Abstract
How various layers of epigenetic repression restrict somatic cell nuclear reprogramming is poorly understood. The transfer of mammalian somatic cell nuclei into Xenopus oocytes induces transcriptional reprogramming of previously repressed genes. Here, we address the mechanisms that restrict reprogramming following nuclear transfer by assessing the stability of the inactive X chromosome (Xi) in different stages of inactivation. We find that the Xi of mouse post-implantation-derived epiblast stem cells (EpiSCs) can be reversed by nuclear transfer, while the Xi of differentiated or extraembryonic cells is irreversible by nuclear transfer to oocytes. After nuclear transfer, Xist RNA is lost from chromatin of the Xi. Most epigenetic marks such as DNA methylation and Polycomb-deposited H3K27me3 do not explain the differences between reversible and irreversible Xi. Resistance to reprogramming is associated with incorporation of the histone variant macroH2A, which is retained on the Xi of differentiated cells, but absent from the Xi of EpiSCs. Our results uncover the decreased stability of the Xi in EpiSCs, and highlight the importance of combinatorial epigenetic repression involving macroH2A in restricting transcriptional reprogramming by oocytes.
Keywords: epiblast stem cells, inactive X chromosome, macroH2A, nuclear reprogramming, Xenopus oocytes
Introduction
The differentiated state of somatic cells is remarkably stable, but can nevertheless be reversed by certain experimental procedures. These include transcription factor overexpression (induced pluripotent stem (iPS) cells), cell fusion and nuclear transfer (Gurdon and Melton, 2008). As cells become progressively more differentiated during development, their nuclei become increasingly resistant to reprogramming after transfer to eggs or oocytes (Pasque et al, 2010). Since different rates of gene reactivation are seen when the nuclei of different cell types are used, the epigenetic state of genes in somatic nuclei before transfer is likely to be an important factor influencing resistance to reprogramming (Halley-Stott et al, 2010). Here, we analyse the relationship between the epigenetic state of genes and reprogramming efficiency by using the easily traceable mammalian inactive X chromosome (Xi) as a tool.
The use of other reprogramming procedures can lead, in some instances, to reactivation of the Xi, such as nuclear transfer to eggs (Eggan, 2000), the generation of iPS cells (Maherali et al, 2007) and cell fusion (Takagi et al, 1983). Several nuclear transfer experiments in the mouse revealed epigenetic defects of the Xi in nuclear transfer embryos, and established that proper X regulation is critical for successful reprogramming, emphasizing the importance of understanding this process (Bao et al, 2005; Nolen et al, 2005; Inoue et al, 2010). However, these reprogramming systems are not suitable for analysing precise molecular processes.
Our experimental system involves the transplantation of multiple mammalian somatic cell nuclei into the germinal vesicle (GV) of the Xenopus oocytes in first meiotic prophase. Under these conditions, most genes, including pluripotency genes, but also some cell-type-specific genes, are transcriptionally activated directly from their quiescent state in somatic cells (Byrne et al, 2003; Biddle et al, 2009). Importantly, transcriptional reprogramming of previously repressed genes occurs within 2 days at 18°C in the absence of cell division.
X chromosome inactivation (XCI) has been widely used to study epigenetic regulation of gene expression and the establishment of heterochromatin (Brockdorff, 2002; Heard and Disteche, 2006; Payer and Lee, 2008; Leeb et al, 2009). The Xi provides a clear example of the stable and irreversible state of gene repression during cell differentiation. In the mouse, one of the two X chromosomes becomes epigenetically inactivated during early development to achieve dosage compensation (Lyon, 1961). Imprinted XCI is maintained in the extraembryonic lineage, while random XCI is induced in somatic cells as they start to differentiate from the epiblast. Initiation of XCI is induced by Xist RNA coating of the Xi (Clemson et al, 1996), creating a silent compartment in which active marks on chromatin are lost and repressive ones are acquired. Xist RNA coating of the Xi recruits Polycomb repressive complexes (PRC), which catalyse the deposition of repressive histone modifications such as H3K27 trimethylation (H3K27me3) and ubiquitination of H2AK119 (ubH2A) (Plath et al, 2003; Silva et al, 2003; de Napoles et al, 2004). Initiation of XCI is followed by maintenance of the repressed state, through the synergistic action of several repressive mechanisms (Csankovszki et al, 2001). These include incorporation of the repressive histone variant macroH2A (mH2A) (Costanzi and Pehrson, 1998), followed by DNA methylation (Blewitt et al, 2008). While the Xi of differentiated cells is believed to be very stable, the stability of the Xi in cells of the early mouse embryo such as post-implantation-derived epiblast stem cells (EpiSCs) is totally unknown so far (Tesar et al, 2007; Hayashi and Surani, 2009). Female EpiSCs have a nuclear domain of H3K27me3 typical of the Xi, but also express pluripotency genes (Guo et al, 2009). It was demonstrated that during early XCI, Xist-induced gene repression shifts from a Xist-dependent (XD) and reversible, to a stable, Xist-independent (XI) state (Wutz and Jaenisch, 2000). The timing at which this switch occurs in the embryo is not known. Therefore, one possibility is that the Xi of EpiSCs may be reversible and dependent on Xist RNA.
In this study, we test the stability of the Xi of EpiSCs and somatic or extraembryonic cells and we aim to identify the mechanisms that may restrict reprogramming following nuclear transfer to Xenopus oocytes. We ask which epigenetic marks correlate with the irreversible, or reversible states of the Xi. We then identify those epigenetic marks characteristic of a repressed X chromosome that are, or are not, reversed by nuclear transfer to oocytes. Finally, we test the extent to which Xist-mediated silencing is reversed in oocytes by using a Xist-inducible system.
This analysis is of interest for three reasons. First, it identifies epigenetic marks that help to ensure the stability of repressed states. This facilitates the maintenance of cell commitment and restricts lineage potential during cell differentiation. Second, the identification of mechanisms that prevent the efficient reversal of gene expression from differentiated cell nuclei transplanted into oocytes may help improve the success of nuclear reprogramming and hence, ultimately, cell replacement strategies. Third, it identifies the decreased stability of the Xi in EpiSCs, as opposed to the irreversibility of the Xi of other cell types, and may reflect a poised developmental potential towards the germline.
Results
The inactive X chromosome of differentiated cells is remarkably resistant to transcriptional reprogramming by Xenopus oocytes
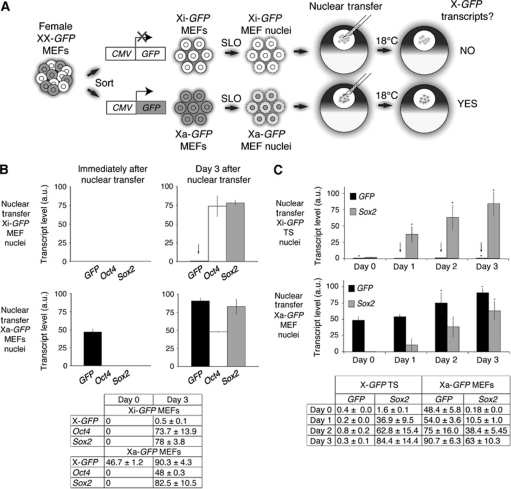
We first tested if the Xi of differentiated cells is reactivated after nuclear transplantation to Xenopus oocytes. To this end, we followed the expression of a CMV-GFP reporter (X-GFP) located on the active, or on the inactive X chromosome of differentiated cell nuclei (Figure 1A). We derived mouse embryonic fibroblasts (MEFs) carrying X-GFP on one of the two X chromosomes (Supplementary Figure S1A and B). X-GFP expression is known to reflect X chromosome states during mouse embryogenesis (Hadjantonakis et al, 2001). Due to random XCI, the X-GFP reporter is subjected to inactivation when it is located on the Xi, but remains active when it is located on the active X chromosome (Xa). We separated by flow cytometry MEFs in which the X-GFP reporter is located on the Xi from ones in which it is on the Xa (Supplementary Figure S1B–D). To determine if gene reactivation occurs on the Xi, a pure population of sorted Xi-GFP MEFs was permeabilized, the resulting nuclei transplanted into the GV of Xenopus oocytes and incubated for several days (Figure 1A). Transcriptional activity of the X-GFP reporter, and of the autosomal genes Oct4 and Sox2 was analysed in samples collected immediately and 3 days after nuclear transfer. Transcriptional reactivation of X-GFP, Oct4 and Sox2 was measured by quantitative RT–PCR (qRT–PCR). While pluripotency genes Oct4 and Sox2 were efficiently reactivated 3 days after nuclear transfer, Xi-GFP was resistant to reprogramming by oocytes (Figure 1B, arrow). Surprisingly, Xi-GFP of MEF nuclei remained repressed even several days after nuclear transfer.
Figure 1.
The inactive X chromosome of differentiated somatic cells is remarkably resistant to reprogramming by Xenopus oocytes. (A) Nuclear transfer experimental scheme. Female MEFs with an X-linked CMV-GFP transgene on the active (Xa) or on the inactive (Xi) X chromosome were sorted, permeabilized with Streptolysin O (SLO) and the resulting nuclei transplanted into the germinal vesicles (GVs) of stage V Xenopus oocytes. Transplanted oocytes were incubated at 18°C and samples were collected at several time points for transcriptional analysis. Transcriptional reactivation of X-GFP was assayed by qRT–PCR. (B) The Xi of MEFs is resistant to transcriptional reprogramming by oocytes. qRT–PCR analysis of GFP (black), Oct4 (white) and Sox2 (grey) expression in transplanted nuclei immediately and 3 days after nuclear transfer. The arrow highlights maintenance of Xi-GFP repression. P<0.05, n=3, error bars are mean±s.d. The table shows transcript levels mean±s.d. a.u. represents arbitrary unit. (C) The imprinted Xi of trophoblast stem (TS) cells is resistant to transcriptional reprogramming by oocytes. Quantitative analysis of GFP (black) and Sox2 (grey) expression in transplanted Xi-GFP TS and Xa-GFP MEFs nuclei. Arrows highlight maintenance of imprinted Xi-GFP silencing. P<0.05 for GFP, except samples marked *P<0.06. For Sox2, P<0.05, except samples marked *P<0.1, n=3, error bars are mean±s.d. The table shows transcript levels mean±s.d. a.u. represents arbitrary unit.
We detected reactivation of silent autosomal Oct4-GFP transgenes in transplanted MEF nuclei, indicating that the resistance to reactivation is not a general effect of all silenced transgenes (Supplementary Figure S2A). The absence of reactivation from the Xi 3 days after nuclear transfer contrasted with the strong expression of X-GFP from the Xa in transplanted MEF nuclei, with a 100-fold difference in transcript levels of the same gene in different epigenetic states (Figure 1B). X-GFP remained highly expressed from the Xa of transplanted MEFs immediately after transfer, and transcript levels increased two-fold over 3 days, indicating high transcriptional activity of Xa-GFP in oocytes (Figure 1B). This suggested that while the oocyte is permissive for X-GFP expression from the Xa, there is a strong resistance to its reprogramming from the Xi. The complete absence of reactivation from the Xi was unexpected, given that many repressed genes, including cell-type-specific genes such as MyoD were found to be transcriptionally reactivated after somatic cell nuclear transfer to Xenopus oocytes (Biddle et al, 2009).
To address whether the resistance seen is unique to the Xi of differentiated somatic cells, we transplanted extraembryonic trophoblast stem (TS) cell nuclei carrying the inactive X-GFP on the paternal Xi and followed its expression after nuclear transfer. Xi-GFP TS cells contained an inactivated X-GFP, resulting from imprinted XCI (Kalantry et al, 2006) (Supplementary Figure S2B). After nuclear transfer of Xi-GFP TS cell nuclei, Xi-GFP repression was maintained, and no reactivation of Xi-GFP was detected, indicating that resistance to reprogramming also occurs for the imprinted Xi (Figure 1C, arrows). Together, our results demonstrate that the random Xi of differentiated cells (Xi diff) and the imprinted Xi of TS cells are particularly resistant to reprogramming by Xenopus oocytes, unlike many other genes, which always show reactivation following nuclear transfer.
The Xi of EpiSCs can be reactivated by nuclear transfer to Xenopus oocytes
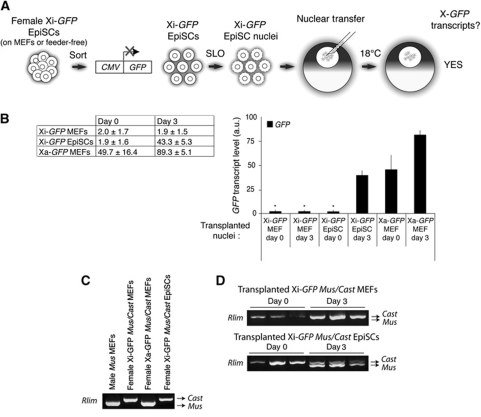
We hypothesized that if resistance to Xi(diff) gene reactivation is under the regulation of epigenetic modifications, the Xi of cells that are less differentiated might carry less repressive marks and be reactivated after nuclear transfer to oocytes. To test this, we used EpiSCs, derived from mouse post-implantation epiblast—the least differentiated cell type known to have undergone XCI (Brons et al, 2007; Tesar et al, 2007). Female EpiSCs have one of their two X chromosomes inactivated, while expressing the autosomal pluripotency genes Oct4, Sox2 and Nanog. The stability of the Xi of EpiSCs is not known so far. We asked if the Xi of EpiSCs (Xi Epi) can be reactivated after nuclear transfer of EpiSC nuclei, again following Xi-GFP expression. We derived X-GFP EpiSCs from E6.5 epiblasts and established Xi-GFP EpiSC lines. We confirmed that female EpiSCs had undergone XCI, and contained an Xi, while expressing pluripotency markers (Supplementary Figures S3 and S4B). To eliminate occasional differentiating EpiSCs or feeder cells from the cultures, we used flow cytometry to separate undifferentiated EpiSCs expressing the pluripotent marker SSEA1 from differentiating, SSEA1-negative cells (Supplementary Figure S5). We transplanted sorted Xi-GFP EpiSCs nuclei to oocyte GVs as depicted in Figure 2A. We also transplanted Xi-GFP and Xa-GFP MEF nuclei (SSEA1 negative) for comparison. While Xi-GFP (diff) of MEF nuclei was not reactivated, the Xi-GFP (Epi) of EpiSC nuclei was strongly reactivated 3 days after nuclear transfer, to a level comparable to that of Xa-GFP MEF nuclei on day 0 (Figure 2B). This indicated that the Xi of EpiSCs is not resistant to reprogramming by oocytes, unlike the Xi of differentiated cells. Similar results were obtained when we transplanted the nuclei of feeder-free EpiSCs cultured on fibronectin (not shown). To test whether endogenous X-linked genes are also reactivated from transplanted EpiSCs nuclei, we carried out allele-specific RT–PCR by exploiting a known polymorphism in X-linked gene Rlim (Huynh and Lee, 2003). We derived Xi-GFP MEFs and EpiSCs from embryos obtained by crossing X-GFP Mus musculus and Mus castaneus mice. Figure 2C shows that restriction enzyme sites present in the musculus, but not the castaneus allele allow to identify the expression origin of the RT–PCR product. We transplanted Xi-GFP MEF and Xi-GFP EpiSC nuclei into oocyte GVs and assayed Rlim expression on day 0 and day 3 after nuclear transfer. Three days after nuclear transfer, monoallelic Rlim expression was detected from transplanted Xi-GFP MEF nuclei, while biallelic expression was detected from transplanted Xi-GFP EpiSCs (Figure 2D). Therefore, Rlim can be reactivated from the Xi after nuclear transfer. These results suggest that the epigenetic inactivation of the Xi in EpiSCs is much less resistant to reprogramming by oocytes than the Xi of differentiated cells.
Figure 2.
The Xi of EpiSCs can be reactivated by nuclear transfer to Xenopus oocytes. (A) Schematic representation of Xi-GFP EpiSCs nuclear transfer experiments. Undifferentiated female EpiSCs cultured on feeders were sorted from differentiating cells by flow cytometry of SSEA1-positive, GFP-negative EpiSCs. After SLO permeabilization, Xi-GFP EpiSC nuclei were transplanted to oocyte GV, and the resulting oocytes were cultured for 3 days. (B) Xi-GFP of EpiSC nuclei can be reactivated after nuclear transfer. Quantitative RT–PCR of X-GFP expression after nuclear transfer. Time points and types of transplanted nuclei are indicated. Transcript levels are shown in table±s.e.m. P<0.05, except samples marked *P<0.2, n=3, error bars show s.e.m. a.u. represents arbitrary unit. (C) Rlim allele-specific RT–PCR. Validation of allele-specific Rlim RT–PCR on cells derived from embryos resulting from a cross between X-GFP Musculus and Castaneus mice (maternal genotype denoted first). MEFs and EpiSCs were derived from embryos genotyped for sex (Ube1 expression) and X-GFP transgene expression and sorted by flow cytometry based on GFP expression. (D) Rlim can be reactivated after nuclear transfer. Allele-specific Rlim RT–PCR of Xi-GFP mus/cast MEFs and EpiSCs, immediately after (day 0) or on day 3 after nuclear transfer.
DNA methylation before nuclear transfer does not correlate with reversibility of the Xi
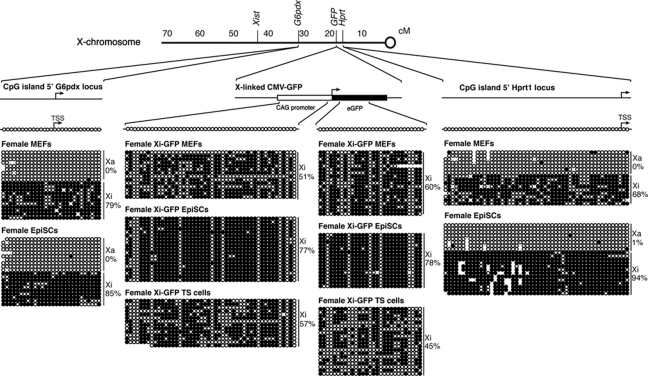
We next sought to identify the epigenetic differences between the Xi of EpiSCs and MEFs that may explain the differences in reactivation of the Xi following nuclear transfer. Because DNA methylation is a known repressor of gene expression, and of nuclear reprogramming, we determined the DNA methylation status of the Xi in these cells before nuclear transfer. We assayed the DNA methylation state of two X-linked genes adjacent to the X-GFP reporter in these cell lines, namely G6pdx and Hprt1; as well as regulatory and coding regions of the X-GFP transgene itself (Supplementary Figure S1A). All these genes are subjected to XCI and repressed in these cells. Bisulphite analysis revealed that the regulatory regions of G6pdx, Hprt1 and X-GFP were fully methylated on the Xi alleles of both female MEFs and EpiSCs (Figure 3), while the Xa alleles were unmethylated. Therefore, DNA methylation alone on the Xi in donor nuclei fails to explain the difference between Xi(diff) and Xi(Epi) reversibility following nuclear transfer. This raised the possibility that other differences in donor nuclei such as histone modifications may be responsible for the effect seen in gene reactivation following nuclear transfer. We conclude that resistance to reprogramming of the Xi by oocytes does not correlate with DNA methylation of Xi(diff) or Xi(Epi).
Figure 3.
DNA methylation does not correlate with reversibility of the Xi before nuclear transfer. Bisulphite analysis of G6pdx, X-GFP and Hprt1 promoter and coding regions in female MEFs, EpiSCs and TS cells. All regulatory regions tested are fully methylated on the Xi of all cell types (black circles), and unmethylated on the Xa allele (open circles). The proportion of methylated CG residues is indicated. No circles represent mutated or missing CpGs.
H3K27me3 does not correlate with reversibility of the Xi before and after nuclear transfer
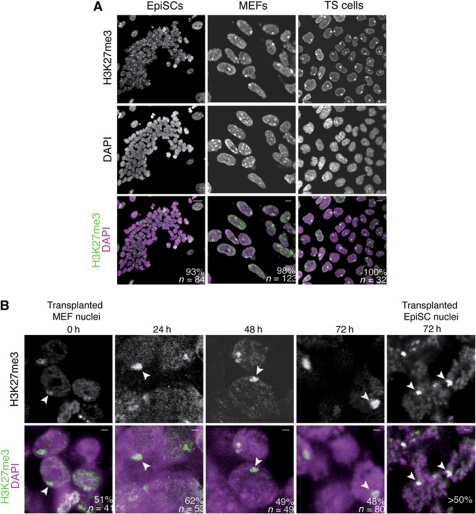
We aimed to find chromatin modifications that correlate with the reversible Xi of EpiSCs or the irreversible Xi of MEFs and TS cells. We determined enrichment of Polycomb-induced marks on the Barr body of the Xi by immunofluorescence. This analysis is facilitated because the condensed chromatin of the Xi can be easily seen by immunofluorescence against H3K27me3 as a bright nuclear macrodomain, often localized at the nuclear periphery (Silva et al, 2003). We determined the proportion of nuclei in which specific staining of the Xi is seen in cells before nuclear transfer. H3K27me3 was enriched on the Xi in all cell types examined (Figure 4A). In agreement with previous studies (de Napoles et al, 2004; Rougeulle et al, 2004; Guo et al, 2009), H3K27me3 was enriched on the Xi in 93% of female EpiSC nuclei (n=84), on 98% of female MEF nuclei (n=132) and on 100% of female TS cell nuclei (n=32). During initiation of XCI, H3K27me3 is deposited by the PRC2 catalytic subunit Ezh2 (Silva et al, 2003). We found that Ezh2 is enriched on the Xi of EpiSC nuclei (73%, n=100), and of TS cell nuclei (100%, n=53), but not on the Xi of MEF nuclei (0%, n=100) (Supplementary Figure S3). The presence of Ezh2 on the Xi of EpiSCs is in agreement with the idea that the Xi of EpiSCs may represent an earlier stage in XCI compared with MEFs (de Napoles et al, 2004; Kohlmaier et al, 2004).
Figure 4.
H3K27me3 does not correlate with reversibility of the Xi before and after nuclear transfer. (A) Immunofluorescence of X-GFP female EpiSCs, MEFs and TS cells against H3K27me3. Confocal images of H3K27me3 immunostainings (green) counterstained with DAPI (magenta) show that H3K27me3 is enriched on the Xi of female EpiSCs grown on feeders (93%, n=84), the Xi of MEFs (98%, n=123) and the Xi of TS cells (100%, n=32). Scale bars=10 μm. Images are projected Z-sections. (B) H3K27me3 is maintained on the Xi after nuclear transfer. Immunofluorescence of transplanted female MEFs nuclei against H3K27me3 (green). A high proportion (>48%) of female nuclei retain an H3K27me3-labelled Xi up to 72 h after nuclear transfer (arrowheads). The proportion of nuclei carrying an H3K27me3-labelled Xi is shown. n=number of nuclei. DAPI is shown in magenta. Scale bars=2 μm. Images are single Z-sections.
We next tested whether the H3K27me3 mark is reversed on the Xi following nuclear transfer using immunofluorescence of transplanted nuclei. Female MEF and EpiSC nuclei were transplanted to oocyte GVs and fixed, immediately, or at various time points, after nuclear transfer. Immunostaining against H3K27me3 in fixed GVs containing transplanted MEF nuclei revealed that the mark is maintained on the Xi of transplanted nuclei 3 days after nuclear transfer (Figure 4B). H3K27me3 was also maintained on the Xi of transplanted female EpiSCs (Figure 4B). Therefore, H3K27me3 is not reversed on the Xi following nuclear transfer to Xenopus oocytes. Since H3K27me3 is maintained on the Xi of both MEF and EpiSC nuclei, this repressive mark does not explain the resistance of Xi(diff) to reactivation.
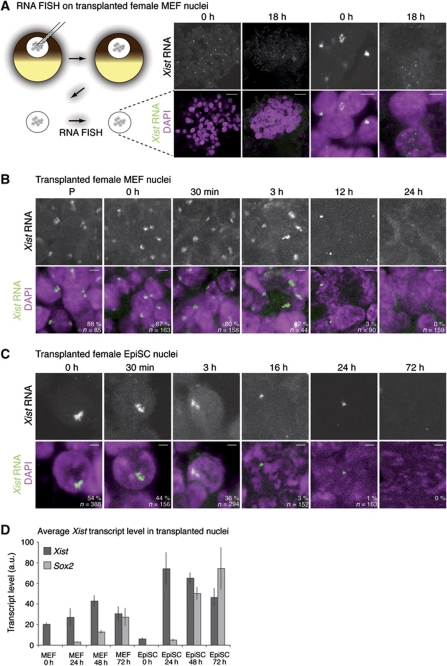
The long noncoding RNA Xist dissociates from chromatin of the Xi after nuclear transfer
During initiation of XCI, the long noncoding RNA Xist induces gene inactivation on the chromosome from which it is produced, by recruiting the machinery necessary for silencing (Heard and Disteche, 2006). Because X reactivation is associated with the removal of Xist RNA, we investigated Xist RNA localization on the Xi in nuclei of female somatic cells transplanted into oocytes. Moreover, the fate of long noncoding RNAs has not previously been described following somatic cell nuclear transfer to Xenopus oocytes. We followed the localization of Xist RNA before and after nuclear transfer by fluorescent RNA in situ hybridization (RNA FISH). RNA FISH against Xist identified a single Xist RNA cloud localized to the Xi of untransplanted female MEFs and EpiSCs (Supplementary Figure S4A and B, respectively). Strikingly, Xist RNA coating of the Xi was lost in transplanted MEF nuclei 18 h after nuclear transfer, although it was fully localized to the Xi immediately after transfer (Figure 5A). A detailed time course revealed that nuclear transfer did not induce obvious changes to this pattern within 3 h after transfer (Figure 5B). However after this, Xist RNA was gradually lost, and was fully delocalized from the Xi after 12 h. Whereas over 80% of transplanted MEF nuclei contained an Xist RNA cloud on their Xi within 3 h after transfer (n=44–163), none of the transplanted MEF nuclei had Xist RNA on their Xi 12 h (3%) and 24 h (0%) after nuclear transfer (n=90 and 158; Figure 4B). In some instances, Xist RNA dispersion was seen, with multiple Xist RNA FISH punctate signals distributed throughout transplanted nuclei, reminiscent of those observed in mitotic cells (Figure 5A (18 h) and Supplementary Figure S5B, high magnification panels). Loss of Xist RNA from the Xi was also observed with similar kinetics in the nuclei of transplanted female EpiSCs (Figure 5C), with near complete loss of the Xist RNA cloud from the Xi 24 h after nuclear transfer (Figure 5C). Visualization of the Xi chromosome territory by H3K27me3 immunofluorescence revealed no obvious change in the shape of the Xi, suggesting that Xist delocalization is not due to changes in Xi organization (Figure 4B). Together, these results show that the long noncoding RNA Xist is dispersed from the Xi domain of both Xi(diff) and Xi(Epi) after somatic cell nuclear transfer to oocyte GVs.
Figure 5.
The long noncoding RNA Xist dissociates from chromatin of the Xi after nuclear transfer. (A) RNA FISH for Xist RNA (green) on transplanted female MEF nuclei. Oocyte GVs containing transplanted nuclei were dissected, fixed and subjected to RNA FISH against Xist RNA. Confocal images reveal that the Xist RNA cloud of female MEFs (0 h) is lost from the Xi 18 h after nuclear transfer. Note the presence of punctate Xist RNA FISH signal dispersed throughout the nucleus of some of the 18 h transplanted nuclei. DAPI is shown in magenta. Low (scale bars=25 μm) and high (scale bars=5 μm) magnification pictures are shown. P denotes permeabilized nuclei. Images are projected Z-sections. (B, C) Xist RNA is lost from the Xi after nuclear transfer of female MEFs (B) and female EpiSCs (C). Xist RNA FISH of nuclear transfer female MEFs and EpiSCs. Samples were collected and fixed at indicated time points. The Xist RNA cloud characteristic of the Xi is maintained up to 3 h after nuclear transfer, then decreases to give a pinpoint signal at 12 and 16 h, and is completely lost from transplanted nuclei by 24–48 h after nuclear transfer. The proportion of nuclei with a Xist RNA cloud is indicated. DAPI is shown in magenta. n=number of nuclei. Scale bars=5 μm in (B) and 2 μm in (C). Images are projected Z-sections. (D) Xist expression levels in transplanted female MEF and EpiSC nuclei. qRT–PCR analysis of Xist (dark grey) and Sox2 (light grey) expression in transplanted nuclei. Xist transcript levels increase after nuclear transfer. Error bars are s.e.m. a.u. represents arbitrary unit.
To determine if Xist RNA dispersion is due to the discontinuation of its synthesis, we quantitated Xist transcript levels after nuclear transplantation. qRT–PCR of transplanted nuclei showed that Xist transcripts accumulate in oocytes transplanted with MEFs and EpiSCs nuclei (Figure 5D). Therefore, the dispersal of Xist RNA from chromatin of the Xi occurs even though Xist transcripts accumulate in the oocyte. Because Xist splicing is required for localization to the Xi, we suspected that Xist might be aberrantly spliced after nuclear transfer. We examined Xist splicing in transplanted MEF and EpiSC nuclei. Remarkably, Xist transcripts were efficiently spliced after nuclear transfer (Supplementary Figure S4C). In conclusion, the resistance of the Xi toward reprogramming in MEF nuclei transplanted in oocytes does not depend on chromosome-associated Xist RNA. This suggested that nuclear transfer to Xenopus oocytes induces reactivation of genes whose repression is maintained by Xist RNA.
Nuclear transfer can reverse Xist-induced, Xist-independent stable gene repression
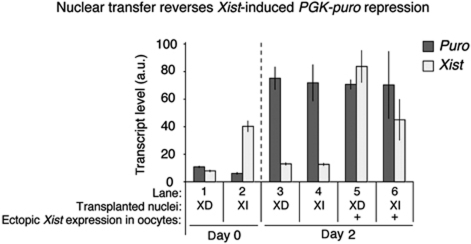
By using an independent system, allowing controlled Xist expression, we tested whether nuclear transfer to oocyte reactivates genes that are maintained in a repressed state in a Xist RNA-dependent or -independent manner. An inducible Xist expression system in ES cells triggers silencing of a PGK-puromycin reporter (PGK-puro) in cis (Wutz and Jaenisch, 2000). This system has been shown to induce reversible, XD PGK-puro repression in ES cells or stable silencing upon combined Xist induction and retinoic acid (RA) ES cells differentiation (Supplementary Figure S6; Wutz and Jaenisch, 2000; Leeb and Wutz, 2007). This is based on PGK-puro reactivation upon removal of Xist after a period during which repression has been triggered by Xist. We tested reactivation of repressed PGK-puro from XD or stable XI cells, by nuclear transfer to Xenopus oocytes. Expression analysis showed a strong reactivation of PGK-puro expression from both types of transplanted nuclei (Figure 6, lanes 1–4). This means that the epigenetically stable repression of PGK-puro induced by Xist during RA differentiation of ES cells is efficiently reprogrammed by Xenopus oocytes. Therefore, we reasoned that the resistance of the Xi of MEFs to reactivation by Xenopus oocytes must be acquired late in the XCI process, even after epigenetically stable, XI repression is induced. This prompted us to examine the incorporation of the repressive histone variant mH2A, a known late event of XCI (see below).
Figure 6.
Nuclear transfer reverses epigenetically stable, Xist-induced and Xist-independent gene repression. Reversibility of PGK-puro silencing following nuclear transfer of clone 36 cells. To obtain the Xist-dependent (XD) PGK-puro repressed state, clone 36 ES cells were induced to express Xist for 4 days. To obtain the Xist-independent (XI), stable PGK-puro repressed state, clone 36 ES cells were induced to differentiate with RA for 4 days while being induced with Xist at the same time. The nuclei of XD and XI PGK-puro repressed cells were transplanted to oocytes. Biological triplicates were collected immediately or 2 days after nuclear transfer. Nuclei induced to ectopically express Xist after nuclear transfer, within the GV is indicated (+). Transcriptional analysis of puro (dark grey) and Xist (light grey) expression by qRT–PCR of oocytes transplanted with nuclei obtained as described in Supplementary Figure S6B is shown. P<0.05, n=3. Error bars are s.e.m. a.u. represents arbitrary unit.
We also tested whether ectopic Xist expression induced from transplanted nuclei could prevent reactivation of repressed PGK-puro following nuclear transfer. Ectopic Xist expression did not prevent PGK-puro reactivation from transplanted nuclei (Figure 6, lanes 5 and 6). We conclude that noncoding RNA Xist-mediated repression is reversed efficiently from transplanted nuclei, and is not affected by continuous Xist expression after nuclear transfer to Xenopus oocytes. Therefore, XD silencing is not a candidate for the irreversible silencing of Xi(diff) following transfer to oocyte.
macroH2A correlates with irreversible Xi and is maintained after nuclear transfer
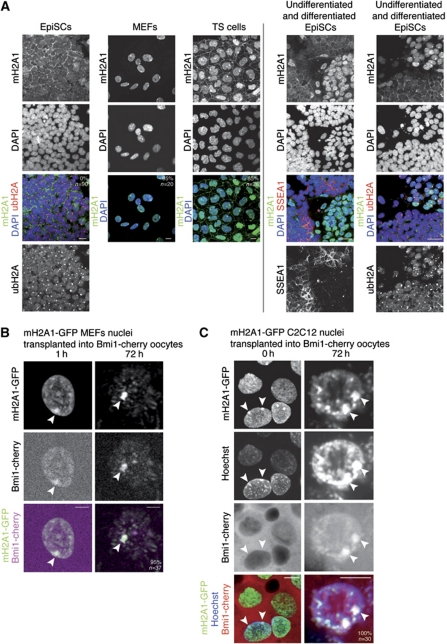
A known late event of XCI is the incorporation of the repressive H2A histone variant macroH2A1 (mH2A1) (Costanzi and Pehrson, 1998), occurring after gene silencing has been induced (Rasmussen et al, 2001). We determined enrichment of mH2A1 on the Xi by immunofluorescence. Consistent with previous work (Rasmussen et al, 2000; Kalantry et al, 2006), we found that mH2A1 is enriched on the Xi of MEFs (95%, n=20) and TS cells (85%, n=26) (Figure 7A). However, mH2A1 was completely absent from the Xi in EpiSCs (0%, n=90), thereby correlating with the resistance of Xi reprogramming to oocyte transfer previously observed (Figure 7A). Thus, incorporation of mH2A1 correlates with a switch from a reversible state (Xi(Epi), no mH2A1) to an irreversible state (Xi(diff), mH2A1 positive) of the Xi. EpiSCs are known to show a high degree of cellular heterogeneity and spontaneously differentiate into somatic lineage such as endoderm (Hayashi and Surani, 2009; Gillich and Hayashi, 2011). Interestingly, we observed that mH2A1 became enriched then incorporated on the Xi of spontaneously differentiated EpiSCs, revealed by loss of the pluripotency marker SSEA1 (Figure 7A, right column). In addition, a mH2A1 nuclear domain was absent from 91% of the nuclei of the Xist-inducible cell line induced for 4 days with Xist and RA, further correlating with the ability to reactivate after nuclear transfer (Supplementary Figure S6D). We conclude that resistance to reprogramming of the Xi(diff) by oocytes is correlated with chromatin changes occurring during XCI, such as the incorporation of mH2A1.
Figure 7.
mH2A correlates with stable Xi and is maintained after nuclear transfer to Xenopus oocytes. (A) Immunostaining of female EpiSCs, MEFs and TS cells against mH2A1 (green), ubH2A (red) and SSEA1 (red). Undifferentiated female EpiSCs do not exhibit accumulation of mH2A1 on the ubH2A-labelled Xi. mH2A1 is induced in differentiated EpiSCs, marked by loss of the pluripotency marker SSEA1 (right panel, left column). mH2A1 is incorporated in chromatin of the Xi in differentiated EpiSCs, as shown by co-localization with Xi marker ubH2A (right panel, right column). Note that the Xi of undifferentiated EpiSCs is stained with ubH2A only. mH2A1 was found accumulated in 95% of female MEFs, 85% of female TS cells and 0% of female EpiSCs. DAPI is shown in blue. Scale bars=10 μm. Images are projected Z-sections. (B, C) mH2A1-GFP remains associated with heterochromatic regions in transplanted nuclei and reveals chromatin reorganization. Projections of confocal images of mH2A1-GFP MEF (B) and sable mH2A1-GFP C2C12 (C) nuclei transplanted into oocytes preloaded with Bmi1-cherry by mRNA injection. Note the persistence of mH2A1-GFP on the Xi (arrowheads), bound by Bmi1-cherry imported from the oocyte, and the appearance of mH2A1-GFP-labelled pericentric heterochromatin foci. Scale bars=10 μm. Images are projected Z-sections.
We wished to test if the histone variant mH2A1, present only on the irreversible Xi(diff), was reversed or maintained on the Xi after nuclear transfer. We followed mH2A1.2-GFP (mH2A1-GFP) on the Xi(diff) of transplanted nuclei of differentiated cells. Because the mH2A antibody binds to an unknown epitope in the GV, immunofluorescence of transplanted nuclei was not possible. Thus, we generated a female C2C12 cell line stably expressing mH2A1-GFP. C2C12 cells are known to contain two Xi (Håkelien et al, 2008; Casas-Delucchi et al, 2011). Accordingly, mH2A1-GFP localized to chromatin and was enriched on the two Xi of C2C12 cell nuclei (Supplementary Figure S7). Immunostaining confirmed co-localization of mH2A1-GFP with H3K27me3 on the two fully inactive Xi of mH2A1-GFP C2C12 cells (Supplementary Figure S7C). Next, we followed mH2A1-GFP from C2C12 nuclei, and from MEF nuclei transplanted into oocytes to see if it is lost from the Xi. As a positive marker for the Xi in transplanted nuclei, we used PRC protein Bmi1 fused to cherry, which became localized to the Xi of transplanted nuclei when expressed in oocytes by mRNA injection (Hernández-Muñoz et al, 2005). We transplanted mH2A1-GFP MEF or mH2A1-GFP C2C12 nuclei into the GV of oocytes preloaded with Bmi1-cherry, and followed mH2A1-GFP and Bmi1-cherry localization by confocal microscopy. This was possible through the isolation of oil GV and real-time monitoring of mH2A1-GFP in transplanted nuclei (Jullien et al, 2010). Time-lapse imaging over the first 12 h after nuclear transfer revealed general nuclear swelling together with a major reorganization of chromatin (Supplementary Movie S1). Although Bmi1-cherry was initially present only in the GV plasm, it became localized to transplanted nuclei and enriched on the Xi within a few hours after transfer (Figure 7B and C). Most importantly, whereas a decrease in overall nuclear mH2A1-GFP was observed within 12 h after transfer, mH2A1-GFP was maintained in heterochromatin of the Xi up to 72 h, co-localizing with Bmi1-cherry on the Xi in 100% of MEF and C2C12 nuclei examined 3 days after nuclear transfer (n=37 and 30, respectively; Figure 7B and C, arrowheads; Supplementary Movie S2). mH2A1-GFP also remained associated with other heterochromatin regions in transplanted nuclei. In conclusion, in vivo real-time monitoring of mH2A1-GFP in transplanted nuclei reveals the unexpected continuous association of this repressive histone variant with heterochromatin of the Xi of differentiated cells after nuclear transfer to oocyte GV. This suggests that mH2A1 is not only involved in stable repression of the Xi, but also in resistance towards reprogramming.
macroH2A depletion from donor nuclei improves reprogramming efficiency
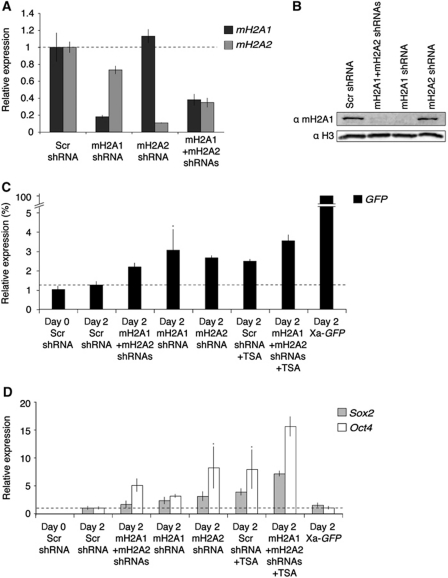
To test if incorporation of mH2A into chromatin restricts transcriptional reactivation after nuclear transfer, we established Xi-GFP MEF lines stably expressing shRNAs against mH2A1 (mH2A1.1 and mH2A1.2), macroH2A2 (mH2A2), control scramble sequence or both mH2A1 and mH2A2 (Figure 8A and B). mH2A depletion alone did not induce reactivation of Xi-GFP, Sox2 or Oct4 before nuclear transfer (Supplementary Figure S8A and B), except for a 2.5-fold increase over background in Oct4 transcripts upon co-depletion of mH2A1 and mH2A2. We transplanted the nuclei of mH2A depleted and control Xi-GFP MEFs to oocyte GV and analysed transcriptional reactivation 2 days after nuclear transfer (Figure 8C and D). mH2A knockdown was not sufficient for full reactivation of Xi-GFP, when compared with Xa-GFP transcript levels from transplanted Xa-GFP MEFs. However, mH2A depletion led to a significant, 1.7- to 2.4-fold increase over background in detected GFP transcripts (Figure 8C). This increase was comparable to the increase seen in transplanted oocytes grown in the presence of the histone deacetylase (HDAC) inhibitor Trichostatin A (TSA) (two-fold; Figure 8C). Moreover, depletion of mH2A1 and mH2A2 together with TSA treatment resulted in the combined effect of mH2A knockdown and TSA alone namely a 2.8-fold increase in GFP transcripts. We conclude that mH2A is not the only factor contributing to Xi reversibility, yet mH2A does restrict transcriptional reprogramming by oocytes. To address whether mH2A may be a more general restriction to gene reactivation, we analysed transcript levels of pluripotency genes Sox2 and Oct4 after nuclear transfer of mH2A depleted cells. Strikingly, the effect of mH2A depletion was even more pronounced, with a 1.6- to 3.1-fold and a 3.1- to 8.2-fold increase in Sox2 and Oct4 reactivation, respectively (Figure 8D). Sox2 and Oct4 reactivation were increased 3.9- and 7.9-fold by TSA alone, and 7.2- and 15.6-fold by TSA together with mH2A1 and mH2A2 co-depletion. We conclude that mH2A contributes to resistance to transcriptional reprogramming.
Figure 8.
mH2A depletion improves reprogramming by nuclear transfer. (A) qRT–PCR analysis of mH2A1 and mH2A2 expression following shRNA-mediated mH2A RNAi. (B) Western analysis of mH2A1 in shRNA expressing Xi-GFP MEFs. (C, D) qPCR analysis of GFP (black), Sox2 (grey) and Oct4 (white) expression in transplanted Xi-GFP MEFs nuclei subjected to mH2A RNAi and/or TSA treatment. P<0.05 except samples marked *P<0.06 in (C), or *P<0.08 in (D), n=3. Error bars are s.e.m. Note the differences in y axis.
Discussion
In this study, we have analysed the relationship between the epigenetic state of genes before nuclear transfer and the efficiency of transcriptional reprogramming by Xenopus oocytes by using the Xi as a tool. One outcome of our analysis is that the epigenetic state of repressed genes in somatic nuclei before nuclear transfer is an important determinant for the efficiency of transcriptional reprogramming. Based on nuclear transfer of X-GFP MEF nuclei, there is a 100-fold difference in the reprogramming of the same gene in two different epigenetic states. This difference is mainly due to a remarkable resistance of the Xi of differentiated cells to reprogramming by Xenopus oocytes. Another striking outcome is that although the stability of XCI in EpiSCs has been unknown so far, we find that the Xi of EpiSCs can be reactivated by nuclear transfer to Xenopus oocytes, unlike the one of differentiated cells. The difference between the Xi(Epi) and Xi(diff) reflects a shift from a reversible to an irreversible repressed state, correlated with the acquisition of the repressive histone variant mH2A1. Collectively, our results show that the Xi of EpiSCs is less stable than that of more differentiated cells, and represents an earlier stage of XCI. This is supported by the presence of Ezh2 on the Xi of EpiSCs, indicative of the initiation phase of XCI (Silva et al, 2003). This could reflect the known higher developmental potential of EpiSCs and of their Xi, which needs to become reactivated during development of the germline, induced from post-implantation epiblast. We propose that the Xi of EpiSCs is poised for reactivation in the germline. We believe that some, but clearly not all, of the molecular mechanisms leading to X reactivation in the ICM may be operative in Xenopus oocytes. The absence of Xi reactivation in transplanted TS cell nuclei may also reflect differences in the mechanisms of maintenance of XCI between imprinted and random X inactivation.
Although we did not find any differences between the DNA methylation state of the Xi between EpiSCs, MEFs and TS cells, the extent to which DNA methylation contributes to Xi repression in our experiments is not known. DNA methylation is a known barrier to reprogramming (Simonsson and Gurdon, 2004; Mikkelsen et al, 2008). Yet, methylated DNA is perfectly well transcribed in Xenopus oocytes until it becomes chromatinized, recruits methyl-DNA-binding protein and HDACs (Jones et al, 1998). Therefore, DNA methylation alone does not restrict transcription. Random XCI occurs in embryos in the absence of Dnmt1, although maintenance of XCI is severely compromised (Sado et al, 2000). To determine the role of non-DNA methylation processes in restricting gene reactivation, it will be interesting to test reactivation from the Xi devoid of DNA methylation. Since we did not observe any difference between DNA methylation states of Xi(Epi) and Xi(diff), additional mechanisms must be responsible for resistance to reprogramming of Xi(diff).
Unexpectedly, reactivation of Xi-GFP from EpiSCs occurred while H3K27me3 is maintained on the Xi. This was surprising given that H3K27me3 is considered a repressive mark. However, there is no direct evidence to suggest that H3K27me3 directly inhibits transcription. In addition, mutation of the repeat A region of Xist prevents gene silencing while still allowing recruitment of PRC2 and deposition of H3K27me3 (Wutz et al, 2002). It is also possible that the mediators of the Polycomb system are not fully effective in the transcriptionally permissive environment of the Xenopus oocyte. Indeed, high H3K27me3 levels are maintained on pluripotency gene regulatory regions after nuclear transfer, concomitant with their transcriptional reactivation in oocytes (Murata et al, 2010). In addition, recent evidence points toward noncatalytically related functions of the PRC system (Eskeland et al, 2010). Our results therefore suggest that H3K27me3 is permissive to transcription in the Xenopus oocyte GV. Also surprising was the maintenance of H3K27me3 on the Xi in the absence of Xist RNA. Conditional deletion of Xist has been reported to lead to loss of H3K27me3 on the Xi (Plath et al, 2004). How quickly this occurs after Xist deletion is unknown and our results suggest that loss of H3K27me3 after Xist RNA delocalization may require cell division, which does not occur in our system.
Dispersion of Xist RNA from the Xi after nuclear transfer occurred with a concomitant increase in Xist transcription, but without defects in Xist splicing. This suggests that nuclear transfer to oocytes disrupts noncoding RNA interactions with chromatin, in agreement with mouse oocyte nuclear transfer studies (Bao et al, 2005). This has interesting implications especially given the emerging roles of long noncoding RNAs in setting up specific chromatin states (Guttman et al, 2009; Koziol and Rinn, 2010). We suggest the loss of noncoding RNA interactions with chromatin as a possible fundamental principle by which nuclear transfer to oocytes leads to transcriptional reprogramming. This could be caused by a passive or an active mechanism. It is possible that dilution of Xist RNA may occur after nuclear transfer, due to nuclear swelling (Gurdon, 1968). However, Xist delocalization is more likely to be caused by the loss of a crucial factor required for Xist localization to the Xi. This could be the recently identified SafA (Hasegawa et al, 2010), required for chromosomal localization of Xist, or SATB1 (Agrelo et al, 2009), whose expression induces dispersed Xist RNA signals in lymphocytes. Alternatively, high Aurora B activity in injected oocytes (Murata et al, 2010) could result in Xist RNA delocalization, as reported in human mitotic cells (Hall et al, 2009). Xist RNA delocalization could also reflect evolutionary changes in the use of a common basal mechanism, as some Xenopus laevis interspersed repeat containing RNAs, homologous to mammalian Xist, are translocated to the Xenopus oocyte germ plasm (Kloc et al, 1993).
Reactivation of Xist-induced, XI repressed PGK-puro transgene in RA differentiated ES cells suggested that the irreversibility of the Xi(diff) is induced late during XCI. This prompted us to examine the H2A histone variant mH2A. mH2A is enriched on the Xi (Mietton et al, 2009) and is a known repressor of transcription (Angelov et al, 2003; Doyen et al, 2006). Genome-wide analysis of mH2A distribution indicates that it is depleted from most active genes (Changolkar et al, 2010) and enriched on repressed chromatin (Buschbeck et al, 2009; Gamble et al, 2010; Barzily-Rokni et al, 2011). We found mH2A to be upregulated and subsequently enriched on the Xi upon differentiation of EpiSCs, consistent with a global increase in mH2A upon ES cell differentiation (Dai and Rasmussen, 2007). Upon nuclear transfer, mH2A was not loss from the Xi, despite Xist RNA delocalization. Since conditional deletion of Xist leads to mH2A delocalization (Csankovszki et al, 1999), mH2A probably also requires cell division in order to be lost from the Xi after Xist RNA dispersal. Interestingly, mH2A is rapidly removed from pronuclei after fertilization (Nashun et al, 2010), as well as from transplanted nuclei after somatic nuclear transfer to egg (Chang et al, 2010).
mH2A knockout mice are fertile and viable (Changolkar et al, 2007, 2010), and double mH2A1/mH2A2 knockouts mouse embryos are said to appear normal (Buschbeck and Di Croce, 2010). mH2A is therefore not required to induce XCI and gene silencing in general. However, a role for mH2A in maintenance of XCI is demonstrated by our nuclear transfer experiments, in which transcriptional reactivation is more efficient in the absence of mH2A, demonstrating that mH2A restricts reprogramming and helps maintain the repressed state of genes after silencing has been acquired during cellular differentiation. This is in agreement with increased Xi-GFP reactivation from MEFs depleted of mH2A and treated with inhibitors of DNA methylation or HDACs inhibitors (Csankovszki et al, 2001; Hernández-Muñoz et al, 2005; Barzily-Rokni et al, 2011). Our experiments suggest that this type of combinatorial repression further stabilized by mH2A may be a more general phenomenon since transcriptional reprogramming of Oct4 and Sox2 was also enhanced in the absence of mH2A.
Insights into how mH2A may mechanistically restrict reprogramming are suggested by several biochemical and in vivo studies. In vitro, mH2A impedes transcription factor binding (Angelov et al, 2003), has lower affinity for SWI/SNF complexes (Chang et al, 2008) and prevents VP16-induced p300-mediated histone acetylation and transcriptional activation (Doyen et al, 2006). In addition, mH2A is thought to interact with HDAC1 and HDAC2 (Chakravarthy et al, 2005). mH2A containing nucleosomes are more stable than canonical H2A nucleosomes as suggested by an increased salt resistance (Abbott et al, 2005). By FRAP, mH2A shows reduced mobility compared with H2A (Gaume et al, 2011). mH2A may restrict nuclear reprogramming by one or a combination of these mechanisms.
Very interestingly, loss of mH2A has been linked to melanoma progression (Kapoor et al, 2010), as well as lung and possibly breast cancer recurrence (Sporn et al, 2009). Probably the most important outcome of our work is that the mechanisms that restrict nuclear reprogramming may also prevent cancer progression.
Materials and methods
Nuclear transfer and Xenopus oocytes preparation
Oocytes were prepared as previously described (Halley-Stott et al, 2010) and injected using a Drummond Nanoject microinjector. All experiments were performed at 18°C. Donor nuclei were permeabilized as described (Halley-Stott et al, 2010).
Cell culture
MEFs were derived from E13.5 embryos hemizygous for the X-GFP transgenic allele (Hadjantonakis et al, 2001). For allele-specific RT–PCR, embryos resulting from X-GFP Mus Musculus musculus crossed with Mus musculus castaneus mice were used to derive MEFs. Embryos were individually genotyped for sex and X-GFP transgene transmission, or sexed by inspecting gonads for the pattern of X-GFP expression. Gonads were removed before processing the embryos for MEF isolation. MEFs were cultured in MEF medium (DMEM (Gibco; 41965-062) supplemented with 10% FBS, 200 μM GlutaMAX™-I Supplement (Gibco; 35050-038), 100 μg/ml penicillin/streptomycin)). X-GFP Mus/Cast MEFs were immortalized using Addgene plasmid 21826 and sorted by flow cytometry. EpiSCs were derived from female E6.5 X-GFP epiblast (129/SvEv female crossed with transgenic X-GFP male mice or Mus musculus castaneus crossed with transgenic X-GFP male mice) as described previously (Bao et al, 2009). EpiSCs were cultured in chemically defined medium (Brons et al, 2007) supplemented with recombinant human activin A (20 ng/ml; Peprotech; 120-14) and bFGF (12 ng/ml; Invitrogen; 13256-029) on MEFs. EpiSCs were passaged every 2 days using collagenase (Invitrogen; 17104-019). For feeder-free culture, EpiSCs were maintained in N2B27 medium (Stem Cell Sciences; SCS-SF-NB-02) in activin and bFGF on fibronectin (Millipore; FC010). EpiSCs were passaged using Accutase (PAA; L11-007) every 2 days. X-GFP TS cells (Kalantry et al, 2006) were cultured on Mitomycin C-treated MEFs in RPMI1640 (Gibco; 31800-022), 10% FCS (LabTech; 4-101-500), 200 mM L-Glutamine (Gibco; 25030-032), 100 mM sodium pyruvate (Gibco; 11360-039), 100 μg/ml penicillin/streptomycin, 10 mM betamercaptoethanol, 2.5 ng/ml recombinant FGF4 (Preprotech, London, UK; cat 100-31), 100 pg/ml Heparin (Sigma; H3149). To establish feeder-free cultures, feeders were removed gradually over four passages and resulting TS cells cultured in feeder-conditioned medium (Tanaka, 2006). ES cells were cultured in ES medium (GMEM (Gibco; 21710) supplemented with 20% ES grade FCS, MEM nonessential amino acids (Gibco; 11140), MEM sodium pyruvate (Gibco; 11360) 0.1 mM betamercaptoethanol and LIF). For the generation of mH2A1-GFP C2C12 cells, pCS2+ mH2A1-GFP plasmid was co-transfected with a selectable puromycin or G418 resistance plasmid using Lipofectamine (Invitrogen). Cells were selected based on resistance to puromycin and mH2A1-GFP expression, before single clone expansion. All cells were cultured in 5%CO2/95% air at 37°C.
RNAi
pSuper.retro.puro vectors encoding shRNAs (Supplementary data) were transfected into plat E cells and the resulting viruses were used to infect X-GFP MEFs. Infected cells were selected with 2 μg/ml puromycin.
Flow cytometry
For flow cytometry of X-GFP MEFs, cells were treated with trypsin, filtered and resuspended at 10–20 × 106/ml. Cells were sorted using the Dako MoFlo high-speed cell sorter or FACSAria (BD Biosciences). Undifferentiated EpiSCs were labelled using anti-SSEA1 antibody (FAB2155P; R&D Systems), as described (Hayashi et al, 2008).
Confocal analysis
Confocal analysis was carried out on a Zeiss 510 META confocal LSM microscope equipped with argon (458/477/488/514 nm lines) and HeNe (543 nm) lasers or on a Olympus FV1000 Upright microscope equipped with solid state (405 nm), argon (458/488/515 nm lines), solid state (559 nm) and solid state (635 nm) lasers using the × 60 objective. The noise of all images was removed by using the despeckle function of ImageJ. Z-sections were then projected on a single plane by using the ImageJ standard deviation function under Z-project.
qRT–PCR
RNA extraction, cDNA synthesis and qPCR were performed as described (Halley-Stott et al, 2010). Primers used are listed in Supplementary data. Standard curve was obtained by diluting Oct4-GFP or clone 36 ES cell cDNA. Allele-specific RT–PCR of Musculus/Castaneus X-linked genes has been described (Huynh and Lee, 2003).
Bisulphite analysis
Bisulphite treatment was performed on 800 ng of gDNA from Xi-GFP MEFs and Xi-GFP EpiSCs using the Epitect Bisulfite Kit (Qiagen; 59104). Nested PCR for regions of the mouse Hprt and G6pdx were performed using bisulphite-specific primers on 0.5 μl of template. The primers used are listed in Supplementary data. The PCR fragments, cloned into pGEM-T easy vector (Promega), contained 53 CpGs for Hprt1 promoter and 28 CpGs for G6pdx promoter.
In vitro transcription
All cDNAs of interest were cloned into pCS2+ vectors, linearized and transcribed in vitro as described (Biddle et al, 2009). The mouse ORF of Bmi1 and mH2A1.2 were cloned into pENTR vectors and recombined into pCS2+ cherry/eGFP-HA destination vectors as C-terminal fusions using the Gateway system (Invitrogen). A measure of 10 ng of mRNA were injected into stage V oocytes and cultured at 18°C.
RNA FISH
GV containing transplanted nuclei were dissected and immediately fixed in 4% PFA/1 × PBS overnight at 4°C. RNA FISH was carried out as described (Panning, 2004), with slight modifications. DIG-labelled Xist RNA probes were synthesized from five different Xist cDNA PCR products as described (Nolen et al, 2005). A full protocol can be found in Supplementary data available at The EMBO Journal Online.
Statistical analysis
Statistical differences were assessed with the unpaired Student's t-test. Data are presented as mean±s.e.m., and P-values <0.05 were considered statistically significant. Additional information about immunohistochemistry, antibodies, primers and bisulphite is included in Supplementary data.
Supplementary Material
Acknowledgments
We thank our colleagues for critical reading of the manuscript. We particularly thank Azim Surani, Anton Wutz, Terry Magnuson, Anne Ferguson-Smith, Josè Silva and Joost Gribnau for reagents and stimulating discussions; Caroline Lee, William Mifsud and Katsuhiko Hayashi for derivation of MEFs and flow cytometry; Edith Heard, Barbara Panning and Jeannie Lee for advice on RNA FISH and allele-specific RT–PCR; Fabio Rossi for advice on bisulphite analysis of CAG promoters; Rachael Walker for flow cytometry; Alex Sossick for imaging assistance, David Ron for SV40 LTa plasmid. This work was supported by The Wellcome Trust (RG54943), (081277), (RG44593). VP was also supported by a Wallonie-Bruxelles International. World ‘Bourse d’Excellence’.
Author contributions: VP designed the research; VP, AG and JBG performed the research; VP, AG and NG analysed the data; and VP and JBG wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbott DW, Chadwick BP, Thambirajah AA, Ausio J (2005) Beyond the Xi: macroH2A chromatin distribution and post-translational modification in an avian system. J Biol Chem 280: 16437–16445 [DOI] [PubMed] [Google Scholar]
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A (2009) SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell 16: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S (2003) The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Bao S, Miyoshi N, Okamoto I, Jenuwein T, Heard E, Azim Surani M (2005) Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep 6: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA (2009) Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461: 1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzily-Rokni M, Friedman N, Ron-Bigger S, Isaac S, Michlin D, Eden A (2011) Synergism between DNA methylation and macroH2A1 occupancy in epigenetic silencing of the tumor suppressor gene p16(CDKN2A). Nucleic Acids Res 39: 1326–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle A, Simeoni I, Gurdon JB (2009) Xenopus oocytes reactivate muscle gene transcription in transplanted somatic nuclei independently of myogenic factors. Development 136: 2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669 [DOI] [PubMed] [Google Scholar]
- Brockdorff N (2002) X-chromosome inactivation: closing in on proteins that bind Xist RNA. Trends Genet 18: 352–358 [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Buschbeck M, Di Croce L (2010) Approaching the molecular and physiological function of macroH2A variants. Epigenetics 5: 118–123 [DOI] [PubMed] [Google Scholar]
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L (2009) The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16: 1074–1079 [DOI] [PubMed] [Google Scholar]
- Byrne JA, Simonsson S, Western PS, Gurdon JB (2003) Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr Biol 13: 1206–1213 [DOI] [PubMed] [Google Scholar]
- Casas-Delucchi CS, Brero A, Rahn HP, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC (2011) Histone acetylation controls the inactive X chromosome replication dynamics. Nat Commun 2: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K (2005) Structural characterization of the histone variant macroH2A. Mol Cell Biol 25: 7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Gao S, Sung LY, Corry GN, Ma Y, Nagy ZP, Tian XC, Rasmussen TP (2010) Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell Reprogram 12: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ (2008) MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry 47: 13726–13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR (2007) Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol 27: 2758–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Singh G, Cui K, Berletch JB, Zhao K, Disteche CM, Pehrson JR (2010) Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol Cell Biol 30: 5473–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601 [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R (2001) Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R (1999) Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22: 323–324 [DOI] [PubMed] [Google Scholar]
- Dai B, Rasmussen TP (2007) Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cell 25: 2567–2574 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S (2006) Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol 26: 1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K (2000) X-chromosome inactivation in cloned mouse embryos. Science 290: 1578–1581 [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL (2010) The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume X, Monier K, Argoul F, Mongelard F, Bouvet P (2011) In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem Res Int 2011: 187624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillich A, Hayashi K (2011) Switching stem cell state through programmed germ cell reprogramming. Differentiation (doi: 10.1016/j.diff.2011.1001.1003) [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A (2009) Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB (1968) Changes in somatic cell nuclei inserted into growing and maturing amphibian oocytes. J Embryol Exp Morphol 20: 401–414 [PubMed] [Google Scholar]
- Gurdon JB, Melton DA (2008) Nuclear reprogramming in cells. Science 322: 1811–1815 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Cox LL, Tam PP, Nagy A (2001) An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis 29: 133–140 [DOI] [PubMed] [Google Scholar]
- Håkelien AM, Delbarre E, Gaustad KG, Buendia B, Collas P (2008) Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp Cell Res 314: 1869–1880 [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Pageau G, Lawrence JB (2009) AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol 186: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley-Stott RP, Pasque V, Astrand C, Miyamoto K, Simeoni I, Jullien J, Gurdon JB (2010) Mammalian nuclear transplantation to germinal vesicle stage Xenopus oocytes—a method for quantitative transcriptional reprogramming. Methods 51: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes S, Tang F, Surani M (2008) Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Surani MA (2009) Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development 136: 3549–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hernández-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Lee JT (2003) Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426: 857–862 [DOI] [PubMed] [Google Scholar]
- Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP, Tian XC, Yang X, Ishino F, Abe K, Ogura A (2010) Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330: 496–499 [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191 [DOI] [PubMed] [Google Scholar]
- Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB (2010) Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci USA 107: 5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T (2006) The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol 8: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E (2010) The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Spohr G, Etkin LD (1993) Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science 262: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A (2004) A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL (2010) RNA traffic control of chromatin complexes. Curr Opin Genet Dev 20: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Steffen PA, Wutz A (2009) X chromosome inactivation sparked by non-coding RNAs. RNA Biol 6: 94–99 [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A (2007) Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 178: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L. Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S (2009) Weak, but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol 29: 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Kouzarides T, Bannister AJ, Gurdon JB (2010) Histone H3 lysine 4 methylation is associated with the transcriptional reprogramming efficiency of somatic nuclei by oocytes. Epigenetics Chromatin 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F (2010) Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137: 3785–3794 [DOI] [PubMed] [Google Scholar]
- Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, Bartolomei MS, Latham KE (2005) X chromosome reactivation and regulation in cloned embryos. Dev Biol 279: 525–540 [DOI] [PubMed] [Google Scholar]
- Panning B (2004) X inactivation in mouse ES cells: histone modifications and FISH. Meth Enzymol 376: 419–428 [DOI] [PubMed] [Google Scholar]
- Pasque V, Miyamoto K, Gurdon JB (2010) Efficiencies and mechanisms of nuclear reprogramming. Cold Spring Harb Symp Quant Biol (advance online publication 3 November 2010; doi: 2010.1101/sqb.2010.2075.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B (2004) Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 167: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TP, Mastrangelo MA, Eden A, Pehrson JR, Jaenisch R (2000) Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J Cell Biol 150: 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TP, Wutz AP, Pehrson JR, Jaenisch RR (2001) Expression of Xist RNA is sufficient to initiate macrochromatin body formation. Chromosoma 110: 411–420 [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E (2004) Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol 24: 5475–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E (2000) X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225: 294–303 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N (2003) Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Simonsson S, Gurdon J (2004) DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol 6: 984–990 [DOI] [PubMed] [Google Scholar]
- Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG (2009) Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28: 3423–3428 [DOI] [PubMed] [Google Scholar]
- Takagi N, Yoshida MA, Sugawara O, Sasaki M (1983) Reversal of X-inactivation in female mouse somatic cells hybridized with murine teratocarcinoma stem cells in vitro. Cell 34: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tanaka S (2006) Derivation and culture of mouse trophoblast stem cells in vitro. Methods Mol Biol 329: 35–44 [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R (2002) Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.