Abstract
Pax5 is a critical regulator of B-cell commitment. Here, we identified direct Pax5 target genes by streptavidin-mediated ChIP-chip analysis of pro-B cells expressing in vivo biotinylated Pax5. By binding to promoters and enhancers, Pax5 directly regulates the expression of multiple transcription factor, cell surface receptor and signal transducer genes. One of the newly identified enhancers was shown by transgenic analysis to confer Pax5-dependent B-cell-specific activity to the Nedd9 gene controlling B-cell trafficking. Profiling of histone modifications in Pax5-deficient and wild-type pro-B cells demonstrated that Pax5 induces active chromatin at activated target genes, while eliminating active chromatin at repressed genes in committed pro-B cells. Pax5 rapidly induces these chromatin and transcription changes by recruiting chromatin-remodelling, histone-modifying and basal transcription factor complexes to its target genes. These data provide novel insight into the regulatory network and epigenetic regulation, by which Pax5 controls B-cell commitment.
Keywords: B-cell development, chromatin-modifying enzymes, epigenetic regulation, in vivo biotinylation, Pax5 target genes
Introduction
Transcription factors, chromatin regulators and cell signalling are critically involved in cell fate decisions during development. B lymphopoiesis is an ideal system to study the interplay of these processes in the context of lineage commitment. Haematopoietic stem cells (HSCs) develop into B cells by sequential differentiation via lymphoid progenitor cell stages known as LMPPs, ALPs, BLPs and pre-pro-B cells (Inlay et al, 2009). The entry of pre-pro-B cells into the B-cell lineage is controlled by the transcription factors E2A, Ebf1 and Pax5. The helix-loop-helix protein E2A and the early B-cell factor Ebf1 specify the B-cell lineage by activating the expression of B-lymphoid genes in pre-pro-B cells (Lin et al, 2010; Treiber et al, 2010). Pax5 subsequently controls B-cell commitment at the transition to the pro-B cell stage by restricting the developmental potential of lymphoid progenitors to the B-cell pathway (Nutt et al, 1999).
Within the haematopoietic system, Pax5 is exclusively expressed from the pro-B to the mature B cell stage (Fuxa and Busslinger, 2007), where it controls the differentiation, function and identity of B lymphocytes (Cobaleda et al, 2007b). Notably, the conditional loss of Pax5 results in the conversion of mature B cells into functional T cells by dedifferentiation to uncommitted progenitors in the bone marrow (Cobaleda et al, 2007a). Loss of the B-cell phenotype upon conditional Pax5 inactivation highlights an important role of Pax5 in the maintenance of B-cell commitment throughout B lymphopoiesis (Mikkola et al, 2002; Cobaleda et al, 2007a). Importantly, Pax5 has also been associated with human B-cell tumours. Frequent inactivation of one of the two PAX5 alleles identified PAX5 as a haploinsufficient tumour suppressor gene in B-cell precursor acute lymphoblastic leukaemia (B-ALL; Mullighan et al, 2007). Moreover, chromosomal translocations have implicated PAX5 as an oncogene in the generation of a subset of B-ALL and non-Hodgkin lymphomas (Cobaleda et al, 2007b).
At the transcriptional level, Pax5 fulfills a dual role by repressing B-lineage-inappropriate genes and simultaneously activating B-cell-specific genes at B-cell commitment (Nutt et al, 1999). Gene expression analyses have identified 110 genes that are repressed by Pax5 in wild-type pro-B cells compared with uncommitted Pax5–/– pro-B cells (Delogu et al, 2006; Pridans et al, 2008). Conversely, Pax5 activates 170 genes in committed pro-B cells (Schebesta et al, 2007; Pridans et al, 2008). The Pax5-repressed and Pax5-activated genes code for key regulatory and structural proteins involved in transcriptional control, receptor signalling, adhesion, migration and immune function (Delogu et al, 2006; Schebesta et al, 2007; Pridans et al, 2008). However, little is yet known about the direct effects of Pax5 in gene activation and repression, as only a few genes have so far been identified as direct targets of Pax5 (Holmes et al, 2006; Schebesta et al, 2007). Consequently, the regulatory network, by which Pax5 controls B-cell commitment, remains to be determined.
Epigenetic regulation of chromatin states also strongly influences cell fate choices, which has been best documented for the differentiation of embryonic stem (ES) cells. In pluripotent ES cells, a bivalent chromatin state is established at key regulatory genes of different lineage programs by the Trithorax group protein MLL and the Polycomb group protein Ezh2, which introduce active H3K4me3 and repressive H3K27me3 histone marks at promoter chromatin, respectively (Bernstein et al, 2006; Boyer et al, 2006). Genes with bivalent chromatin are not expressed in ES cells due to the presence of the H3K27me3 mark, but are poised to be activated upon loss of H3K27me3 during ES cell differentiation (Schuettengruber et al, 2007). In contrast to ES cells, little is known about epigenetic reprogramming during B-cell commitment and about a possible involvement of Pax5 in this process, as only nine Pax5-regulated genes have so far been shown to undergo chromatin activation in committed pro-B cells (Schebesta et al, 2007).
Here, we have systematically characterized the direct and indirect effects of Pax5 regulation by ChIP-chip mapping of Pax5-binding sites in committed pro-B cells using a high-density oligonucleotide array that contained 1306 annotated genes, including the previously identified Pax5-activated and Pax5-repressed genes. The role of Pax5 in the epigenetic regulation of these genes was determined by ChIP-chip profiling of active and repressive histone modifications in Pax5-deficient and committed pro-B cells. Notably, Pax5 was shown to rapidly induce chromatin changes by recruiting chromatin-remodelling and histone-modifying complexes to its target genes. Together, these comprehensive ChIP-chip analyses have provided novel important insight into the regulatory network and epigenetic regulation, by which Pax5 directly controls B-cell commitment at the onset of B lymphopoiesis.
Results
In vivo biotinylation of the endogenous Pax5 protein
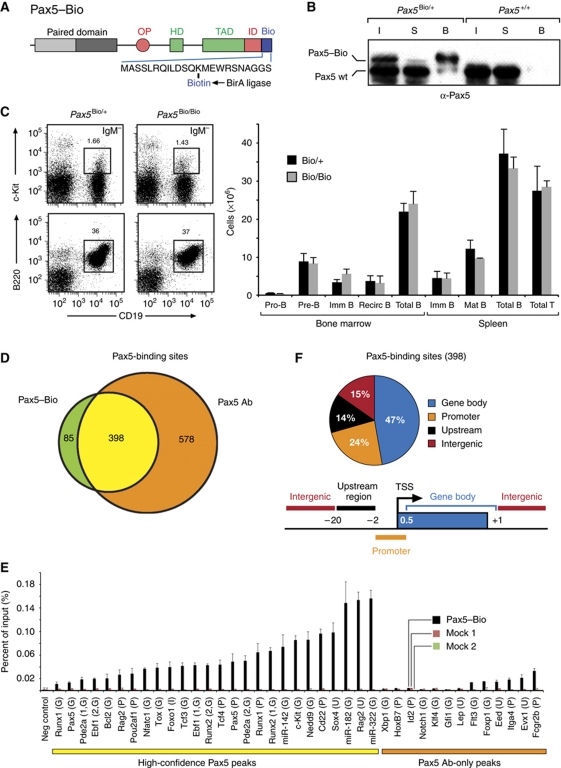
To identify Pax5 target genes with high confidence, we adapted an antibody-independent method for protein precipitation by generating a knock-in mouse carrying a biotin acceptor sequence at the C-terminus of Pax5 (Figure 1A; Supplementary Figure S1 available online), which can be biotinylated in vivo by co-expression of the Escherichia coli biotin ligase BirA (de Boer et al, 2003). Additionally, we inserted an IRES-BirA expression cassette in the 3′ untranslated region of the Pax5 gene. Consequently, the Pax5Bio allele gave rise to simultaneous expression of the biotin-tagged Pax5 protein and its modifying enzyme BirA. The Pax5–Bio protein was indeed fully biotinylated in B lymphocytes, as it was quantitatively precipitated from a nuclear extract of Pax5Bio/+ pro-B cells by pulldown with streptavidin beads (Figure 1B). Flow cytometric analysis furthermore demonstrated that pro-B, pre-B, immature B and mature B cells were present at similar numbers in the bone marrow and spleen of homozygous Pax5Bio/Bio mice compared with control Pax5Bio/+ littermates (Figure 1C; Supplementary Figure S2). The normal B-cell development in Pax5Bio/Bio mice, therefore, demonstrates that the C-terminal addition of a biotin acceptor sequence has not altered the in vivo function of Pax5.
Figure 1.
Identification of Pax5-binding sites by streptavidin pulldown of in vivo biotinylated Pax5 protein. (A) Schematic diagram of the Pax5–Bio protein with its C-terminal biotin acceptor sequence. OP, octapeptide; HD, partial homeodomain; TAD, transactivation domain; ID, inhibitory domain. For gene targeting, see Supplementary Figure S1. (B) Efficient precipitation of in vivo biotinylated Pax5 protein by streptavidin pulldown. The biotinylated Pax5–Bio protein was precipitated with streptavidin beads from a nuclear extract of Pax5Bio/+ pro-B cells. The bound fraction (B), supernatant (S) and input nuclear extract (I) were analysed by immunoblot analysis with an anti-Pax5 antibody. (C) Normal B-cell development in Pax5Bio/Bio mice. Bone marrow and spleen of 6-week-old littermates of the indicated genotypes (n=4 each) were analysed by flow cytometry to determine the absolute cell numbers of the different B-cell types (left). Representative flow cytometric data (right) are shown for pro-B cells (c-Kit+CD19+IgM−) and total B cells (B220+CD19+) of the bone marrow. See Supplementary Figure S2 for further flow cytometric data and the definition of all B-cell types. (D) Venn diagram indicating the number of shared and unique Pax5-binding sites that were detected in pro-B cells by antibody (Ab)- and Bio-ChIP-chip analysis. (E) Validation of Pax5-binding sites by streptavidin-mediated ChIP analysis of Pax5Bio/Bio Rag2–/– pro-B cells. Input and precipitated DNA were quantified by real-time PCR with primer pairs amplifying Pax5-binding regions of the indicated genes, and the amount of precipitated DNA is shown as percentage relative to input DNA (Pax5–Bio). Control ChIP experiments were performed with pro-B cells of a Rosa26BirA/BirA mouse using streptavidin beads (mock 1) or with Pax5Bio/Bio pro-B cells using protein A-coupled Dynabeads (mock 2). The average values and standard deviations of two independent experiments are shown together with the location of the Pax5-binding sites of the indicated genes in the gene body (G), promoter (P), upstream sequence (U) or intergenic region (I), as defined below. (F) Pie diagram showing the percentage of the 398 high-confidence Pax5-binding sites that are located in the upstream region (−20 to −2 kb relative to the TSS), promoter (−2 to +0.5 kb), gene body (+0.5 to 1 kb past the 3′ end) or intergenic region (upstream of −20 kb and downstream of 1 kb beyond the 3′ end). The result of one of two representative antibody- and Bio-ChIP-chip experiments is shown.
Mapping of Pax5-binding sites
For ChIP-chip analysis, we designed a high-resolution oligonucleotide tiling array, which contained 1306 annotated genes including 102 Pax5-activated and 68 Pax5-repressed genes as well as regulatory genes of different haematopoietic lineages (Supplementary Table S1). Although this microarray contained only 1.6% of the mouse genome, it was highly enriched in Pax5-regulated genes and was thus ideal for providing informative data about these genes. For ChIP-chip experiments, we used bone marrow Pax5+/+, Pax5Bio/Bio and Pax5–/– pro-B cells on a Rag2-deficient background, which were cultured ex vivo for only 4–5 days in the presence of IL-7 and OP9 cells. For mapping of Pax5-binding sites, we took advantage of the high-affinity biotin–streptavidin interaction by performing streptavidin-mediated chromatin precipitation of Pax5Bio/Bio pro-B cells followed by hybridization of the precipitated DNA onto our custom-made DNA microarray (referred to as Bio-ChIP-chip). For comparison, we also used a polyclonal anti-Pax5 antibody for ChIP-chip analysis. Peak-finder analysis of the microarray data identified 483 and 976 Pax5 peaks for the Bio-ChIP-chip and antibody-ChIP-chip experiments, respectively (Figure 1D). As the majority of the Pax5 peaks obtained with Bio-ChIP-chip overlapped with the Pax5 peaks detected by antibody-ChIP-chip, we considered the common 398 Pax5 peaks to represent high-confidence Pax5-binding sites (Figure 1D). Consistent with this notion, all 26 randomly selected Pax5-binding sites from this high-confidence class could be validated by regular Bio-ChIP analysis in contrast to the confirmation of only 6 out of 13 Pax5 peaks from the antibody-only class (Figure 1E). We, therefore, used the common 398 Pax5-binding sites (Supplementary Table S2) for further analysis and assigned them to a given gene, if they were located within the upstream region, promoter or gene body of this gene (Figure 1F). Based on these criteria, we identified 241 direct Pax5 target genes that could be classified as known Pax5-activated, known Pax5-repressed or newly identified genes.
Identification of activated Pax5 target genes
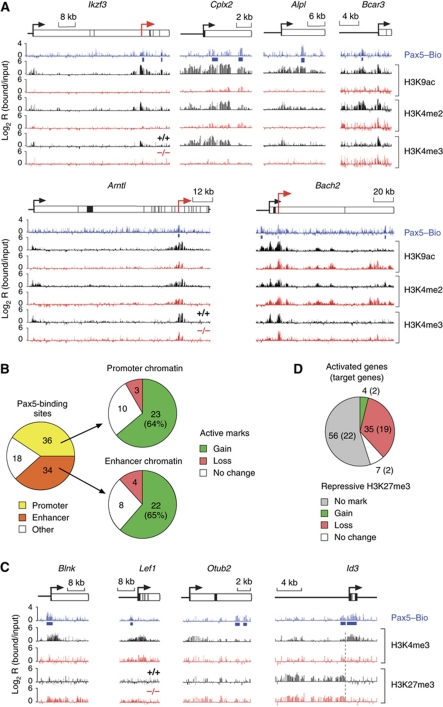
Figure 2A shows representative patterns of Pax5 binding for activated Pax5 target genes. Whereas some Pax5 peaks were detected only with the Pax5 antibody, high-confidence Pax5-binding sites were detected by both antibody- and Bio-ChIP-chip analyses in the genes encoding the transcription factors Aiolos (Ikzf3), Bach2 and E2F2 or the non-muscle myosin heavy-chain Myh10 (Figure 2A). In total, 88 high-confidence Pax5 peaks were found in the gene body, promoter or upstream region of known Pax5-activated genes with the majority of Pax5-binding sites being located within the gene body (Figure 2B; Supplementary Table S3). These Pax5-binding sites identified 45 direct target genes among the 102 activated genes that were present on the microarray (Figure 2C; Supplementary Table S4). These direct Pax5 target genes code for proteins of distinct function, whereby the three largest classes consist of 11 transcriptional regulators, 9 cell surface or adhesion receptors and 8 intracellular signal transducers (Figure 2D and E). Given the fact that Pax5 directly activates several transcription factor genes, it is likely that these transcription factors mediate the regulation of the 57 indirectly activated genes (Figure 2C).
Figure 2.
Identification of activated Pax5 target genes. (A) ChIP-chip mapping of Pax5-binding sites in pro-B cells. Antibody-ChIP-chip (Pax5 Ab) or Bio-ChIP-chip (Pax5–Bio) analysis was used to detect the binding of Pax5 to its target genes in Rag2–/– or Pax5Bio/Bio Rag2–/– pro-B cells, respectively. A schematic diagram of four selected target genes indicates the TSS (arrow), exons (black boxes) and introns (white boxes) together with a scale bar (in kb). The logarithmic ratio (log2) of the hybridization intensities between precipitated and input DNA (bound/input) is shown as a vertical bar for each oligonucleotide on the microarray. Horizontal bars below the ChIP tracks identify significant Pax5 peaks that reached values above the threshold of the peak-finder analysis in both Ab- and Bio-ChIP-chip analyses (blue) or only in the Ab-ChIP-chip experiment (red). The exons of Ikzf3 are numbered. (B) Location of Pax5-binding sites. A pie diagram indicates the percentage of Pax5-binding sites in the promoter (−2 to +0.5 kb relative to TSS), gene body (+0.5 to 1 kb beyond 3′ end) or upstream regions (−2 to −10 and −10 to −20 kb) of activated Pax5 target genes. (C) Direct versus indirect regulation of Pax5-activated genes. Direct Pax5 target genes were defined by the presence of Pax5-binding sites. (D) List of direct Pax5 target genes that are activated in pro-B cells. The colour code refers to the gene functions listed in (E). (E) Pie diagram indicating the different functional classes of activated Pax5 target genes.
Epigenetic control of activated Pax5 target genes
We next mapped active histone modifications in Pax5+/+ and Pax5–/– pro-B cells by ChIP-chip analysis with antibodies that recognized acetylated lysine 9 (H3K9ac), dimethylated lysine 4 (H3K4me2) or trimethylated lysine 4 (H3K4me3) on histone H3. Representative results of these ChIP-chip analyses are shown in Figure 3A. In Pax5–/– pro-B cells, active histone modifications were largely absent at the Pax5 target genes Ikzf3, Cplx2 and Alpl as well as in the upstream region of Bcar3. In Pax5+/+ pro-B cells, active chromatin was, however, strongly induced at the Pax5-binding sites and adjacent regions of these genes (Figure 3A) consistent with strong activation of their expression by Pax5 (Schebesta et al, 2007). In contrast, Arntl and Bach2 already carried active histone marks in Pax5–/– pro-B cells, and Pax5 induced active chromatin only at the upstream promoter of Arntl and at a putative upstream enhancer of Bach2 in Pax5+/+ pro-B cells (Figure 3A), consistent with a five-fold transcriptional activation of both genes by Pax5 (Schebesta et al, 2007). Hence, these histone modification changes highlight a correlation between chromatin activation and transcriptional induction by Pax5.
Figure 3.
Chromatin changes at activated Pax5 target genes. (A) Induction of active chromatin by Pax5. Pax5+/+ (+/+, black) and Pax5–/– (–/–, red) pro-B cells on a Rag2-deficient background were used for ChIP-chip analysis with antibodies detecting H3K4me2, H3K4me3 and H3K9ac. Pax5 binding was determined by Bio-ChIP-chip analysis of Pax5Bio/Bio Rag2–/– pro-B cells (Pax5–Bio, blue). Blue bars denote significant Pax5 peaks identified by peak-finder analysis. Red arrows denote newly identified putative promoters. (B) Statistical evaluation of chromatin changes at promoter and enhancer regions of activated Pax5 target genes. Promoters and putative enhancers were defined by the chromatin signature H3K4me3+ and H3K9ac+ H3K4me2+ H3K4me3–, respectively. ‘Other’ refers to other assortments of active histone marks, which primarily corresponded to the combination H3K9ac– H3K4me2+ H3K4me3–. Pax5-dependent gain, loss or no change of active histone marks was evaluated for Pax5-binding sites at promoter and enhancer positions in Pax5+/+ Rag2–/– pro-B cells compared with Pax5–/– Rag2–/– pro-B cells. (C) Conversion of repressive to active chromatin at promoter regions of Pax5-activated genes. Active H3K4me3 and repressive H3K27me3 modifications as well as Pax5-binding sites were mapped by ChIP-chip analysis of the respective pro-B cell type. (D) Pie diagram describing the changes of repressive chromatin (H3K27me3) at activated genes in Pax5+/+ Rag2–/– pro-B cells compared with Pax5–/– Rag2–/– pro-B cells. All 102 Pax5-activated genes present on the microarray were evaluated. The result obtained with direct Pax5 target genes is shown in brackets.
Promoters and putative enhancer of transcribed genes can be distinguished by their chromatin signatures. Active promoters typically contain the three active histone marks H3K4me2, H3K4me3 and H3K9ac (Bernstein et al, 2005; Kim et al, 2005) (Figure 3A; Supplementary Figure S3). In contrast, enhancers lack the modification H3K4me3 (Heintzman et al, 2007, 2009; Schebesta et al, 2007) (Supplementary Figure S3), as exemplified by the putative enhancers (containing Pax5-binding sites) upstream of the Bcar3 and Bach2 genes (Figure 3A). For evaluation of our chromatin data, we therefore defined active promoters by the presence of the promoter-specific H3K4 trimethylation (H3K4me3+) and putative enhancers by the histone modification code H3K4me2+ H3K9ac+ H3K4me3– (Supplementary Figure S3). Based on these criteria, Pax5 bound to a similar number of active promoters (36) and putative enhancers (34) at its activated target genes in Pax5+/+ pro-B cells, whereas 18 Pax5 peaks were present in genomic regions containing neither of these two chromatin signatures (Figure 3B; Supplementary Table S5). Surprisingly, by defining regulatory elements according to their chromatin code, we detected more promoters with Pax5-binding sites (36; Figure 3B) compared with the RefSeq annotation (13; Figure 2B), which is explained by the identification of previously unannotated putative promoters as exemplified by Arntl, Bach2 and Ikzf3 (Figure 3A, red arrows). Comparison of Pax5–/– and Pax5+/+ pro-B cells revealed that Pax5 binding correlated with an increase of active chromatin at the majority of Pax5-binding sites in both promoter (64%) and enhancer (65%) regions, whereas active chromatin remained unchanged at ∼25% of the Pax5 peaks at both regulatory elements (Figure 3B). We conclude, therefore, that Pax5 primarily induces active chromatin upon interaction with its binding sites in promoter or enhancer regions of activated target genes.
The Polycomb repressive complex 2 (PRC2) has an important role in maintaining gene repression by establishing the histone modification H3K27me3 (Schuettengruber et al, 2007). By mapping this repressive histone mark, we investigated whether PRC2 contributes to chromatin silencing of Pax5-activated genes before B-cell commitment. In Pax5–/– pro-B cells, the Blnk gene was enriched for the repressive mark H3K27me3, while lacking the active histone modification H3K4me3 (Figure 3C). Upon B-lineage commitment, Pax5 binding correlated with conversion of the repressive state (H3K4me3– H3K27me3+) to active chromatin (H3K4me3+ H3K27me3–) at the Blnk promoter (Figure 3C). A similar conversion of chromatin states was also observed at the Lef1 and Otub2 genes in contrast to the Id3 gene, where the repressive H3K27me3 mark was only present in its upstream region in both pro-B cell types (Figure 3C). Systematic evaluation of H3K27me3 enrichment demonstrated that 56 (55%) of the Pax5-activated genes including 22 direct target genes contained no repressive H3K27me3 marks in either Pax5–/– or Pax5+/+ pro-B cells (Figure 3D; Supplementary Table S6). Notably, almost half (19/45; 42%) of the directly Pax5-activated genes were PRC2 targets in Pax5–/– pro-B cells and lost their H3K27me3 modification in Pax5+/+ pro-B cells (Figure 3D; Supplementary Table S6). Thus, PRC2 appears to maintain a subset of activated Pax5 target genes in a repressed state, presumably to prevent aberrant expression before Pax5-dependent activation at B-cell commitment.
Functional validation of a predicted enhancer by transgenic analysis
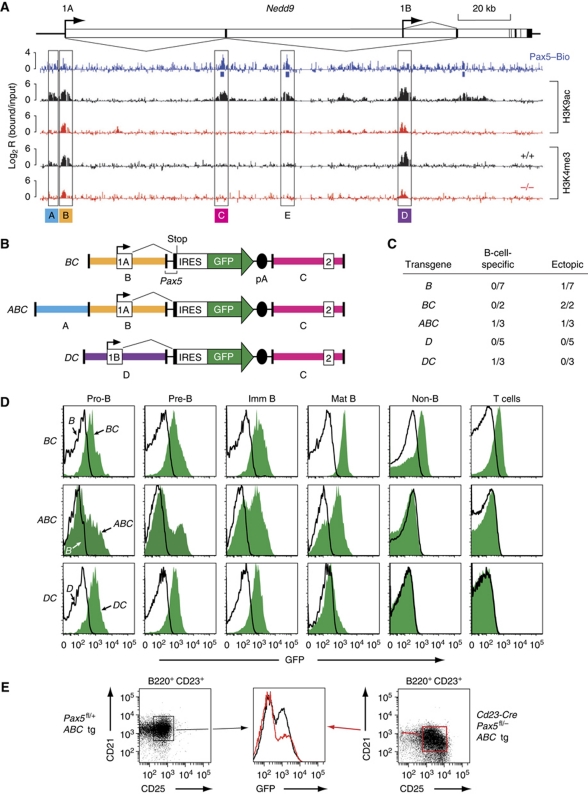
Although the unbiased prediction of putative enhancers is an important aspect of chromatin profiling, we wanted to experimentally verify the importance of Pax5 in enhancer regulation by transgenic analysis in vivo. For this, we selected the Nedd9 gene, which codes for an important adaptor protein in integrin signalling (Kim et al, 2006) and is essential for B-cell trafficking in peripheral lymphoid tissues (Seo et al, 2005). Chromatin profiling of the entire Nedd9 locus in Pax5+/+ pro-B cells resulted in the identification of one enhancer without Pax5 binding (A), two enhancers with Pax5-binding sites (C and E) and two promoters (B and D) that give rise to different Nedd9 mRNAs by alternative splicing (Figure 4A). Active chromatin was detected at the two Nedd9 promoters also in Pax5–/– pro-B cells (Figure 4A). However, the two active marks H3K9ac and H3K4me2 were absent at all three enhancers (A, C and E) in Pax5–/– pro-B cells, suggesting that Pax5 stringently controls the function of these enhancers (Figure 4A; unpublished data). Consistent with this hypothesis, transcription from both Nedd9 promoters is strongly Pax5 dependent in pro-B cells (Supplementary Figure S4A).
Figure 4.
Transgenic characterization of a Pax5-dependent enhancer of the Nedd9 gene. (A) Chromatin profiling of the Pax5 target gene Nedd9, which is shown with its exon–intron structure, two promoters and alternative splicing pattern. Active histone modifications (H3K9ac and H3K4me3) and Pax5-binding sites were mapped in Pax5+/+ (+/+, black), Pax5–/– (–/–, red) or Pax5Bio/Bio (Pax5–Bio, blue) pro-B cells on a Rag2-deficient background. The locations of the DNA fragments used for the assembly of transgenes are shown. (B) Schematic diagram of the Nedd9 transgenes BC, ABC and DC. The backbone of the transgenes consisted of the 3′ intronic sequences and 5′ part of exon 2 of Pax5 linked to stop codons in all three reading frames, an internal ribosome entry sequence (IRES), a green fluorescent protein (GFP) gene and SV40 polyadenylation (pA) sites. The mm8 sequence coordinates for the cloned DNA fragment of the Nedd9 gene on mouse chromosome 13 are 41 503 763–41 500 531 (fragment A), 41 500 360–41 495 609 (fragment B), 41 440 609–41 436 311 (fragment C) and 41 372 020–41 366 820 (fragment D). (C) Statistical evaluation of the GFP expression patterns of the different Nedd9 transgenes. The number of transgenic lines with a B-cell-specific or ectopic expression pattern is shown relative to the total number of lines obtained for each transgenic construct. (D) GFP expression of the transgenic lines. GFP expression was analysed by flow cytometry in pro-B (CD19+c-Kit+), pre-B (CD19+CD25+IgM–), immature B (CD19+IgM+IgD–), mature B (CD19+IgM–IgD+) and non-B cells (B220–CD19–) cells from the bone marrow and unfractionated T cells from the thymus. GFP expression (green surface) is shown for one ectopic BC line as well as for the B-cell-specific ABC and DC lines (see panel (C)). The non-expressing lines B and D served as negative controls (black line). (E) Pax5-dependent GFP expression of the transgenic ABC line in mature B cells. Follicular B cells (B220+CD23+CD21+) of a transgenic ABC Cd23-Cre Pax5fl/− mouse (black) and control ABC Pax5fl/+ littermate (red) were analysed by flow cytometry for CD25 and GFP expression. The representative analysis of one of four transgenic ABC Cd23-Cre Pax5fl/− and control mice is shown.
To investigate whether the enhancer C could confer B-cell-specific expression to a green fluorescent protein (GFP) reporter construct, we cloned five different Gfp transgenes (Figure 4B and C), established the corresponding transgenic mouse lines and analysed their GFP expression in haematopoietic cells by flow cytometry (Figure 4C). The Gfp constructs B and D, which contained only the upstream (B) or downstream (D) Nedd9 promoter, did not give rise to B-cell-specific GFP expression in any transgenic line, indicating that their expression requires enhancer activity (Figure 4C and D). Moreover, construct BC resulted in ectopic GFP expression in all bone marrow B and non-B cells and thymic T cells of two transgenic lines (Figure 4C and D). In contrast, one of the transgenic ABC lines expressed GFP only during B-cell development in a gradually increasing manner from pro-B cells to mature B cells (Figure 4C and D), and thus recapitulated the endogenous activity of the distal promoter B giving rise to the Nedd9 transcript 1A (Supplementary Figure S4B). Furthermore, one transgenic DC line expressed GFP equally in pro-B, pre-B and immature B cell, but not in mature B cells or any other haematopoietic cell type (Figure 4C and D), which reflects the endogenous activity of the proximal promoter D giving rise to the Nedd9 transcript 1B (Supplementary Figure S4B). Together, these data indicate that the enhancer C confers correct B-cell-specific activity to the proximal promoter D and, in conjunction with enhancer A, to the distal promoter B of the Nedd9 gene.
To demonstrate dependency of the Nedd9 enhancer C on Pax5, we crossed the transgene ABC into Cd23-Cre Pax5fl/– mice, which initiate Cre-mediated deletion of the floxed (fl) Pax5 allele in immature splenic B cells (Kwon et al, 2008). As surrogate markers for Pax5 deletion, we analysed the expression of the cell surface proteins CD21 and CD25 (Horcher et al, 2001). CD21 expression was downregulated and CD25 expression was strongly upregulated in follicular B cells (B220+CD23+CD21+) of transgenic ABC Cd23-Cre Pax5fl/– mice compared with a control ABC Pax5fl/+ littermates, thus indicating efficient Pax5 inactivation in these splenic B cells (Figure 4E). Importantly, GFP expression was significantly reduced in Pax5-deficient follicular B cells of the ABC Cd23-Cre Pax5fl/– mice compared with control B cells in four independent experiments (Figure 4E; data not shown). In summary, element C of the Nedd9 gene functions as a Pax5-dependent B-cell-specific enhancer in vivo, which validates the prediction of active enhancers based on their chromatin signature.
Chromatin changes at Pax5-repressed genes
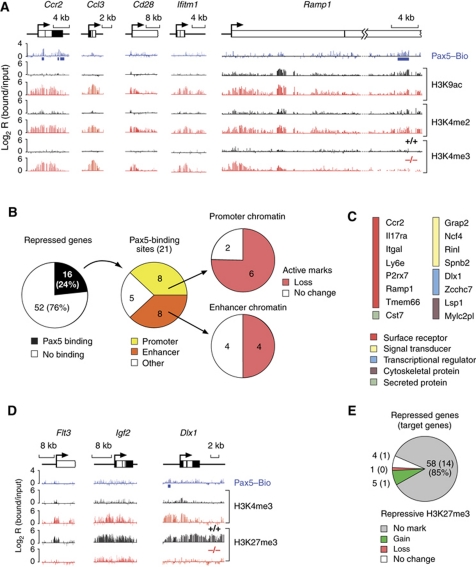
An inverse pattern of active chromatin marks was revealed for the Pax5-repressed genes, which contained active histone modifications in Pax5–/–pro-B cells (Figure 5A) consistent with their expression in the absence of Pax5 (Delogu et al, 2006). Importantly, this active chromatin was lost or reduced in Pax5+/+ pro-B cells (Figure 5A), indicating a direct or indirect involvement of Pax5 in the removal of active histone marks at Pax5-repressed genes. We could only detect Pax5 binding at 16 genes (24%) of all 68 Pax5-repressed genes, which suggests that Pax5 represses most genes indirectly in committed pro-B cells (Figure 5B; Supplementary Table S7). Interestingly, 11 of the 16 repressed Pax5 target genes code for cell surface receptors and intracellular signal transducers, demonstrating that Pax5 directly represses inappropriate signalling pathways at B-cell commitment (Figure 5C). As shown by statistical evaluation of the chromatin data, Pax5 bound with equal frequency to promoter or enhancer regions of the repressed target genes, where its binding predominantly correlated with a loss of active chromatin (Figure 5B; Supplementary Table S7). Notably, the chromatin at the Pax5-repressed genes Flt3, Igf2 and Dlx1 was converted from an active state (H3K4me3+H3K27me3–) in Pax5–/– pro-B cells to a repressive state (H3K4me3–H3K27me3+) in Pax5+/+ pro-B cells (Figure 5D). However, silencing by PRC2 was the exception as 58 genes (85%) of the Pax5-repressed genes did not acquire the repressive histone modification H3K27me3 in committed pro-B cells (Figure 5E; Supplementary Table S8). We conclude, therefore, that Pax5 represses gene expression at B-cell commitment primarily by eliminating active histone modifications.
Figure 5.
Chromatin changes at repressed Pax5 target genes. (A) Loss of active chromatin at repressed Pax5 target genes in committed pro-B cells. Active histone modifications (H3K9ac, H3K4me2 and H3K4me3) and Pax5-binding sites were mapped in Pax5+/+ (+/+, black), Pax5–/– (–/–, red) or Pax5Bio/Bio (Pax5–Bio, blue) pro-B cells on a Rag2-deficient background. Blue bars denote significant Pax5 peaks. (B) Evaluation of chromatin changes at repressed direct Pax5 target genes. See legend of Figure 3B for further explanations. (C) List of direct Pax5 target genes that are repressed in committed pro-B cells. The colour code indicates the function of each gene. Tmem66 is also known as 1810045K07Rik and Rinl as 5830482F20Rik. (D) Gain of the repressive H3K27me3 modification at Pax5-repressed genes. Pax5-binding sites as well as active H3K4me3 and repressive H3K27me3 modifications were mapped by ChIP-chip analysis of the respective pro-B cell type. (E) Absence of the repressive H3K27me3 modification at most Pax5-repressed genes in committed Pax5+/+ Rag2–/– pro-B cells. All 68 Pax5-repressed genes present on the microarray were evaluated. The result obtained with direct Pax5 target genes is shown in brackets.
Identification of novel Pax5 target genes
So far, we have analysed direct Pax5 target genes that were previously shown to be repressed or activated by Pax5 in committed pro-B cells. In addition to these genes, we also identified 178 novel target genes containing 216 Pax5-binding sites (Supplementary Figure S5A) by assigning all 398 high-confidence Pax5 peaks (Figure 1D) to their corresponding genes. These novel genes thus constitute the majority (74%) of all Pax5 target genes identified on our microarray (Supplementary Figure S5A; Supplementary Table S9). However, many Pax5-binding sites in promoters (62) and enhancers (16) of these genes showed no significant changes of active chromatin from Pax5–/– progenitors to Pax5+/+ pro-B cells (Supplementary Figure S5A and B). Hence, these target genes may not be transcriptionally regulated by Pax5, which was verified by RT–PCR analysis for five genes of this class (Supplementary Figure S6). In contrast, the gain of active chromatin at promoters (34) and enhancers (20) identified novel target genes that are activated by Pax5 in committed pro-B cells, as shown for the genes encoding the terminal deoxynuleotidyltransferase (Dntt) and signalling adaptor Grb7 (Supplementary Figures S5A, C and S6). Finally, the loss of active chromatin at few promoters (11) and enhancers (13) led to the identification of novel repressed target genes, as exemplified by the tyrosine kinase gene Zap70 and the transcription factor genes Runx2 and Tox (Supplementary Figures S5A, D and S6).
Rapid and direct induction of chromatin changes by Pax5
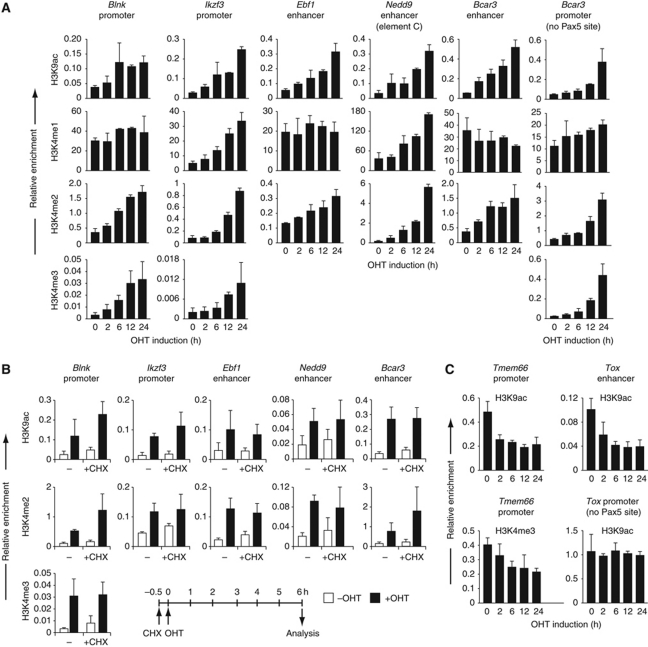
To gain further insight into the epigenetic regulation by Pax5, we next studied the kinetics of Pax5-dependent chromatin changes by using a post-translational induction system, which is based on the hormone-inducible Pax5–oestrogen receptor (ER) fusion protein (Nutt et al, 1998). As a control, we used a Prd–ER fusion protein consisting of only the N-terminal DNA-binding Paired (Prd) domain of Pax5 fused to the hormone-binding domain of the ER (Supplementary Figure S7A). Pax5–/– pro-B cells expressing the Pax5–ER or Prd–ER protein (referred to as KO-Pax5–ER or KO-Prd–ER pro-B cells) were stimulated with the oestrogen analogue 4-hydroxytamoxifen (OHT) for up to 24 h, and the increase of active histone modifications at activated target genes was determined by ChIP analysis at different time points (Figure 6A; Supplementary Figure S7B). Importantly, the Prd–ER protein did not increase the H3K9ac modification at the Blnk and Ikzf3 promoters in KO-Prd–ER pro-B cells within 24 h of OHT treatment, indicating that the paired domain of Pax5 and the hormone-binding domain of ER were unable to induce this active chromatin mark (Supplementary Figure S7B). However, in KO-Pax5–ER pro-B cells, the abundance of the H3K9ac modification was already increased 2 h after OHT stimulation and continued to increase up to 24 h at the Pax5-binding sites in the promoters of Blnk and Ikzf3 and enhancers of Ebf1, Nedd9 and Bcar3 (Figure 6A), which demonstrates that the central and C-terminal sequences of Pax5 are essential for inducing active chromatin at Pax5-binding sites. Interestingly, the Blnk promoter as well as the Ebf1 and Bcar3 enhancers already carried high levels of H3K4me1 before Pax5–ER activation (Figure 6A). All three regulatory elements rapidly gained the H3K4me2 mark and the Blnk promoter additionally the H3K4me3 modification within 2 h of Pax5 activation, which suggests that mono-methylation may be a prerequisite for di- and tri-methylation by H3K4 methyltransferases. In contrast, Pax5 rapidly induced H3K4 mono-methylation at the Ikzf3 promoter and Nedd9 enhancer, where the H3K4me2 mark at both elements and additionally the H3K4me3 modification at the Ikzf3 promoter increased with delayed kinetics in response to Pax5–ER activation (Figure 6A). Notably, the Bcar3 promoter, which lacks a Pax5-binding site, gained the active H3K4me2, H3K4me3 and H3K9ac marks with a considerable delay compared with the rapid increase of H3K4me2 and H3K9ac at the Bcar3 enhancer (Figure 6A). These data indicate that Pax5 first induces active chromatin at the Pax5-binding site in the Bcar3 enhancer, from where active marks may subsequently spread to the promoter (Figure 6A). Notably, the repressive H3K27me3 mark was diminished at the Blnk and Ikzf3 promoters as well as the Bcar3 enhancer (Supplementary Figure S7C) concomitant with the observed increase of active histone modifications in response to Pax5-ER activation (Figure 6A). In summary, Pax5 rapidly induces active chromatin and eliminates repressive histone marks at the Pax5-binding sites of activated target genes.
Figure 6.
Rapid induction of chromatin changes by Pax5. (A) Rapid induction of active chromatin at Pax5-binding sites in promoters and enhancers of Pax5 target genes. Pax5–/– pro-B cells expressing the Pax5–oestrogen receptor (ER) fusion protein (KO-Pax5–ER pro-B cells) were treated for the indicated time with 4-hydroxytamoxifen (OHT, 1 μM) before ChIP analysis with histone modification-specific antibodies. Input and precipitated DNA were quantified by real-time PCR with primer pairs amplifying the Pax5-binding regions of the indicated genes and the promoter of the ubiquitously expressed control gene coding for the TATA-binding protein (TBP). The amount of precipitated DNA was determined as percentage relative to input DNA for each region analysed and is shown as relative enrichment at the target site compared with the Tbp promoter by dividing the percentage of precipitated DNA at the Pax5-binding site (target ChIP/target input) by the percentage of precipitated DNA at the Tbp promoter (Tbp ChIP/Tbp input). The average values and standard deviations of two independent experiments are shown. (B) Pax5-dependent induction of active chromatin in the absence of protein synthesis. Where indicated, the KO-Pax5–ER pro-B cells were pre-incubated with cycloheximide (CHX; 25 μg/ml) for 30 min before the addition of OHT (1 μM) for 6 h, as schematically indicated below. The ChIP analysis was performed and evaluated as described in (A). Average values and standard deviations of two independent experiments are shown. Black and white bars refer to the presence or absence of OHT treatment, respectively. (C) Rapid reduction of active chromatin at the Pax5-binding sites of the repressed Pax5 target genes Tmem66 and Tox. The ChIP data were evaluated as described in (A).
To further investigate the induction of active chromatin by Pax5, we took advantage of the fact that the KO-Pax5–ER pro-B cells constitutively express the Pax5–ER protein in an inactive form before OHT stimulation. The Pax5–ER protein can therefore be activated by OHT addition even in the presence of cycloheximide, which inhibits protein synthesis (Supplementary Figure S8A). Under these conditions, the Pax5–ER protein was still able to induce the H3K4me2 and H3K9ac modifications at the Blnk and Ikzf3 promoters and Ebf1, Nedd9 and Bcar3 enhancers within 6 h of OHT treatment (Figure 6B). Hence, these chromatin changes are induced by Pax5 rather than by the expression of a Pax5-regulated gene. As Pax5 activates the transcription of all five target genes (Schebesta et al, 2007), it is conceivable that the process of transcription itself instead of Pax5 binding is responsible for the observed chromatin changes. To rule out this possibility, we blocked RNA polymerase II-mediated transcription by adding α-amanitin for 8 h to KO-Pax5–ER pro-B cells (Supplementary Figure S8B), which did not interfere with cell viability (Supplementary Figure S8C). In the absence of gene transcription, Pax5–ER was still capable of inducing H3K4me2 and H3K9ac marks at the Blnk and Ikzf3 promoters and Ebf1, Nedd9 and Bcar3 enhancers within 6 h of OHT treatment (Supplementary Figure S8D). Finally, Pax5–ER activation rapidly reduced active chromatin at the promoter of the repressed target gene Tmem66 and at the enhancer of the repressed Tox gene in contrast to the Tox promoter lacking a Pax5-binding site (Figure 6C). Together, these data demonstrate that Pax5 rapidly induces chromatin changes at its target genes in a transcription- and protein synthesis-independent manner.
Pax5 rapidly recruits chromatin regulators to its target genes
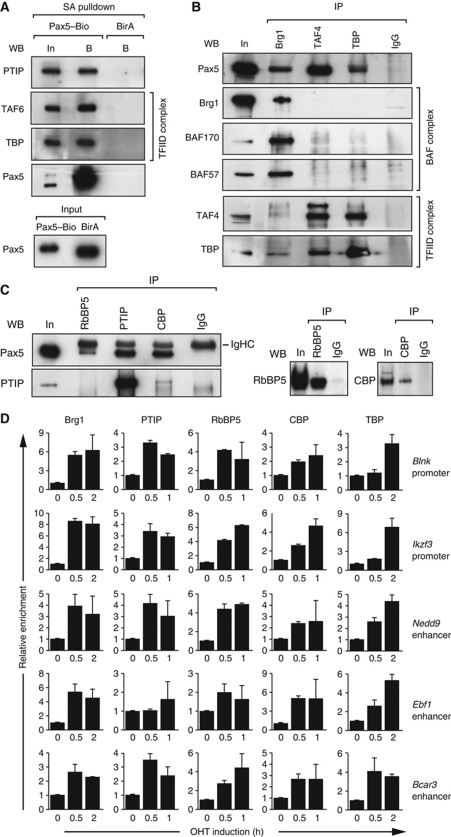
To identify Pax5-interacting proteins mediating the observed chromatin changes, we took advantage of the in vivo biotinylated Pax5–Bio protein, which facilitates the one-step purification of putative Pax5-interacting complexes from pro-B cell nuclear extracts by streptavidin pulldown followed by identification of the precipitated proteins by mass spectrometry. To control for the specificity of the streptavidin-mediated precipitation, we used wild-type pro-B cells additionally expressing the E. coli biotin ligase BirA. In the context of this study, we identified several TAF subunits of the basal transcription factor TFIID (D’Alessio et al, 2009), two components of the BAF chromatin-remodelling complex (Ho and Crabtree, 2010) and three proteins of the NCoR1 corepressor complex (Perissi et al, 2010) that were specifically enriched in the Pax5–Bio sample compared with the control BirA sample (Supplementary Figure S9). Previous transient transfection studies in the 293T kidney cell line indicated that Pax5 can interact with the TATA-binding protein (TBP; Eberhard and Busslinger, 1999), the ATPase Brg1 of the BAF complex (Barlev et al, 2003) and the histone acetyltransferase CBP (Emelyanov et al, 2002). Moreover, PTIP is known to interact not only with the related transcription factor Pax2 (Lechner et al, 2000) but also with the MLL3- and MLL4-containing H3K4 methyltransferase complex (Cho et al, 2007; Patel et al, 2007). We, therefore, investigated by streptavidin pulldown and co-immunoprecipitation experiments whether Pax5 interacts with these protein complexes under physiological conditions in pro-B cells. As shown in Figure 7A, PTIP, TBP and TAF6 were specifically detected in the streptavidin-bound fraction of nuclear extracts obtained from Pax5Bio/Bio pro-B cells expressing the biotinylated Pax5–Bio protein, but not in the corresponding fraction of the BirA extract (Figure 7A). Hence, PTIP, TBP and TAF6 co-precipitated with the Pax5–Bio protein. Importantly, a Brg1 antibody co-precipitated Pax5 together with BAF57 and BAF170 from pro-B cell nuclear extracts, thus demonstrating that Pax5 interacts with the BAF complex (Figure 7B). Pax5 was furthermore co-precipitated with antibodies detecting TBP, TAF4, CBP, PTIP, the subunit RbBP5 of the MLL-containing H3K4 methyltransferase complex (Figure 7B and C) and NCoR1 (Figure 8A). The interaction of Pax5 with PTIP was furthermore confirmed by co-immunoprecipitation in transiently transfected 293T cells (Supplementary Figure S10). Together, these data demonstrate that Pax5 interacts with the chromatin-remodelling BAF complex, PTIP and its associated H3K4 methyltransferase complex, the histone acetyltransferase CBP, the basal transcription factor TFIID and the histone deacetylase 3 (HDAC3)-containing NCoR1 complex in committed pro-B cells.
Figure 7.
Pax5 rapidly recruits histone-modifying, chromatin-remodelling and basal transcription factor complexes to activated Pax5 target genes. (A) Co-precipitation of PTIP, TAF6 and TBP with Pax5–Bio. Abelson murine leukaemia virus (Ab-MLV)-transformed pro-B cells of the Pax5Bio/Bio (Pax5–Bio) or control Rosa26BirA/BirA (BirA) genotype were used for streptavidin (SA) pulldown of nuclear extracts. The input (In) fraction (1/100) and streptavidin-bound (B) precipitate were analysed by western blotting (WB) with antibodies (Abs) detecting the indicated proteins. Pax5 was present in similar amounts in both input fractions. (B, C) Co-immunoprecipitation of Pax5 from nuclear extract of Ab-MLV-transformed Pax5Bio/Bio (B) and Rag2–/– (C) pro-B cells with Brg1, TAF4, TBP, RbBP5, PTIP or CBP antibodies. In panel (B), Pax5 was visualized in the immunoprecipitate (IP) by western blotting with a biotinylated rat anti-Pax5 mAb (detected with streptavidin-coupled horse radish peroxidase). Input (In; 1/100) and rabbit IgG were used as controls. Only one tenth of the immunoprecipitated fractions were used for western blotting with the Brg1 antibody. In panel (C), Pax5 was detected with unlabelled rat anti-Pax5 mAb, which was visualized with an anti-rat IgG Ab that crossreacted with the heavy-chain (IgHC) of the rabbit IgG Abs (left). Only one tenth of the immunoprecipitated fractions were used for western blotting with RbBP and CBP antibodies (right). (D) Rapid recruitment of histone-modifying, chromatin-remodelling and basal transcription factor complexes to activated Pax5 target genes. KO-Pax5–ER pro-B cells were treated for up to 2 h with 4-hydroxytamoxifen (OHT, 1 μM) before ChIP with antibodies precipitating the indicated proteins. Input and precipitated DNA were quantified by real-time PCR with primer pairs amplifying the Pax5-binding regions of the indicated genes and the control Tbp promoter. The enrichment of precipitated DNA at the target sites relative to the Tbp promoter was determined as described in the legend of Figure 6A. The relative enrichment at time point 0 was set to 1. The average values and standard deviations of two independent experiments are shown.
Figure 8.
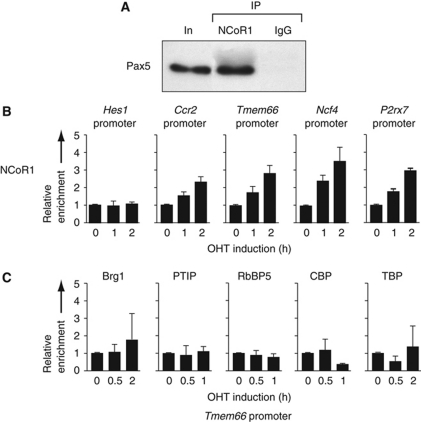
Rapid recruitment of the NCoR1 complex to Pax5-binding sites of repressed target genes. (A) Interaction of Pax5 with the NCoR1 corepressor complex. Co-immunoprecipitation of Pax5 from nuclear extracts of Rag2–/– pro-B cells with NCoR1 antibodies. Pax5 was visualized in the immunoprecipitate (IP) by western blotting with a biotinylated rat anti-Pax5 mAb (detected with streptavidin-coupled horse radish peroxidase). Input (In; 1/200) and rabbit IgG were used as controls. (B) NCoR1 recruitment to repressed Pax5 target genes. KO-Pax5–ER pro-B cells were treated for the indicated time with 4-hydroxytamoxifen (OHT, 1 μM) before ChIP with an NCoR1 antibody. Input and precipitated DNA were quantified by real-time PCR with primer pairs amplifying the Pax5-binding regions of the indicated repressed Pax5 target genes and an inactive intergenic region on chromosome 1. The enrichment of precipitated DNA at the target sites relative to the chromosome 1 region was determined as described in the legend of Figure 6A. The relative enrichment at time point 0 was set to 1. The average values and standard deviations of two independent experiments are shown. The Hes1 promoter lacking Pax5-binding sites served as negative control. (C) No recruitment of activating protein complexes to the promoter of the repressed target gene Tmem66 upon Pax5–ER activation. KO-Pax5–ER pro-B cells were treated with OHT (1 μM) before ChIP with Brg1, PTIP, RbBP5, CBP and TBP antibodies and PCR analysis as described in (B). The average values and standard deviations of two independent experiments are shown.
The Pax5–ER induction system is ideal for studying the recruitment of Pax5-interacting complexes to Pax5 target genes, as the ligand OHT prevents the binding of co-activator proteins to the ER by inducing an inactive conformation of the ligand-binding domain (Shiau et al, 1998; Nettles and Greene, 2005). We confirmed this notion by demonstrating that the Prd–ER fusion protein was unable to recruit the Brg1 protein of the BAF complex to the Blnk and Ikzf3 promoters in OHT-treated KO-Prd–ER pro-B cells (Supplementary Figure S7B). To study protein recruitment by Pax5–ER, we stimulated the KO-Pax5–ER pro-B cells with OHT for only a short time period (up to 2 h) followed by ChIP analysis with specific antibodies at different time points (Figure 7D). Within 30 min of Pax5 activation, the Brg1 protein strongly accumulated at the Pax5-binding sites of the Blnk and Ikzf3 promoters and Ebf1, Nedd9 and Bcar3 enhancers, suggesting that Pax5-dependent recruitment of the BAF complex induces rapid chromatin remodelling at these elements (Figure 7D). PTIP and the subunit RbBP5 of the MLL-containing H3K4 methyltransferase complex were also rapidly recruited to all five regulatory elements (Figure 7D). Hence, Pax5 likely recruits the H3K4 methyltransferase complex via its partner protein PTIP, which provides a molecular explanation for the rapid induction of H3K4 methylation at activated Pax5 target genes. Finally, the Pax5-dependent accumulation of CBP and TBP at all five regulatory elements indicates that Pax5 is also involved in the recruitment of histone acetyltransferase and basal transcription factor complexes to activated target genes (Figure 7D).
Importantly, Pax5–ER activation did not result in recruitment of any of the above Pax5-interacting complexes to the promoter of the repressed Pax5 target gene Tmem66 (Figure 8C). Instead, the NCoR1 corepressor complex with its associated HDAC3 activity (Perissi et al, 2010) was rapidly recruited to the promoters of the repressed target genes Tmem66, Ccr2, Ncf4 and P2rx7 (Figure 8B). In summary, Pax5 does not only interact with chromatin-remodelling, histone-modifying and basal transcription factor complexes, but also rapidly recruits them to regulatory elements to coordinate the epigenetic and transcriptional control of Pax5 target genes.
Discussion
Developmental cell fate decisions are controlled by the interplay of transcription factors and epigenetic modifiers, which together determine cellular identity (Schuettengruber et al, 2007). We have previously demonstrated that the transcription factor Pax5 controls B-cell identity by activating B-cell-specific genes and simultaneously repressing B-lineage-inappropriate genes at B-cell commitment (Nutt et al, 1999; Delogu et al, 2006; Schebesta et al, 2007). Here, we have characterized the direct and indirect effects of Pax5-mediated gene regulation by ChIP-chip identification of genomic Pax5-binding sites. Large-scale analysis of histone modifications furthermore revealed that Pax5 regulates gene expression by controlling the chromatin state at its target genes. Pax5 orchestrates these chromatin and transcription changes by recruiting chromatin-remodelling, histone-modifying and basal transcription factor complexes to its target genes. Together, these data identified Pax5 as a critical epigenetic regulator of gene transcription in early B-cell development.
By combining in vivo biotinylation of Pax5 with streptavidin-mediated chromatin precipitation, we identified high-confidence Pax5-binding sites in Pax5Bio/Bio pro-B cells by ChIP-chip analysis. In vivo biotinylation was previously used for mapping genomic binding sites of transcription factors, although these experiments were performed with stably transfected cell lines ectopically expressing the biotin-tagged proteins (de Boer et al, 2003; Kim et al, 2008; Soler et al, 2010). In contrast, we generated, to our knowledge, the first knock-in mouse expressing a biotin-tagged transcription factor. The Pax5Bio/Bio mouse expresses biotinylated Pax5 protein at all B-cell developmental stages and undergoes normal B lymphopoiesis, indicating that the C-terminal biotin tag does not interfere with Pax5 function. One important advantage of in vivo biotinylation is the extremely high affinity between streptavidin and biotin that allows for high-stringency washing of the streptavidin–Pax5–Bio precipitate in 3% sodium dodecyl sulphate, which is not possible with standard antibody–antigen-based methods. The different washing stringencies used for Bio- and antibody-ChIP analyses may also explain why the Bio-ChIP-chip experiment detected fewer Pax5 peaks that, however, largely belonged to the high-confidence class of Pax5-binding sites.
Systematic analysis of Pax5-binding sites identified 44% of the previously known Pax5-activated genes (Schebesta et al, 2007) as direct targets of Pax5. The largest functional class of these genes codes for transcription factors including Ebf1, Lef1, Tcf4 (Tcf7l2), Aiolos (Ikzf3), Id3, Bach2, Irf4 and Irf8, which regulate different aspects of B-cell development and function (Matthias and Rolink, 2005). Hence, Pax5 directly establishes a small network of transcription factors, which are likely to control the remaining 56% of indirectly Pax5-activated genes at B-cell commitment. In addition, Pax5 directly activates the expression of many cell surface receptors and signal transducers implicated in B-cell signalling and migration and, at the same time, directly represses the transcription of several cell surface receptors and signal transducers that contribute to cell signalling and migration in other haematopoietic lineages. Hence, Pax5 redirects the signalling and migration potential of haematopoietic progenitors to the B-cell pathway at lineage commitment.
Chromatin profiling identified a critical role for Pax5 in controlling the chromatin state and thus transcriptional regulation of its target genes. Pax5 thereby fulfills opposing functions at activated and repressed Pax5 target genes, as it induces active chromatin at promoters and enhancers of activated target genes, while eliminating active chromatin at the regulatory elements of repressed target genes. Previously, we demonstrated that Pax5 is able to bind Groucho corepressors as part of a large histone deacetylase complex (Eberhard et al, 2000). However, we could not investigate the recruitment of Groucho complexes to Pax5 target genes due to the lack of ChIP-grade antibodies detecting mouse Groucho proteins. Instead, we have shown Pax5-dependent recruitment of the NCoR1 corepressor complex with its associated HDAC3 activity to repressed Pax5 target genes, which likely eliminates active histone acetylation marks at these genes in committed pro-B cells. The post-translational Pax5–ER induction system combined with the identification of Pax5-interacting proteins has also provided important molecular insight into the epigenetic function of Pax5 at its activated target genes. Pax5 was thus shown to rapidly induce H3K4 methylation and H3K9 acetylation at enhancers and promoters of activated target genes even in the absence of protein synthesis or RNA polymerase II-mediated transcription. Moreover, we could demonstrate that Pax5 interacts in pro-B cells with the basal transcription factor complex TFIID, the chromatin-remodelling BAF complex, the histone acetyltransferase CBP and the PTIP protein, which is known to recruit the MLL-containing H3K4 methyltransferase complex to chromatin (Cho et al, 2007; Patel et al, 2007). Importantly, these Pax5-interacting complexes are rapidly recruited to enhancers and promoters of activated target genes in response to Pax5 activation. Hence, Pax5 activates regulatory elements at its target genes by orchestrating epigenetic and transcriptional changes through direct recruitment of chromatin-remodelling, histone-modifying and basal transcription factor complexes.
The repressive modification H3K27me3 was detected at 42% of the activated Pax5 target genes in Pax5–/– progenitors and was subsequently lost upon Pax5-mediated induction of active chromatin in committed pro-B cells. Hence, the H3K27-methylating PRC2 contributes to the silencing of Pax5-activated genes before B-cell commitment similar to its role in suppressing distinct lineage programs in ES cells (Schuettengruber et al, 2007). Interestingly, however, the loss of active chromatin at most Pax5-repressed genes was not accompanied by induction of the repressive H3K27me3 mark in committed pro-B cells, indicating that the PRC2 complex is not involved in maintaining the transcriptional silencing of Pax5-repressed genes. Consequently, the lack of PRC2-mediated silencing may facilitate the rapid reactivation of Pax5-repressed genes upon Pax5 loss in pro-B and mature B cells (Delogu et al, 2006) and could thus contribute to the observed developmental plasticity of B lymphocytes (Mikkola et al, 2002; Cobaleda et al, 2007a).
Enhancers regulate tissue-specific gene transcription by controlling promoters from distant locations, and yet their systematic identification has only recently become feasible by the discovery that enhancers are characterized by specific chromatin signatures (Heintzman et al, 2007, 2009; Schebesta et al, 2007). However, few enhancers identified by their chromatin code have been tested for activity and then only in transiently transfected cell lines (Heintzman et al, 2007). Here, we have described an important role for Pax5 in establishing active chromatin at enhancers of activated target genes. Moreover, we functionally characterized a novel enhancer of the Nedd9 gene, which was able to confer B-cell-specific activity to a Gfp reporter gene in transgenic mice. Importantly, the B-cell specificity of this Nedd9 enhancer and associated induction of active chromatin was dependent on Pax5, demonstrating that this enhancer is a functional target of Pax5. Nedd9 functions as an adaptor protein in integrin signalling (Kim et al, 2006), has an critical role in B-cell migration (Seo et al, 2005) and is expressed in several tissues including the myeloid, T- and B-cell lineages of the haematopoietic system (Schebesta et al, 2007). Our finding that a Pax5-dependent B-lymphoid enhancer controls Nedd9 expression in B cells predicts that other cell type-specific transcription factors and enhancers may regulate Nedd9 transcription in other haematopoietic lineages and different tissues. By activating B-lymphoid enhancers, Pax5 can thus impart B-cell specificity to genes that are broadly expressed in other cell lineages.
Materials and methods
Mice
The generation of the Pax5Bio/Bio and Nedd9 transgenic mice is described in Supplementary data. All animal experiments were carried out according to valid project licenses, which were approved and regularly controlled by the Austrian Veterinary Authorities.
In vitro culture of lymphoid progenitors
B220+ pro-B cells were isolated from the bone marrow by MACS sorting and were cultured on OP9 cells in IL-7-containing IMDM medium for 4–5 days as described (Nutt et al, 1997).
Antibodies
Rabbit polyclonal antibodies recognizing the following histone tail modifications were used: H3K4me1 (ab8895) from Abcam; H3K4me2 (07-030), H3K4me3 (07-473) and H3K9ac (07-352) from Upstate Biotechnology; and H3K27me3 from T Jenuwein (MPI Freiburg). The following antibodies were used for immunoblot and/or ChIP analyses: anti-Brg1 (Santa Cruz (sc-17796X)), anti-BAF57 (G Crabtree), anti-BAF170 (G Crabtree), anti-CBP (Santa Cruz (A-22, sc-369X)), anti-hERα (Santa Cruz (HC-20, sc-543)), anti-NCoR1 (Abcam (ab24552)), anti-Pax5 (rabbit polyclonal; Adams et al, 1992), anti-Pax5 (rat mAb clone 1H9 from S Nutt), anti-PTIP (K Ge), anti-RbBP5 (Bethyl Laboratories (A300-109A)), anti-TAF4 (L Tora), anti-TAF6 (L Tora) and anti-TBP (L Tora).
Chromatin precipitation by streptavidin pulldown
Pax5Bio/Bio Rag2–/– pro-B cells (50 × 106) were fixed with 1% formaldehyde in culture medium for 10 min at room temperature followed by quenching with 0.125 M glycine for 5 min. The cells were washed twice with ice-cold PBS and lysed in 1 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0) for at least 30 min on ice. The crosslinked chromatin was sheared to an average size of 500 bp by eighth 30-s pulses using a Biorupter™ sonicator (Diagenode). The lysate was centrifuged to remove cell debris, and the chromatin was quantified in a UV spectrophotometer. The chromatin (400 μg) was diluted 10-fold in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.10, 167 mM NaCl) and incubated for 2 h at RT with magnetic streptavidin beads. The Pax5–Bio-containing chromatin complexes bound to the streptavidin beads were isolated by magnetic sorting and washed with the following highly stringent buffers: 2 × with 0.5 M LiCl, 1 mM EDTA, 1% Nonidet P-40, 1% Na-deoxycholate; 3 × with 10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 3% SDS and 2 × with TE (20 mM Tris–HCl, pH 8.0, 2 mM EDTA). The protein–DNA complexes were eluted from the beads in 500 μl elution buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1% SDS, 100 mM NaHCO3, 200 mM NaCl) at 65°C overnight in the presence of proteinase K (500 μg/ml). Genomic DNA was isolated from the precipitated material by phenol extraction and ethanol precipitation.
ChIP-chip analysis
Pro-B cells were subjected to chromatin immunoprecipitation with purified antibodies (Schebesta et al, 2007) or to chromatin pulldown with streptavidin beads. Genomic DNA prepared from the chromatin-immunoprecipitated material and sheared input chromatin was subjected to T7-based linear amplification as described (Schebesta et al, 2007) or to exponential amplification with the Whole Genome Amplification (WGA) kit (Sigma). The amplified DNA was sent to NimbleGen Systems for probe preparation and hybridization to a custom-made 50-mer oligonucleotide microarray, which contained the non-repetitive genomic sequences of 1306 genes at 100 bp resolution. The hybridization data were normalized based on the assumption that there should be on average no change between bound (immunoprecipitated) and input hybridization signals. We, therefore, corrected for different amounts of hybridized bound and input material, labelling efficiency and other experimental variations by calculating a robust average of the log2 (bound/input) intensities across the experimental probes using a one-step Tukey biweight function. The resulting average was subtracted from all the log2 (bound/input) intensities effectively centering the log2 (bound/input) ratios at 0.
Nuclear extract preparation and co-precipitation analysis
Pro-B cells (1–2 × 108) were lysed in a buffer consisting of 10 mM Tris pH 8.0, 0.32 M Sucrose, 50 mM KCl, 20 mM NaCl, 3 mM CaCl2, 2 mM magnesium acetate (MgAc), 0.1% NP-40, 1 mM DTT, 2 mM 6-aminocaproic acid (6AA), 0.15 mM spermine, 0.5 mM spermidine, 0.5 μM MSF and 0.1% protease inhibitor cocktail, and the nuclei were collected by centrifugation for 5 min at 500 g. Pelleted nuclei were then lysed in a buffer consisting of 10 mM HEPES pH 7.9, 20% glycerol, 2.0 mM MgAc, 0.36 M KCl, 0.2% CHAPS, 10 mM NaF, 0.5 mM DTT, 2 mM 6-aminocaproic acid, 0.5 μM PMSF and 0.1% protease inhibitor cocktail. The BenzonaseR endonuclease (1.8 μl with 250 units/μl; Merck) was immediately added to 800 μl of the nuclei suspension corresponding to 2 × 108 cells followed by incubation at 4°C for 45 min and centrifugation at 16 100 g for 10 min. The protein content of the nuclear extract was measured to be ∼15 mg/ml by Bradford assay (Bio-Rad). The extract was diluted with three volumes of the 0.36 M KCl lysis buffer (see above) before streptavidin pulldown or immunoprecipitation. Dynal beads were blocked with 2 mg/ml BSA in PBS for 2 h at room temperature. Nuclear extracts were incubated with streptavidin Dynal beads (M280; Invitrogen) or with purified antibodies at 4°C overnight. In the latter case, protein A Dynal beads (Invitrogen) were subsequently added for another 3 h. The beads were washed five times in 20 mM Tris pH 8, 250 mM KCl, 2 mM MgCl2, 10% glycerol, 1 mM DTT, 2 mM 6-aminocapronic acid, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM sodium pyrophosphate and 0.1% protease inhibitor cocktail by removing the supernatant by magnetic sorting. The precipitated proteins were resuspended in 2 × SDS sample buffer, eluted from the beads by boiling and separated by SDS–PAGE followed by western blotting.
Accession numbers
The ChIP-chip microarray data discussed in this paper are available at the GEO repository at NCBI under the accession numbers GSE27215 and GSE28373.
Supplementary Material
Acknowledgments
We thank G Stengl and G Schmauß for FACS sorting; C Theussl for blastocyst injection; K Mechtler and his team for mass spectrometry analysis; J Choudhary for statistical evaluation of mass spectrometry results; and G Crabtree, K Ge, T Jenuwein, S Nutt and L Tora for providing antibodies. This research was supported by Boehringer Ingelheim, the Austrian GEN-AU initiative (financed by the Bundesminsterium für Bildung und Wissenschaft) and the European Union Sixth Framework Programme FP6 (funding the EuTRACC project). Anja Ebert was the recipient of a long-term EMBO fellowship.
Author contributions: SM performed the ChIP-chip mapping of chromatin modifications; AE identified and validated Pax5-binding sites by ChIP-chip analysis; GS generated the Pax5Bio/+ mouse; JM identified and validated the Pax5-interacting protein complexes; QS generated the Nedd9 transgenic mice and analysed them together with SM and AE; IT and MJ performed the bioinformatics analysis of the ChIP-chip data together with SM and AE; HT performed all Pax5–ER induction experiments; MB planned the project and designed the experiments together with SM, AE, GS, JM and HT, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbánek P, Maurer-Fogy I, Busslinger M (1992) Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev 6: 1589–1607 [DOI] [PubMed] [Google Scholar]
- Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, Robert F, Tora L, Kouzarides T, Birshtein BK, Berger SL (2003) A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol 23: 6944–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cho Y-W, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M (2007a) Conversion of mature B cells into T cells by dedifferentation to uncommitted progenitors. Nature 449: 473–477 [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M (2007b) Pax5: the guardian of B cell identity and function. Nat Immunol 8: 463–470 [DOI] [PubMed] [Google Scholar]
- D’Alessio JA, Wright KJ, Tjian R (2009) Shifting players and paradigms in cell-specific transcription. Mol Cell 36: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 100: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M (2006) Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity 24: 269–281 [DOI] [PubMed] [Google Scholar]
- Eberhard D, Busslinger M (1999) The partial homeodomain of the transcription factor Pax-5 (BSAP) is an interaction motif for the retinoblastoma and TATA-binding proteins. Cancer Res 59: 1716s–1724s [PubMed] [Google Scholar]
- Eberhard D, Jiménez G, Heavey B, Busslinger M (2000) Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J 19: 2292–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov AV, Kovac CR, Sepulveda MA, Birshtein BK (2002) The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J Biol Chem 277: 11156–11164 [DOI] [PubMed] [Google Scholar]
- Fuxa M, Busslinger M (2007) Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J Immunol 178: 8222–8228 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR (2010) Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes ML, Carotta S, Corcoran LM, Nutt SL (2006) Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev 20: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcher M, Souabni A, Busslinger M (2001) Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity 14: 779–790 [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL (2009) Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23: 2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, Flotte TJ, Duncan LM, Granter SR, Chin L (2006) Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 125: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M (2008) Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28: 751–762 [DOI] [PubMed] [Google Scholar]
- Lechner MS, Levitan I, Dressler GR (2000) PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin. Nucleic Acids Res 28: 2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C (2010) A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias P, Rolink AG (2005) Transcriptional networks in developing and mature B cells. Nat Rev Immunol 5: 497–508 [DOI] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M (2002) Reversion of B cell commitment upon loss of Pax5 expression. Science 297: 110–113 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui C-H, Relling MV, Evans WE, Shurtleff SA, Downing JR (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764 [DOI] [PubMed] [Google Scholar]
- Nettles KW, Greene GL (2005) Ligand control of coregulator recruitment to nuclear receptors. Annu Rev Physiol 67: 309–333 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401: 556–562 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Morrison AM, Dörfler P, Rolink A, Busslinger M (1998) Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J 17: 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbánek P, Rolink A, Busslinger M (1997) Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev 11: 476–491 [DOI] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR (2007) The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG (2010) Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 11: 109–123 [DOI] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL (2008) Identification of Pax5 target genes in early B cell differentiation. J Immunol 180: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M (2007) Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration and immune function. Immunity 27: 49–63 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by Polycomb and Trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Seo S, Asai T, Saito T, Suzuki T, Morishita Y, Nakamoto T, Ichikawa M, Yamamoto G, Kawazu M, Yamagata T, Sakai R, Mitani K, Ogawa S, Kurokawa M, Chiba S, Hirai H (2005) Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance. J Immunol 175: 3492–3501 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95: 927–937 [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F (2010) The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R (2010) Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity 32: 714–725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.