible manners. Multiple gene defects can be combined or added in sequence through a combination of breeding and inducible systems. Polymorphisms can also be introduced in the germ line or in tissue-specific and inducible manners. Similarly, the effects of microenvironment can be functionally tested through genetic combination with gene alterations in those compartments and through alteration of the animal’s environment. GEMs with more complex genetic combinations and more highly controlled regulation of tumor suppressor genes and oncogenes are now the predominant GEMs being used for mouse modeling of human cancer (reviewed2– 4). In the fields of immunology and inflammation, GEMs have been predominant experimental tools, although the genetic complexity of these GEMs is not as great. Because cancer and inflammation are more intimately related in the GI tract than in any other system, the use of animal modeling has been critical for our understanding of inflammatory bowel disease (IBD) and its relationship to tumorigenesis (reviewed5).
In this review, a brief history of the development of the mouse genetic engineering field will be followed by a discussion of the genetic complexity being introduced into GEM models of human cancer. Brief discussions of the use of GEMs in GI cancers and IBD are then discussed.
History of GEMs
Transgenic Mice
Approaches to overexpress exogenous gene sequences in mice were being investigated in the 1970s. Germ-line transmission was first attained with Maloney Murine Leukemia Virus infection6 and then pronuclear DNA microinjection.7 Transgene expression, however, was elusive until it was discovered that an intron introduced into the β-globin transgene conferred expression.8 Copy-dependent β-globin transgene expression was achieved by inclusion of a distant cis-acting locus control region.9 Excellent reviews on transgenic animals and their usefulness for understanding gene function are available.10,11
Embryonic Stem Cells and Gene Targeting
Mouse teratocarcinoma or embryonal carcinoma cell lines12 were first cultured from spontaneous teratomas that had been shown previously to contain pluripotent cells through their ability to be serially transplantable in the mouse.13,14 Blastocyst injection of these cells produced chimeric animals, but germ-line transmission was too infrequent to be useful as an approach to transgenesis.15,16 The breakthrough came with the isolation of primary pluripotent stem cells (later to be known as embryonic stem [ES] cells) from the inner cell mass of mouse blastocysts.17,18 These cells colonized the germ line with useful efficiency19 and were capable of transmitting transgenes20 with generational stability.21 Homologous gene targeting of an endogenous gene in a mammalian cell was first demonstrated in 1985.22 Gene targeting in ES cells occurred shortly thereafter,23,24 followed by germ-line transmission of some of those cells.25,26 Reviews on the early developments in gene targeting are available.27,28
Variety of Genetic Modifications Now Possible
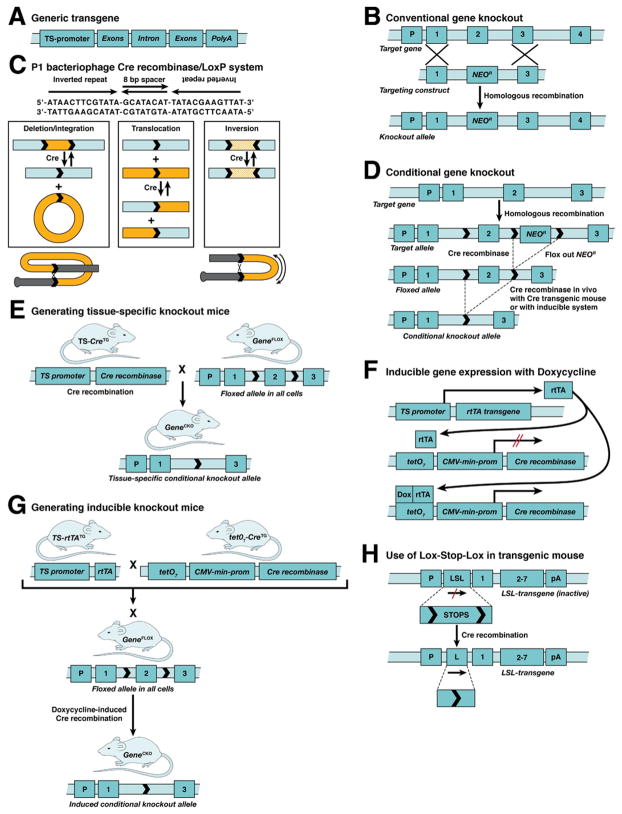
The first gene modifications involved transgenes driven by tissue-specific promoters and gene knockouts in which homologous recombination was used in ES cells to replace or add sequences that eliminated gene function (Figure 1A, B; (reviewed10,11,27,28). A bacteriophage recombination system has been employed to alter gene function in a conditional manner by generating a mouse in which the sequences encoding that function were flanked with recombinase recognition (LoxP) sites (Figure 1C).29 This system has been used for both tissue-specific30 or inducible31,32 gene ablation (Figure 1D–G; reviewed with a wide range of very useful examples33). Variations on this theme have been developed in which transgenes could be conditionally activated by floxing (flanking with LoxP sites) transcriptional stop elements generally called Lox-Stop-Lox (LSL) sites (Figure 1H).34,35 This approach has been used in lineage tracing studies in development (reviewed for pancreas36,37), and for lineage-specific expression of oncogenes in an inducible fashion (reviewed4). Reporter systems that are CRE activated include a LacZ reporter, in which an LSL was targeted into the ROSA26 locus so that the LacZ gene is expressed only in the presence of CRE,38,39 and another in which an LSL EGFP fluorescent reporter was targeted into the ROSA 26 locus.40 Deleter systems in which CRE activity causes removal of the LSL resulting in reporter gene expression include the conventional tissue-specific and doxycycline-inducible Cre transgenes referred to above, and also a tamoxifen-inducible system in which the CRE protein is fused to an estrogen receptor (CRE-ER) such that CRE becomes active only when the fusion protein binds tamoxifen.41,42 Combination of the Cre/LoxP recombinase system with another recombination system from yeast (Flip/FRT43) increases the complexity of conditional gene alterations that can occur. Finally, the development of asymmetric LoxP sites that can irreversibly invert the floxed sequences can be used to conditionally invert sequences on the complementary DNA strand onto the reading strand.44 These can be used to move a wild-type exon into the opposite strand and a mutant exon, for example, with a single nucleotide polymorphism from the opposite strand, into the reading frame.
Figure 1.
Mouse genetic engineering: common procedures. (A) Conventional transgene with promoter, exons, ≥1 intron, and poly A stop signal. (B) Conventional scheme for knocking out a gene in ES cells using homologous recombination. In this scheme, the neomycin resistance selectable marker gene NeoR replaces Exon 2 when homologous recombination occurs. (C) In the CreILoxP recombination system from PI bacteriophage, the CRE recombinase protein recognizes the LoxP recognition sites, and, depending on the orientation of the 8-bp spacer, can delete sequences between or translocate sequences at the LoxP sites if the spacers are in the same orientation, or it can invert the sequences between the sites if the spacers are in opposite orientation. A diagram of the CRE/LoxP reaction is given. (D) Conditional knockout scheme in which both exon 2 and the NeoR marker genes are flanked by LoxP recognition sites (floxed). The NeoR marker gene can be removed in the ES cells or in vivo. In the animal, CRE recombinase is supplied either through breeding with a Cre transgenic (CreTG) mouse in a tissue-specific (TS) manner, as in (E), or an inducible marmer, as in (G). Often, the marker gene is flanked with FLP recognition sites and removed with FLP recombinase (not shown; see text) either in vitro or in vivo. (E) Animal breeding scheme to generate a conditional knockout mouse in a tissue-specific manner where the GeneFLOX mouse is made from the type of construct shown in (D). Upon tissue-specific expression of the Cre transgene, exon 2 is removed only in that tissue. (F) Inducible Cre transgenic scheme.31 In this particular scheme, rtTA is a chimeric gene expression transactivator composed of (i) the DNA binding domain of a mutant tetracycline repressor (tetR) that binds DNA only in presence of the tetracycline analog doxycycline (Dox), and (ii) the gene expression trans-activation domain of the herpes simplex virus VP16 gene. The rtTA transgene is expressed in a tissue-specific manner, but the Cre transgene, being driven by a tandem set of7 tet operators (tet07) coupled to a cytomegalovirus (CMV) minimal promoter, is not expressed until the animal is treated with Doxycycline so that it can complex with the rtTA protein allowing it to bind to the tetR element and transactivate Cre transgene expression. (G) Breeding scheme associated with the inducible scheme (shown in F). It requires genetic combination of the TS-rtTATG and tet07-CreTG doubly transgenic animals with the floxed gene-targeted gene (GeneFLOX). When the genetically combined offspring are treated with doxycycline, the target gene will become ablated. (H) LSL cassette contains transcriptional and translational stop signals that block expression of the transgene. CRE recombinase deletes the LSL sequences so that transgene expression is initiated. The LSL sequences can also be inserted in opposite orientation so that CRE recombination turns off an active transgene.
Engineering Genetic Complexity Into Mouse Models of Human Cancer
Hereditary Cancer
Germ-line mutations such as in p53 (Li-Fraumeni syndrome), BRCA genes (breast cancer), mismatch repair (MMR) genes (hereditary nonpolyposis colorectal cancer; Lynch syndrome) can be introduced into mice with simple targeted gene ablation,45– 47 but they do not always serve as good models for the human disease owing either to embryonic lethality in the homozygous state and low phenotype penetrance in the heterozygous state, as in Brca gene knockout mice,46 or a tumor tissue prevalence that is not representative of humans, as in the MMR-deficient Msh6 knockout mouse.47 Inherited mutations are often the initiators that establish conditions for additional mutations that then direct the tissue prevalence and progression pathway for tumorigenesis. Consequently, it is necessary to generate tissue-specific or inducible ablation alleles to better model human hereditary cancer.
Sporadic Cancer
Most human cancers are sporadic in that they involve an initiating mutation in a cell, with subsequent accumulation of other genetic changes that drive pathways of progression to malignancy.48 In mice, the initial tumor suppressor or oncogenic mutation can be generated through conditional gene targeting or transgenesis. Through tissue-specific and inducible systems these mutations can be combined or added in a sequential manner (reviewed2). An excellent demonstration of this approach has been applied to a set of tumor suppressor and oncogenes to delineate the tumorigenic processes that they affect.49 Cell-specific pRb (retinoblastoma protein) inactivation was introduced by a truncated SV40 T antigen transgene whose product inactivates pRb. When combined with inactivations of Pten, p53, and E2f1 and transgenic Kras activation in astrocytes, prostate, breast, brain, and ovarian cells, a variety of progression pathways for tumorigenesis in each of these cell types was delineated. Knowledge of these cell-type–specific pathways may inform clinical approaches to treatments for each type of cancer.
GI Cancer GEMs
Esophageal Cancer
In Barrett’s esophagus, normal squamous epithelium is replaced by metaplastic columnar epithelium with goblet cells and is associated with a 30- to 125-fold increased risk of esophageal adenocarcinoma.50 –52 Zinc deficiency has been linked to esophageal cancer.53 Squamous epithelial dysplasia of the oral– esophageal tissue was achieved by driving cyclin D1 expression with the Epstein–Barr virus ED-L2 promoter in transgenic mice.54 Combination with a p53 deficiency led to invasive oral–esophageal cancer.55 Further treatment of cyclin D1 transgenic mice or p53−/− mice with a zinc-deficient diet or with the esophageal carcinogen N-nitrosomethylbenzylamine resulted in esophageal cancer.56,57 Apcmin/+, p53−/− and p27−/− mice have undergone esophagojejunostomy with gastric preservation to model jejunoesophageal reflux to determine which genes are important in the development of columnar metaplasia. It was found that loss of either of these genes leads to columnar metaplasia, but only loss of p53 and p27, but not APC, supports tumorigenesis in some of these mice.58,5
Expanded use of GEMs to identify the genes involved in formation of columnar metaplasia and progression to esophageal cancer under various nutritional and physiologic conditions should inform diagnostics and treatment.
Gastric Cancer
In mice, Helicobacter infection can lead to gastric inflammation and hyperplasia, but not to duodenal ulcer or gastric cancer without additional coupling to carcinogen treatment or other genetic alterations.60 Because hypergastrinemia is associated with gastric cancer in humans, a transgenic mouse line with an insulin-promoter–driven gastrin gene was developed and infected with Helicobacter.61 By 20 months of age three quarters of the mice had developed gastric cancer. Another set of models were based on the observations that in human gastric cancer there is decreased BMP, increased WNT signaling, and increased levels of prostaglandin (PG)E2. Several transgenic GEMs were generated all using the cytokeratin K19 promoter to drive transgene expression in the gastric epithelium, and the resulting mice were infected with Helicobacter (reviewed62). Noggin (inhibitor of BMP) transgenics and doubly transgenic Cox2 and Pmes mice (both genes in PGE2 synthetic pathway) did not develop gastric cancer until triply combined. Similarly, although Wnt1 transgenics developed preneoplastic lesions, no adenocarcinomas developed until genetically combined with the double transgenic Cox2 and Pmes mice. These GEM models provide insight into the combinatorial approaches that may be useful in gastric cancer therapy.
The role of inflammation in gastric apoptosis and preneoplasia has been investigated using ionizing radiation and Helicobacter infection in GEMs with epithelial-specific disruption of I-κB-kinase β/nuclear factor κB signaling.63 Increased apoptosis was found in response to cellular stress, and accelerated development of dysplasia occurred in Helicobacter-infected animals. A role for the myeloid-derived suppressor system in enhancing gastritis and initiating gastric carcinogenesis has been shown in a GEM transgenic line with increased interleukin-1β production.64 Nuclear factor κB was found, in part, to mediate activation of the myeloid-derived suppressor cells.
Pancreatic Cancer
The first pancreatic cancer model was an SV40 T-antigen transgene driven by an insulin promoter (RIP-Tag mouse), which developed β-islet tumors.65 Preneoplastic ductal lesions developed in an acinar-cell–specific elastase-promoter– driven KrasG12D transgenic mice,66 consistent with the commonly found oncogenic RAS mutations found in human pancreatic cancer. Elastase-driven Tgfα transgenic mice (acinar cell compartment), when genetically combined with p53−/−, developed pancreatic tumors that often carried additional losses of the bi-allelic Ink4a/Arf locus or Smad4 gene,67 loci commonly mutated in human pancreatic cancer. Pdx1-Cre LSL-KrasG12D double transgenic mice, in which the LSL stop signal is removed with a Cre transgene driven by the pancreas progenitor-cell–specific promoter from the Pdx1 gene, developed intraepithelial neoplasias (PanINs).68 Further genetic dissection of the KrasG12D/Smad4 combination demonstrated that whereas the KrasG12D mutation in pancreatic epithelium (Pdx1/Cre-driven) resulted in PanINs that slowly developed neoplasias, and conditional ablation of Smad4 alone in the same tissue had no effect, combination of the 2 resulted in rapid progression to neoplasias.69 GEMs modeling the increased levels of NOTCH signaling often found in KrasG12D-mutant pancreatic tumors suggested that the combination in mature acinar cells induces initiation and progression of acinar-derived PanINs.70 This study, along with the early Tgfα transgenic study,67 and another study finding that KRAS activation in acinar cells (Elastase-Cre LSL-KrasG12D mice) leads to PanINs without a requirement for chronic exocrine injury,71 reinforce the notion that different pancreatic epithelial cell compartments can transform into Pan-INs. Indeed, insulin-expressing endocrine cells normally refractory to KRAS-activated PanIN formation (RipCre-ER™ LSL-KrasG12D or RipCre-ER™ LSL-KrasG12D Ink4A/Arf flox/flox mice) can, however, transform in the presence of chronic inflammatory stress (cerulein) into the source of exocrine neoplasias.72 In summary, these GEMs are providing important clues as to how KrasG12D pancreatic cells, which very rarely develop neoplasias, can transform into 1 of the most deadly cancers in humans.
Colon Cancer
Although there are many GEM colon cancer models, this discussion is restricted to 2 of the most commonly mutated pathways in human colon cancer, APC/β-catenin and transforming growth factor (TGF)-β pathways. Mutations in the human APC gene are found in nearly 90% of human colon tumors. APC deficiency leads to constitutive WNT signaling through inability of APC to retain β-catenin in the cytoplasm for degradation, resulting in nuclear translocation where it becomes a transcriptional co-activator with lymphoid enhancer factor/T-cell factor (LEF/TCF) transcription factors.73 Mice heterozygous for an N-ethyl-N-nitrosourea–mediated mutation in the mouse Apc gene have multiple intestinal neoplasia (Apcmin/+ mice),74 and tumorigenesis in these mice is independent of colitis.75 Unlike in humans, the tumors are primarily in the small intestine. However, if treated with the carcinogen azoxymethane (AOM),76 or if fed an arginine-rich diet,77 or if genetically combined with Smad3−/− mice,78 the incidence of colon tumors is increased. An in-depth Gastroenterology review of the Apc-based GEM models, including those in which associated mutations found in humans are combined, was recently presented by Taketo and Edelmann.79
TGF-β pathway disruptions are found in up to 30% of human colon tumors and have been modeled in several GEM strains. If maintained on a predominantly 129 genetic background, Tgfb1−/− Rag2−/− mice develop proximal (cecum and proximal colon) mucinous colon cancer without APC or p53 pathway disruptions.80,81 However, if immunodeficient Tgfb1−/− mice are maintained on a C3H background, they do not develop colon cancer.81 Similarly, Smad3−/− mice, if on a primarily 129 genetic background,82– 84 also develop colon cancer with a preference for proximal colon and a requirement for Helicobacter.82,85 It is not yet clear what the required modifier genes are in the 129 strain or why tumorigenesis is dependent upon Helicobacter-induced inflammation. Nor is it clear how TGF-β signaling provides tumor suppressor function. It was originally thought that epithelial growth is uninhibited in the absence of TGF-β. However, studies in the Tgfb1−/− Rag2−/− and Villin-Cre Tgfbr2flox/flox tumor models80,86 indicate that TGF-β tumor suppressor function does not involve growth inhibition.
Expression profiling of tumors from the 4 main mouse colon cancer models (Apcmin/+, AOM-treated, Smad3−/−, and Tgfb1−/− Rag2−/− mice) revealed 2 general profiles (Apcmin/+ and AOM; and Tgfb1−/− Rag2−/−, and Smad3−/−). This classification of colon tumors is consistent with the major pathways disrupted in human colorectal cancer, APC, and TGF-β, and indicated that the APC-deficient tumors represented an earlier embryonic colon expression pattern and less of an inflammatory pattern than that of the TGF-β– deficient tumors.87 These results combined with the fact that both the Smad3−/− and Tgfb1−/− Rag2−/− strains require Helicobacter infection and its associated inflammatory response for tumorigenesis, strongly suggest that the tumor suppressor function of TGF-β signaling involves regulation of inflammation and immunoregulation of the interaction between the gut mucosa and gut microbiome.
Mice with a TGF-β type 2 receptor (Tgfbr2) inactivation in which a Tgfbr2flox allele is combined with a Villin promoter-driven Cre transgene (Villin-Cre; expressed only in intestinal epithelium) develop duodenal adenomas and few intestinal tumors. However, when genetically combined with mice harboring an Apc truncation mutation (Apc1638N/+), the intestinal epithelial-specific loss of TGF-βR2 increased progression of intestinal Apc1638N/+ adenomas to invasive adenocarcinomas.86 Similarly, genetic combination of another engineered truncation mutation (ApcΔ716)88 with a Smad4 knockout allele led to increased malignant progression.89 Finally, conditional ablation of Smad4 in the T-cell compartment through genetic combination of Smad4flox/flox and Cd4-Cre transgenic mice led to epithelial tumors throughout the GI tract from the oral cavity to the rectum, whereas conditional ablation in the intestinal epithelial compartment using a Transthyretin-Cre transgenic mouse did not.90 Both this and a previous study in which the G protein subunit alpha i2 was knocked out, causing a thymocyte deficiency, which led to mucinous adenocarcinoma of the colon,91 indicate the importance of the immunomodulatory microenvironment in tumorigenesis.
Because human APC or TGFβ mutations usually occur in combination with other mutations, GEMs are also modeling this complexity. Genetic combination of ApcMin/+ and Smad3−/− mice increases distal tumor progression and burden over that found in ApcMin/+ mice.78 Villin-Cre LSL-KrasG12D mice exhibit colonic epithelial hyperplasia,92 but when combined with Ink4a/Arf−/− mice, the resulting tumors model aspects of human serrated colon cancer.92 However, when KRAS activation mice are combined with Apc mutant mice (Ah-Cre [β-naphthoflavone-inducible Cyp1a1 cytochrome p450 promoter] LSL-KrasG12D Apcflox/flox mice),93 increased progression and tumor burden occur. With the increasing complexity of these models, colon cancer researchers should be able to better correlate specific gene mutation with phenotypic aspects of colon tumors.
In conclusion, over the past 2 decades GI GEMs have become important tools in modeling human GI diseases. With respect to GI cancers, the variety of genetic manipulations that are now available allow us to introduce single mutations that model simple hereditary cancers, combinations of tumor suppressor knockouts and oncogene transgenics to model tumors with multiple mutations, sequential addition and subtraction of oncogenes and tumor suppressor genes to investigate mechanisms of tumor progression, polymorphisms thought to alter tumor susceptibility, and genetic alterations in microenvironments. With these GEMs, we can also probe the mechanisms underlying the effects of environmental stresses on tumorigenesis. With respect to GI inflammatory diseases GEMs with immunoregulatory defects can be used to model IBDs, and combinatorial genetics allow us to determine which of these pathway disruptions are sufficient and which are contributory. Finally, the IBD models should enable us to dissect the disrupted regulatory pathways underlying dysbiosis and the conversion of commensal to pathogenic bacteria. Finally, in all of these cases, GI GEMs can be used for preclinical testing of diagnostic and treatment therapies for GI diseases, including antibiotic and probiotic approaches.
Acknowledgments
Funding
Supported by grants R01AI067903 and U01CA084291.
Footnotes
Conflicts of interest
The author discloses no conflicts.
References
- 1.Bates MD, Erwin CR, Sanford LP, et al. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology. 2002;122:1467–1482. doi: 10.1053/gast.2002.32975. [DOI] [PubMed] [Google Scholar]
- 2.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 3.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 4.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 5.Westbrook AM, Szakmary A, Schiestl RH. Mechanisms of intestinal inflammation and development of associated cancers: Lessons learned from mouse models. Mutat Res. 2010;705:40–59. doi: 10.1016/j.mrrev.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A. 1976;73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon JW, Scangos GA, Plotkin DJ, et al. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci U S A. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner TE, Hoppe PC, Jollick JD, et al. Microinjection of a rabbit beta-globin gene into zygotes and its subsequent expression in adult mice and their offspring. Proc Natl Acad Sci U S A. 1981;78:6376–6380. doi: 10.1073/pnas.78.10.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosveld F, van Assendelft GB, Greaves DR, et al. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 10.Jaenisch R. Transgenic animals. Science. 1988;240:1468–1474. doi: 10.1126/science.3287623. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Transgenic mice as probes into complex systems. Science. 1989;246:1265–1275. doi: 10.1126/science.2686032. [DOI] [PubMed] [Google Scholar]
- 12.Evans MJ. The isolation and properties of a clonal tissue culture strain of pluripotent mouse teratoma cells. J Embryol Exp Morphol. 1972;28:163–176. [PubMed] [Google Scholar]
- 13.Stevens LC, Little CC. Spontaneous testicular teratomas in an inbred strain of mice. Proc Natl Acad Sci U S A. 1954;40:1080–1087. doi: 10.1073/pnas.40.11.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens LC. Embryonic potency of embryoid bodies derived from a transplantable testicular teratoma of the mouse. Dev Biol. 1960;2:285–297. doi: 10.1016/0012-1606(60)90010-5. [DOI] [PubMed] [Google Scholar]
- 15.Brinster RL. The effect of cells transferred into the mouse blastocyst on subsequent development. J Exp Med. 1974;140:1049–1056. doi: 10.1084/jem.140.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaioannou VE, McBurney MW, Gardner RL, et al. Fate of teratocarcinoma cells injected into early mouse embryos. Nature. 1975;258:70–73. doi: 10.1038/258070a0. [DOI] [PubMed] [Google Scholar]
- 17.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 18.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley A, Evans M, Kaufman MH, et al. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 20.Robertson E, Bradley A, Kuehn M, et al. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 21.Gossler A, Doetschman T, Korn R, et al. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci U S A. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smithies O, Gregg RG, Boggs SS, et al. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 23.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 24.Doetschman T, Gregg RG, Maeda N, et al. Targeted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 25.Thompson S, Clarke AR, Pow AM, et al. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- 26.Koller BH, Hagemann LJ, Doetschman T, et al. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci U S A. 1989;86:8927–8931. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour SL. Gene targeting in murine embryonic stem cells: introduction of specific alterations into the mammalian genome. Genet Anal Tech Appl. 1990;7:219–227. doi: 10.1016/0735-0651(90)90004-y. [DOI] [PubMed] [Google Scholar]
- 28.Koller BH, Smithies O. Altering genes in animals by gene targeting. Annu Rev Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 29.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajewsky K, Gu H, Kuhn R, et al. Conditional gene targeting. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gossen M, Freundlieb S, Bender G, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn R, Schwenk F, Aguet M, et al. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 33.Torres RM, Kühn R. Laboratory protocols for conditional gene targeting. Oxford: Oxford University Press; 1997. [Google Scholar]
- 34.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 35.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu G, Brown JR, Melton DA. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- 37.Herrera Merino PL. Transgenic tagging defining pancreatic pedigrees. Ann N Y Acad Sci. 2004;1014:38–49. doi: 10.1196/annals.1294.004. [DOI] [PubMed] [Google Scholar]
- 38.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 39.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao X, Fujiwara Y, Chapdelaine A, et al. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 41.Metzger D, Clifford J, Chiba H, et al. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danielian PS, Muccino D, Rowitch DH, et al. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 43.O’Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 44.Lam KP, Rajewsky K. Rapid elimination of mature autoreactive B cells demonstrated by Cre-induced change in B cell antigen receptor specificity in vivo. Proc Natl Acad Sci U S A. 1998;95:13171–13175. doi: 10.1073/pnas.95.22.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 46.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 47.Edelmann W, Yang K, Umar A, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 49.Simin K, Hill R, Song Y, et al. Deciphering cancer complexities in genetically engineered mice. Cold Spring Harb Symp Quant Biol. 2005;70:283–290. doi: 10.1101/sqb.2005.70.038. [DOI] [PubMed] [Google Scholar]
- 50.Wijnhoven BP, Tilanus HW, Dinjens WN. Molecular biology of Barrett’s adenocarcinoma. Ann Surg. 2001;233:322–337. doi: 10.1097/00000658-200103000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falk GW. Barrett’s esophagus. Gastroenterology. 2002;122:1569–1591. doi: 10.1053/gast.2002.33427. [DOI] [PubMed] [Google Scholar]
- 52.Drewitz DJ, Sampliner RE, Garewal HS. The incidence of adenocarcinoma in Barrett’s esophagus: a prospective study of 170 patients followed 4.8 years. Am J Gastroenterol. 1997;92:212–215. [PubMed] [Google Scholar]
- 53.Fong LY, Newberne PM. Nitrosobenzylmethylamine, zinc deficiency and oesophageal cancer. IARC Sci Publ; 1978. pp. 503–513. [PubMed] [Google Scholar]
- 54.Nakagawa H, Wang TC, Zukerberg L, et al. The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and fore-stomach. Oncogene. 1997;14:1185–1190. doi: 10.1038/sj.onc.1200937. [DOI] [PubMed] [Google Scholar]
- 55.Opitz OG, Harada H, Suliman Y, et al. A mouse model of human oral-esophageal cancer. J Clin Invest. 2002;110:761–769. doi: 10.1172/JCI15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fong LY, Ishii H, Nguyen VT, et al. p53 deficiency accelerates induction and progression of esophageal and forestomach tumors in zinc-deficient mice. Cancer Res. 2003;63:186–195. [PubMed] [Google Scholar]
- 57.Fong LY, Mancini R, Nakagawa H, et al. Combined cyclin D1 overexpression and zinc deficiency disrupts cell cycle and accelerates mouse forestomach carcinogenesis. Cancer Res. 2003;63:4244–4252. [PubMed] [Google Scholar]
- 58.Fein M, Peters JH, Baril N, et al. Loss of function of Trp53, but not Apc, leads to the development of esophageal adenocarcinoma in mice with jejunoesophageal reflux. J Surg Res. 1999;83:48–55. doi: 10.1006/jsre.1998.5559. [DOI] [PubMed] [Google Scholar]
- 59.Lechpammer M, Xu X, Ellis FH, et al. Flavopiridol reduces malignant transformation of the esophageal mucosa in p27 knockout mice. Oncogene. 2005;24:1683–1688. doi: 10.1038/sj.onc.1208375. [DOI] [PubMed] [Google Scholar]
- 60.Chen D, Stenstrom B, Zhao CM, et al. Does Helicobacter pylori infection per se cause gastric cancer or duodenal ulcer? Inadequate evidence in Mongolian gerbils and inbred mice. FEMS Immunol Med Microbiol. 2007;50:184–189. doi: 10.1111/j.1574-695X.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang TC, Dangler CA, Chen D, et al. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118:36–47. doi: 10.1016/s0016-5085(00)70412-4. [DOI] [PubMed] [Google Scholar]
- 62.Oshima H, Oguma K, Du YC, et al. Prostaglandin E2, Wnt, and BMP in gastric tumor mouse models. Cancer Sci. 2009;100:1779–1785. doi: 10.1111/j.1349-7006.2009.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shibata W, Takaishi S, Muthupalani S, et al. Conditional deletion of IkappaB-kinase-beta accelerates Helicobacter–dependent gastric apoptosis, proliferation, and preneoplasia. Gastroenterology. 2010;138:1022–1034. doi: 10.1053/j.gastro.2009.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu S, Bhagat G, Cui G, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 66.Grippo PJ, Nowlin PS, Demeure MJ, et al. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 67.Wagner M, Greten FR, Weber CK, et al. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 69.Bardeesy N, Cheng KH, Berger JH, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De La OJ, Emerson LL, Goodman JL, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Habbe N, Shi G, Meguid RA, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gidekel Friedlander SY, Chu GC, Snyder EL, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 74.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 75.Dove WF, Clipson L, Gould KA, et al. Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997;57:812–814. [PubMed] [Google Scholar]
- 76.Suzui M, Okuno M, Tanaka T, et al. Enhanced colon carcinogenesis induced by azoxymethane in min mice occurs via a mechanism independent of beta-catenin mutation. Cancer Lett. 2002;183:31–41. doi: 10.1016/s0304-3835(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 77.Yerushalmi HF, Besselsen DG, Ignatenko NA, et al. The role of NO synthases in arginine-dependent small intestinal and colonic carcinogenesis. Mol Carcinog. 2006;45:93–105. doi: 10.1002/mc.20168. [DOI] [PubMed] [Google Scholar]
- 78.Sodir NM, Chen X, Park R, et al. Smad3 deficiency promotes tumorigenesis in the distal colon of ApcMin/+ mice. Cancer Res. 2006;66:8430–8438. doi: 10.1158/0008-5472.CAN-06-1437. [DOI] [PubMed] [Google Scholar]
- 79.Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 80.Engle SJ, Hoying JB, Boivin GP, et al. Transforming growth factor beta1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- 81.Engle SJ, Ormsby I, Pawlowski S, et al. Elimination of colon cancer in germ-free transforming growth factor beta 1-deficient mice. Cancer Res. 2002;62:6362–6366. [PubMed] [Google Scholar]
- 82.Zhu Y, Richardson JA, Parada LF, et al. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 83.Yang X, Letterio JJ, Lechleider RJ, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Datto MB, Frederick JP, Pan L, et al. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maggio-Price L, Treuting P, Zeng W, et al. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006;66:828–838. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Munoz NM, Upton M, Rojas A, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 87.Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oshima M, Oshima H, Kitagawa K, et al. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci U S A. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takaku K, Oshima M, Miyoshi H, et al. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 90.Kim BG, Li C, Qiao W, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 91.Rudolph U, Finegold MJ, Rich SS, et al. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat Genet. 1995;10:143–150. doi: 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 92.Bennecke M, Kriegl L, Bajbouj M, et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell. 2010;18:135–146. doi: 10.1016/j.ccr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 93.Sansom OJ, Meniel V, Wilkins JA, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]