Abstract
Translation of most mRNAs is suppressed under stress conditions. Phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 (eIF2), which delivers initiator tRNA (Met-tRNAi) to the P site of the 40S ribosomal subunit, is responsible for such translational suppression. However, translation of hepatitis C viral (HCV) mRNA is refractory to the inhibitory effects of eIF2α phosphorylation, which prevents translation by disrupting formation of the eIF2–GTP–Met-tRNAi ternary complex. Here, we report that eIF2A, an alternative initiator tRNA-binding protein, has a key role in the translation of HCV mRNA during HCV infection, in turn promoting eIF2α phosphorylation by activating the eIF2α kinase PKR. Direct interaction of eIF2A with the IIId domain of the HCV internal ribosome entry site (IRES) is required for eIF2A-dependent translation. These data indicate that stress-independent translation of HCV mRNA occurs by recruitment of eIF2A to the HCV IRES via direct interaction with the IIId domain and subsequent loading of Met-tRNAi to the P site of the 40S ribosomal subunit.
Keywords: eIF2A, HCV, IRES, stress, translation
Introduction
Translation initiation is the process of assembling elongation-competent 80S ribosomes, in which the initiation codon is base-paired with the anticodon of initiator tRNA (Met-tRNAi) in the ribosomal P site (Jackson et al, 2010). In eukaryotes, eukaryotic translation initiation factor 2 (eIF2), which is composed of three subunits, α, β, and γ (Schmitt et al, 2010), brings Met-tRNAi to the ribosome in the form of the ternary complex eIF2–GTP–Met-tRNAi. Under various stress conditions and during virus infection, translation initiation is globally repressed by phosphorylation of eIF2α. Generally, translation of both cap-dependent mRNAs and internal ribosome entry site (IRES)-dependent mRNAs is subject to regulation by eIF2α phosphorylation. However, some mRNAs, particularly those that must endure stress conditions and/or are needed to overcome the stress conditions, are continuously translated under stress. This suggests that delivery of Met-tRNAi to the ribosome is not the sole responsibility of eIF2.
The translation of a few mRNAs containing an IRES domain is known to continue despite phosphorylation of eIF2α, even though IRES-dependent translation also generally requires a ternary eIF2–GTP–Met-tRNAi complex for delivery of Met-tRNAi to the ribosomal P site. For instance, cricket paralysis virus (CrPV) mRNA translation, which is directed by a specialized IRES element, is not affected by phosphorylation of eIF2α because part of the CrPV IRES functionally mimics initiator tRNA and directs translation from the A site in the absence of a Met-tRNAi positioned at the P site (Pisarev et al, 2005). Hepatitis C viral (HCV) mRNA, which contains another type of IRES element at the 5′ untranslated region and part of the core coding region, is a second such example (Lukavsky, 2009). Translation of HCV mRNA starts from the AUG codon, and the positioning of Met-tRNAi at the P site of the 40S ribosomal subunit is a prerequisite for translation. Nevertheless, translation of HCV mRNA is unaffected by phosphorylation of eIF2α (Robert et al, 2006; Terenin et al, 2008; also see Figure 1). Investigations into the mechanism of continuous translation of HCV mRNA under stress conditions in which eIF2 activity is compromised have been conducted by several groups. Three distinct possible mechanisms have been reported: (a) HCV mRNA directs synthesis of peptides in the absence of any initiation factor (Lancaster et al, 2006). However, the peptide synthesis without initiation factors occurs at high Mg++ concentration but not at physiological Mg++ concentration; (b) Recruitment of the initiator tRNA to a HCV IRES/ribosome complex is facilitated by eIF5B (Pestova et al, 2008; Terenin et al, 2008). The authors showed that eIF3 and eIF5B are sufficient for recruiting the initiator tRNA to a ribosome through reconstitution experiments with purified proteins and ribosomes. The relationship between eIF5B and the initiator tRNA was also demonstrated by a partial suppression of severe slow growth phenotype of fun12Δ yeast strain with a mutation in the eIF5B gene through overexpression of the gene encoding tRNAi (Choi et al, 1998). However, it remains unclear how eIF5B, which does not form a stable complex with Met-tRNAi, in contrast to IF2 (Roll-Mecak et al, 2000), delivers Met-tRNAi to the P site of a ribosome associated with a HCV mRNA. (c) Recruitment of the initiator tRNA to a HCV IRES/ribosome complex is facilitated by eIF2D/ligatin (Dmitriev et al, 2010; Skabkin et al, 2010). The authors showed that eIF2D, which binds to Met-tRNAi and elongator tRNAs, stimulates loading of the tRNAs to the P site of a 40S ribosome in a GTP-independent manner through toe-printing analyses with purified proteins and ribosomes. However, none of these reports showed the physiological role of the suggested proteins (eIF5B and eIF2D) in translation of HCV mRNA using an in vivo or an in cellulo system. Moreover, there was no report showing the importance of these proteins in HCV infection. Therefore, the function of these proteins in translation of HCV mRNA remains to be elucidated. eIF3 (Sizova et al, 1998; Lukavsky, 2009), another canonical translation factor, and RNA-binding proteins, known as IRES trans-acting factors, including polypyrimidine tract-binding protein and heterogeneous nuclear ribonucleoproteins-NSAP1 (hnRNP Q/SYNCRIP), HNRNP-L, and HNRNP-D (Ali and Siddiqui, 1995; Hahm et al, 1998; Kim et al, 2004; Paek et al, 2008), are known to assist in the translation of HCV mRNA. However, the roles of these proteins in the translation of HCV mRNA under stress conditions remain obscure.
Figure 1.
Translation of HCV mRNA is refractory to translational suppression by stresses. (A) Schematic diagram of bicistronic mRNA containing various IRES elements at the inter-cistronic region. RLuc and FLuc denote Renilla luciferase and firefly luciferase, respectively. (B) Relative translational efficiencies of cap-dependent (RLuc) and HCV IRES-dependent (FLuc) genes under various stress conditions. Before transfecting with bicistronic mRNA reporter constructs, cells were pre-treated with the following agents that induce phosphorylation of eIF2α: amino acid-depleted media (AAS; 1 h), 2 μg/ml tunicamycin (Tu; 1 h), 40 μM sodium arsenite (SA; 1 h), or 1000 IU/ml IFN-α (12 h). Cells were harvested 2 h after transfection, and RLuc and FLuc activities in cell lysates were measured using a dual luciferase assay system (Grentzmann et al, 1998). Luciferase activities were normalized to those in mock-treated cells (defined as 1); the relative translational efficiencies of reporter genes are depicted (upper panel). The levels of phosphorylated eIF2α and eIF3b were monitored by western blotting using the indicated antibodies (lower panel). (C) Translation of EMCV or poliovirus mRNAs was susceptible to stresses. Experiments were performed and data were analysed as described in panel (B) except using bicistronic reporter mRNAs containing EMCV IRES (left panel) or Poliovirus IRES (right panel). The experiments were performed three times. Columns and bars represent mean values±s.d. values.
An additional Met-tRNAi-interacting protein, termed eIF2A, was identified in the 1970s as a protein that stimulates Met-tRNAi binding to 40S ribosomal subunits (Merrick and Anderson, 1975). eIF2A homologues are found in organisms from yeasts to mammals. eIF2A is functionally similar to prokaryotic IF2, in that both eIF2A and IF2 catalyse the binding of initiator tRNA to the small ribosomal subunit in an AUG-dependent manner. This is in contrast to the behaviour of eIF2, which forms a ternary complex with Met-tRNAi and GTP and next binds to the small ribosomal subunit, without AUG (Zoll et al, 2002). The eIF2-dependent pathway has an apparent Km for GTP hydrolysis of 1.1 μM, whereas the eIF2A-dependent pathway has an apparent Km for GTP hydrolysis of 12.6 μM. A plausible GTP-binding factor in eIF2A-dependent translation is eIF5B, which has a Km for GTP hydrolysis of ∼10 μM (Zoll et al, 2002; Komar et al, 2005). eIF2A might be utilized in translation when functional eIF2 is limiting (e.g., under stress conditions), even though there have been no reports of eIF2A-dependent translation of any cellular mRNA. eIF2A was recently proposed to participate in the translation of alphavirus 26S mRNA when functional eIF2 is limited by the stress of virus infection (Ventoso et al, 2006), but the molecular basis of the continuous translation of alphavirus mRNA by eIF2A was not explored.
Here, we report that eIF2A has a key role in the translation of HCV mRNA under stress conditions. The continued translation of HCV mRNA under such conditions requires protein–mRNA interactions between eIF2A and the IIId domain stem-loop structure in the HCV IRES. Our results suggest that IRES element-bound eIF2A delivers the initiator tRNA to the P site of the 40S ribosomal subunit, while the IRES element is loaded on the solvent side. This is the first report demonstrating the molecular basis for the translation of a specific mRNA under stress conditions, under which the activity of eIF2 is compromised.
Results
eIF2A is required for translation of HCV mRNA under stress conditions
HCV IRES-dependent translation is refractory to the inhibitory effects of eIF2α phosphorylation under various stress conditions (Robert et al, 2006; Terenin et al, 2008). We confirmed this phenomenon by comparing the translation efficiencies of HCV mRNA and a control capped mRNA under normal and stress conditions (Figure 1). For this purpose, Huh-7 cells were exposed to amino-acid starvation, tunicamycin, interferon-α (IFN-α), or sodium arsenite, which induce phosphorylation of eIF2α through activation of the eIF2 kinases GCN2, PERK, PKR, and HRI, respectively. After such treatments, cells were transfected with a 5′-capped bicistronic mRNA containing the Renilla luciferase (RLuc) gene translated in a cap-dependent manner and the firefly luciferase (FLuc) gene translated under control of a HCV IRES element inserted into the inter-cistronic region, as shown in Figure 1A. HCV IRES-dependent translation was much less affected by the eIF2α phosphorylation-inducing stresses than was cap-dependent translation (Figure 1B, compare grey columns with black columns). The phosphorylation level of eIF2α was reversely correlated with the efficiency of cap-dependent translation (Figure 1B). As in cap-dependent translation, translation of other types of viral RNAs with IRESs (i.e., EMCV and poliovirus IRESs) was strongly inhibited by such treatments (Figure 1C). These data indicate that phosphorylation of eIF2α strongly inhibits cap-dependent translation as well as IRES-dependent translation of other viral mRNAs, whereas HCV IRES-dependent translation is refractory to the inhibitory effects of eIF2α phosphorylation.
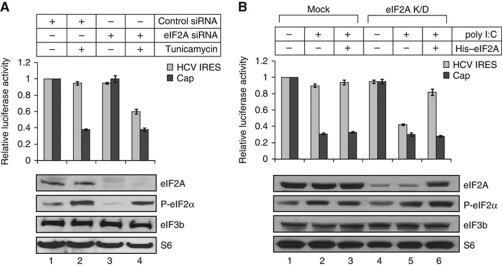
In an effort to identify the protein responsible for the loading of Met-tRNAi onto the 40S ribosomal subunit during HCV IRES-dependent translation under stress conditions, we investigated the role of eIF2A, which had previously been shown to carry Met-tRNAi to the 40S ribosomal subunit (Merrick and Anderson, 1975), albeit via a very poorly characterized mechanism. First, we tested the effect of small interfering RNA (siRNA)-mediated eIF2A knockdown on HCV IRES-dependent translation. Knocking down eIF2A did not affect either cap-dependent translation or HCV IRES-dependent translation under normal conditions (Figure 2A, compare lane 3 with 1). This indicates that eIF2A does not have a major role in cap- or HCV IRES-dependent translation under normal conditions, in which eIF2 remains unphosphorylated. Thus, eIF2 is responsible for loading Met-tRNAi onto the 40S ribosomal subunit unless eIF2 is phosphorylated. As expected, translation of cap-dependent mRNA was greatly reduced by tunicamycin treatment (Figure 2A, lane 2), but was not further inhibited by eIF2A depletion in this setting (Figure 2A, compare black columns in lane 2 and 4). Importantly, HCV IRES-dependent mRNA translation, which was unaffected by tunicamycin treatment alone, was greatly reduced by tunicamycin treatment in eIF2A-depleted cells (Figure 2A, compare grey columns on lanes 2 and 4). This indicates that eIF2A has a key role in translation of HCV mRNA, but not in translation of EMCV, poliovirus, and capped mRNAs, under stress conditions (Supplementary Figure S1). In other words, eIF2A activity is required for translation of specific mRNAs including HCV mRNA under stress conditions. We next investigated the effect of depleting and replenishing of eIF2A on HCV IRES-dependent translation in an in vitro translation system using lysates of 293T cells, with (Figure 2B, lanes 4–6) and without (Figure 2B, lanes 1–3) knockdown of eIF2A. In some of these extracts, in vitro phosphorylation of eIF2α was induced by adding poly(I:C) (Farrell et al, 1977). Elevated levels of eIF2α phosphorylation were observed by western blotting in cell extracts treated with poly(I:C) (Figure 2B, lanes 2, 3, 5, and 6 in panel labelled ‘P-eIF2α’). Cap-dependent translation, but not HCV IRES-dependent translation, was inhibited by phosphorylation of eIF2α in eIF2A-replete extracts (Figure 2B, compare lane 1 with 2). The addition of purified eIF2A to such extracts did not affect translation of cap-dependent or IRES-dependent mRNAs (Figure 2B, compare lane 2 with 3), indicating that eIF2A levels in cell extracts were sufficient for translation of HCV mRNA. Under normal conditions, neither control nor HCV mRNA translation was affected by knockdown of eIF2A (Figure 2A, compare lane 3 with 1). However, translation of both HCV mRNA and control mRNA was inhibited in the eIF2A-depleted lysates under stress conditions, in which eIF2 is phosphorylated (Figure 2B, lane 5). Importantly, translation of HCV mRNA, but not control mRNA, was restored by addition of purified eIF2A to eIF2A-depleted lysates under stress conditions (Figure 2B, compare grey columns on lanes 5 and 6).
Figure 2.
eIF2A is required for translation of HCV mRNA under stress conditions in intact cells and cell-free systems. (A) HCV IRES-dependent translation under stress conditions was monitored with or without knockdown of eIF2A. Huh-7 cells were transfected with control siRNA (lanes 1 and 2) or eIF2A siRNA (lanes 3 and 4). After incubation for 48 h, cells were additionally transfected with bicistronic reporter mRNAs containing HCV IRES, incubated in the presence or absence of tunicamycin for 2 h, and then harvested for analysis. Upper panel: dual luciferase assays; lower panel: western blotting using the indicated antibodies. (B) The effect of depleting and replenishing eIF2A on translation of cap-dependent and HCV IRES-dependent mRNAs was observed using an in vitro translation system. 293T cells transfected with control siRNA (Mock) or eIF2A siRNA (eIF2A K/D) were used in the preparation of cytoplasmic cell extracts for in vitro translation experiments. For in vitro phosphorylation of eIF2α, 5 μg/ml of poly(I:C) was added to the cell extracts (lanes 2, 3, 5, and 6) (Farrell et al, 1977). The effect of eIF2A on translation was monitored by adding purified eIF2A (100 ng) to the in vitro translation systems (lanes 3 and 6). Each experiment was performed three times. Luciferase activities were normalized to those in mock-treated cells (defined as 1). Columns and bars represent mean values±s.d. values.
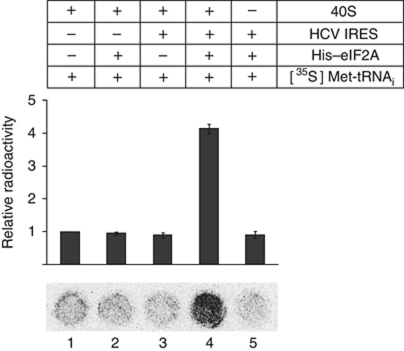
In order to test whether eIF2A delivers Met-tRNAi to 40S ribosome, we performed filter-binding assays with purified eIF2A, 40S ribosomal subunit, HCV mRNA, and 35S-methionine-charged tRNAi ([35S]Met-tRNAi) (Figure 3). The binding of the Met-tRNAi to the 40S ribosome/HCV mRNA complex was greatly stimulated by eIF2A (compare lane 4 with 3 in Figure 3). Removal of either HCV mRNA or 40S ribosomal subunit from the reaction mixtures showed background level of radioactivity (Figure 3, compare lanes 2 and 5 with 1). The data indicate that the delivery of Met-tRNAi onto 40S ribosomal subunit requires both eIF2A and HCV IRES RNA. We also monitored the binding affinities between eIF2A and the initiator tRNAi and elongator tRNAs (leucyl tRNA (tRNALeu) and methionyl tRNA (tRNAm)). eIF2A strongly bound to the initiator tRNAi but only weakly to elongator tRNAs (Supplementary Figure S3A). Both charged Met-tRNAi (Kd=12.4 nM) and uncharged tRNAi (Kd=16.5 nM) bound to eIF2A strongly (Supplementary Figure S3). No detectable binding of eIF2A to Let 7 miRNA was observed. The data suggest that eIF2A facilitates loading of Met-tRNAi, but not elongator tRNAs, to 40S ribosome in a HCV mRNA-dependent manner. Together, these results strongly suggest that eIF2A participates in the translation of HCV mRNA under stress conditions.
Figure 3.
eIF2A facilitates binding of Met-tRNAi to 40S ribosome in a HCV IRES-dependent manner. The binding of [35S]Met-tRNAi to 40S ribosome was monitored by a filter-binding assay. The requirement of HCV IRES RNA and eIF2A was tested by leave-one-out approach. Recombinant His–eIF2A protein purified from E. coli, 40S ribosomal subunit purified from HeLa S3 cell, HCV IRES RNA synthesized by T7 RNA polymerase, and [35S]Met-tRNAi charged by methionyl tRNA synthetase (Pestova and Hellen, 2005) were used in the filter-binding assays. Each experiment was performed three times. Columns and bars represent mean values±s.d. values.
eIF2A binds to the IIId domain stem-loop of the HCV IRES
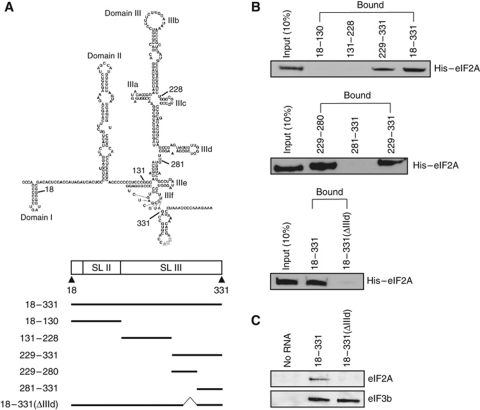
How does eIF2A render HCV mRNA refractory to translational inhibition by eIF2α phosphorylation? Because eIF2A supports the translation of HCV mRNAs but not that of other mRNAs tested (see above), we speculated that eIF2A might distinguish HCV mRNA from other mRNAs through specific eIF2A–mRNA interactions. To test whether eIF2A binds the HCV IRES element, we performed an in vitro RNA pull-down experiment using biotinylated RNAs corresponding to various parts of the HCV IRES (Figure 4A). Purified recombinant His6-tagged eIF2A (His–eIF2A) protein was incubated with biotinylated RNAs and RNA–protein complexes were precipitated using streptavidin-agarose beads. Visualization of co-precipitated proteins by western blotting using an antibody against the His tag revealed that the region between nucleotides (nts) 229 and 331 was sufficient for binding to His–eIF2A (Figure 4B, upper panel). To further define the eIF2A-binding site, we divided the 229–331 fragment into two fragments corresponding to nts 229–280 (IIIc–d) and 281–331 (IIIe–f), and found that only the IIIc–d fragment bound eIF2A (Figure 4B, middle panel). To confirm that the IIId domain was required for eIF2A binding, we created a mutant HCV IRES lacking the IIId domain. As shown in Figure 4B (lower panel), eIF2A binding to this 18–331 (ΔIIId) HCV IRES mutant was severely reduced, indicating that the IIId domain is required for interaction with eIF2A. To test whether eIF2A–HCV IRES binding occurs in the presence of other cellular proteins, we performed biotinylated RNA pull-down experiments using whole-cell extracts of Huh-7 cells. Both eIF2A and eIF3 were co-precipitated with wild-type (WT) HCV RNA corresponding to nts 18–331 (Figure 4C). In contrast, eIF2A failed to interact with the 18–331 (ΔIIId) HCV IRES mutant, whereas eIF3 retained the ability to bind this IIId domain-deletion mutant (Figure 4C). This strongly indicates that, in the cell, eIF2A interacts with HCV RNA through the IIId domain.
Figure 4.
eIF2A binds to the IIId domain of the HCV IRES. (A) The predicted secondary structure of the HCV IRES (upper panel) and RNA segments used in RNA pull-down experiments (lower panel). Biotinylated RNAs corresponding to the lines shown in this panel were synthesized by in vitro transcription. (B) RNA pull-down experiments were performed using biotinylated RNAs and purified His6-tagged eIF2A proteins. The RNA-bound proteins were precipitated with streptavidin-conjugated agarose resin and visualized by western blotting using an anti-His antibody. (C) RNA pull-down experiments were performed using cell extracts and biotinylated 18–331 (WT) and 18–331 (ΔIIId) RNAs corresponding to nts 18–331 of wild-type and IIId domain-deleted HCV IRES, respectively. The RNA-bound proteins were precipitated with streptavidin-conjugated agarose resin and visualized by western blotting using anti-eIF2A and anti-eIF3b antibodies.
eIF2A–HCV IRES interaction is required for HCV IRES-dependent translation under stress conditions
To test whether eIF2A–HCV IRES interaction is required for translation of HCV mRNA under stress conditions, we evaluated the effects of mutations at eIF2A-binding sites on protein–mRNA interaction and translation under normal and stress conditions. For this purpose, monocistronic constructs corresponding to nts 1–331 of WT HCV mRNA and derivatives containing mutations in the IIId domain were generated (Figure 5A). In addition of the ΔIIId derivative in which the IIId domain was completely deleted (described above), we created a derivative in which GGG in the loop of the IIId domain was changed to AAA (IIId-1), and one containing a substitution of A to UC in the bulge of the IIId domain that changes the bulge structure to a stem structure (IIId-2). Using biotinylated RNAs corresponding to these four HCV IRES and whole-cell extracts of Huh-7 cells, we first performed in vitro RNA–protein interaction assays. eIF2A bound equally well to WT and IIId-2 RNAs (Figure 5B). In contrast, eIF2A binding was greatly impaired by the mutations in ΔIIId and IIId-1 (Figure 5B). This indicates that the apical loop region, but not the bulge, of the IIId domain is critical for eIF2A binding. Notably, the UUGGGU sequence that is mutated in the IIId-1 construct is absolutely conserved among all HCV isolates (Smith et al, 1995).
Figure 5.
Interaction between eIF2A protein and the IIId domain of the HCV IRES is required for translation of HCV mRNA under stress conditions. (A) Schematic diagram of monocistronic mRNAs containing wild-type HCV IRES and variants with mutations in the IIId domain. Nucleotide sequences of the IIId domain of the wild-type and mutant RNAs are depicted. Black boxes denote mutations in the IIId domain of the HCV IRES. (B) RNA pull-down experiments using the mutant RNAs were performed as described in the legend to Figure 3C. RNA-bound proteins were precipitated with streptavidin-conjugated agarose resin and visualized by western blotting using anti-eIF2A and anti-eIF3b antibodies. (C) The effect of IIId mutations on HCV IRES-dependent translation under stress conditions. Monocistronic mRNAs with wild-type or mutant HCV IRES sequences were transfected into Huh-7 cells pre-treated with either sodium arsenite (SA; upper panel) or tunicamycin (Tu; lower panel) for 1 h. As a control, the capped mRNA with Renilla luciferase reporter, shown in panel (A), was co-transfected with wild type of mutated HCV mRNA. Cells were harvested 2 h after transfection, and RLuc and FLuc activities in cell lysates were measured using a dual luciferase assay system (Grentzmann et al, 1998). Luciferase activities were normalized to those in mock-treated cells (defined as 1); the relative translational efficiencies of reporter genes are depicted. (D) In vitro translation reactions were performed using eIF2A-depleted or replenished 293T cell lysates (prepared as described in Materials and methods), and control capped mRNA in combination with monocistronic IRES-dependent mRNAs containing the IIId domain mutations shown in panel (A) or wild-type IIId. The effect of eIF2α phosphorylation was assessed by adding 5 μg/ml of poly(I:C) to the translation reaction mixtures. Luciferase activities were normalized to those in mock-treated cells (defined as 1); the relative translational efficiencies of reporter genes are depicted. Translation reactions were performed three times. Columns and bars represent mean values±s.d. values.
Next, we investigated the effects of these IIId domain mutations on HCV mRNA translation under stress conditions. In these experiments, translation was monitored in cells transfected with HCV IRES variant reporter mRNAs after incubating with or without sodium arsenite or tunicamycin for 1 h (Figure 5C, upper and lower panels, respectively). Translational efficiencies of reporter mRNAs under stress conditions, expressed relative to those of the corresponding mRNAs under normal conditions, are depicted in Figure 5C. The translation of WT and IIId-2 mutant HCV mRNAs was minimally reduced (∼20%) under stress conditions. In contrast, translation of ΔIIId and IIId-1 mutant mRNAs was reduced by about 60% under the same stress conditions, a decrease in translation efficiency that approached that of control capped mRNA (up to ∼80%) under stress conditions. These results indicate that the intact sequence of the IIId loop, which is essential for eIF2A interaction, is required for translation of HCV mRNA under stress conditions, implying a critical role for eIF2A–IIId interactions in the continuous translation of HCV mRNA under such conditions. We further analysed the role of eIF2A–IIId interaction in HCV mRNA translation under stress conditions in an in vitro translation system using cytoplasmic extracts from control cells or eIF2A-knockdown cells (Figure 5D). As shown in Figure 2B, translation of WT HCV mRNA was refractory to the inhibitory effects of poly(I:C)-induced phosphorylation of eIF2α, whereas translation of a control capped mRNA was greatly inhibited by the same treatment (Figure 5D, lane labelled ‘IIId-WT’ in upper panel). Translation of the mutant mRNA IIId-2, which interacts with eIF2A, was also refractory to the inhibitory effects of eIF2α phosphorylation. In contrast, translation of ΔIIId and IIId-1 mutant mRNAs was greatly reduced (Figure 5, compare panel D with C), a result similar to that observed in intact cells. This persistence of WT and IIId-2 mutant mRNA translation in the face of eIF2α phosphorylation was greatly reduced when in vitro translation reactions were performed using eIF2A-knockdown cell extracts (Figure 5D, lower panel). Together, the result of translation experiments using intact cells, and those from cell-free systems exploring various aspects of mRNA and eIF2A binding using reporter mRNAs, lead us to conclude that binding of eIF2A to the IIId domain of the HCV IRES has an important role in translation of HCV mRNA under stress conditions.
eIF2A is required for HCV proliferation
HCV induces activation of the eIF2α kinases PKR and PERK by virtue of actions of viral RNA and viral proteins (Tardif et al, 2002; Benali-Furet et al, 2005; Garaigorta and Chisari, 2009). Activation of PKR and PERK leads to eIF2α phosphorylation, which subsequently blocks translation of viral and cellular mRNAs that might inhibit proliferation of HCV. However, the eIF2A-dependent translation of HCV mRNAs described here might allow the production of viral proteins and thus support the proliferation of HCV. To investigate the importance of eIF2A-dependent translation of HCV mRNA in HCV proliferation, we evaluated the effect of eIF2A knockdown during HCV infection. HCV infectivity was quantitatively assessed by western blotting of HCV core protein and by measuring luciferase activity in cells infected with HCV strain JFH/5aRluc containing the Renilla luciferase (RLuc) gene in the viral genome (Kim et al, 2007). As expected, phosphorylation of eIF2α was observed in Huh-7.5.1 cells 36 h after infection with HCV (JFH strain) even though the eIF2α level remained the same (Figure 6A). Importantly, knockdown of eIF2A reduced the infectivity of HCV by 50–60% in Huh-7.5.1 cells (Figure 6B, relative luciferase activity and core level), indicating that eIF2A-dependent translation, which effectively nullifies the innate immune response against HCV infection (i.e., activation of PKR and PERK), is required for efficient proliferation of HCV.
Figure 6.
eIF2A is required for efficient proliferation of HCV. (A) Phosphorylation of eIF2α was induced by HCV infection. Huh-7.5.1 cells were infected with the JFH1 strain of HCV for 36 h, and then the levels of phosphorylated eIF2α and HCV core protein were monitored by western blotting. (B) Knockdown of eIF2A in the host cells reduced the infectivity of HCV. Huh-7.5.1 cells were transfected with control siRNA or eIF2A siRNA. Thirty-six hours after transfection, the cells were inoculated with an infectious HCV strain containing the Renilla luciferase gene (JFH/5aRluc) (Kim et al, 2007), and then the cells were incubated for an additional 48 h. The infectivity of HCV was monitored by luciferase activity assay and western blotting of HCV core. The efficiency of eIF2A knockdown was monitored by western blotting. The luciferase activity in eIF2A-knockdown cells was normalized to that in control cells (defined as 1). Each experiment was performed three times. Columns and bars represent mean values±s.d. values.
eIF2A is redistributed from the nucleus to the cytoplasm in HCV-infected cells
Finally, we monitored eIF2A localization under normal and stress conditions. Under normal conditions, eIF2A was localized mainly in the nucleus and partially in the cytoplasm (Figure 7A). Interestingly, eIF2A was redistributed to the cytoplasm from the nucleus under stress conditions (Figure 7A), and in Huh-7 cells infected with HCV (Figure 7B). In cells exposed to HCV, nuclear exclusion of eIF2A was observed specifically in HCV-infected cells (identified by immunostaining for HCV core protein); surrounding cells that were not infected by HCV showed primarily a nuclear distribution and a partial cytoplasmic distribution of eIF2A, similar to that observed in unstressed cells (Figure 7, compare panel A with B). Merged images of HCV core protein and eIF2A immunostaining photographs showed that the subcellular localizations of both proteins partially overlapped. It is conceivable that some of the cytoplasmically redistributed eIF2A protein in HCV-infected cells participates in the translation of HCV mRNAs colocalized with core proteins, which bind HCV RNAs (Shimoike et al, 2006; Yu et al, 2009).
Figure 7.
Localization of eIF2A in HCV-infected cells. (A) Localization of eIF2A in Huh-7 cells under normal and stress conditions. Huh-7 cells were incubated with or without 100 μM sodium arsenite (SA) for 1 h, and then immunostained for eIF2A using a rabbit anti-eIF2A primary antibody and a FITC-conjugated anti-rabbit IgG secondary antibody. eIF2A protein is shown in green. Bars, 10 μm. (B) Huh-7 cells were incubated for 48 h after inoculating with HCV. eIF2A and HCV core proteins were visualized by immunocytochemistry using mouse monoclonal anti-HCV core protein and rabbit polyclonal anti-eIF2A primary antibodies. FITC-conjugated anti-rabbit IgG and rhodamine-conjugated anti-mouse IgG antibodies were used as secondary antibodies. eIF2A and HCV core proteins are shown in green and red, respectively; HCV core proteins were only detected in HCV-infected cells. Left panel: eIF2A; middle panel: HCV core protein; right panel: merged image of eIF2A and HCV core protein signals. Bars, 10 μm.
Discussion
Regulation of translation initiation by eIF2α phosphorylation, which results in translational repression by blocking eIF2-mediated delivery of initiator tRNA to the P site of 40S ribosomal subunits, is a pivotal physiological mechanism for adapting to various stress conditions, including viral infection (Holcik and Sonenberg, 2005). PKR, one of the several eIF2α kinases and a component of the interferon-mediated antiviral response, participates in the innate immune response that protects host cells from virus infections by blocking translation of viral mRNAs (Samuel, 2001; Garcia et al, 2006). Many viruses have evolved various strategies to nullify this cellular response (Garcia et al, 2006). Previous reports using subgenomic HCV replicon systems, which allow replication of part of the HCV genome but do not support full HCV infection, have demonstrated that certain HCV proteins, namely NS5A and E2, block PKR activity (Gale et al, 1997; Taylor et al, 1999). However, recent reports using infectious HCV clones (Wakita et al, 2005; Zhong et al, 2005) have shown that HCV infection strongly induces both PKR and eIF2α phosphorylations (Garaigorta and Chisari, 2009; Kang et al, 2009), demonstrating that host protein synthesis is suppressed by activated PKR in HCV-infected cells, but HCV IRES-dependent translation is not (Garaigorta and Chisari, 2009). Moreover, the authors of the latter study suggested that eIF2α phosphorylation represses the expression of genes that trigger the interferon response required for protection from virus infection (Garaigorta and Chisari, 2009). In other words, continuous translation of HCV mRNA under eIF2α-phosphorylated conditions contributes to evasion of the host innate immune responses by the infecting virus.
Here, we reconfirmed that eIF2α phosphorylation occurs in HCV-infected cells (Figure 6A) and that HCV IRES-dependent translation is refractory to the inhibitory effects of eIF2α phosphorylation (Figure 1B). We also showed that this phenomenon is HCV IRES specific, because EMCV and polioviral IRES-dependent translation, like cap-dependent translation, was severely hampered under stress conditions (Figure 1C). Two proteins eIF5B and Ligatin/eIF2D were suggested to deliver Met-tRNAi onto 40S ribosome for translation of HCV and related viral mRNAs under stress conditions (Pestova et al, 2008; Terenin et al, 2008; Dmitriev et al, 2010; Skabkin et al, 2010). However, physiological relevance of the discoveries remains to be elucidated since none of the reports showed the effects of depletion of the proteins on HCV mRNA translation in cells or on the infectivity of HCV. We showed that eIF2A, a potential Met-tRNAi carrier, is required for translation of HCV mRNA under stress conditions by using a siRNA against eIF2A (Figure 2), and that eIF2A substitutes for eIF2 in delivering Met-tRNAi to the 40S ribosomal subunit (Figure 3). Notably, eIF2A is required for efficient proliferation of HCV (Figure 6B), indicating that eIF2A-dependent translation is important for translation of HCV mRNA during HCV infection which triggers eIF2α phosphorylation (Figure 6A).
How does HCV mRNA, unlike the majority of mRNAs, utilize eIF2A in translation under stress conditions?
As noted above, eIF2A is responsible for the persistent translation of HCV mRNA under stress conditions. The specificity of eIF2A activity towards HCV mRNA appears to be attributable to the direct interaction of eIF2A with the apical loop region of the IIId domain of the HCV IRES. The GGG motif in the IIId domain loop, in particular, was critical for both eIF2A interaction and continuous translation of HCV mRNA under stress conditions (Figure 5; Supplementary Figure S2). These data strongly suggest that under such conditions, eIF2A associated with the IIId domain of the HCV IRES delivers Met-tRNAi to the P site of a 40S ribosomal subunit that is associated with the same IRES element (Figure 8B). It is intriguing that the IIId domain also participates in direct binding of 40S ribosome to the HCV IRES (Kolupaeva et al, 2000; Lukavsky et al, 2000; Kieft et al, 2001). Mutations in the IIId domain reduced IRES-dependent translation consistently with the previous reports (Supplementary Figure S2C; Jubin et al, 2000; Lukavsky et al, 2000; Kieft et al, 2001). Further translational inhibition of HCV mRNAs containing mutations in the apical loop of domain IIId (35–45% of translation level under normal conditions) was observed when cells were treated with sodium arsenite inducing eIF2α phosphorylation (Supplementary Figure S2, lanes 2–5). However, much less translational inhibition by the eIF2α phosphorylation was observed from mutant IIId-2 maintaining eIF2A-binding capability even though ribosome-binding capability is partially hampered. In fact, similar levels of translational resistance to the eIF2α phosphorylation were observed from the WT HCV IRES (75%) and the mutant IIId-2 IRES (72%) (Supplementary Figure S2B, lanes 1 and 6). According to the cryo-EM structure of HCV IRES–40S ribosome complex, the stem region of the IIId domain takes part in the interaction with 40S ribosome, but the apical loop region seems to be exposed to solvent on the backside of the shoulder of 40S ribosome (Spahn et al, 2001; Figure 8). eIF2A is likely to bind to the solvent-exposed apical loop for delivering Met-tRNAi to 40S ribosome. The existence of a quaternary complex composed of Met-tRNAi–eIF2A–HCV IRES–40S ribosome was shown by a filter-binding assay (Figure 3). This indicates that an eIF2A protein and a 40S ribosomal subunit can associate with the IIId domain of HCV IRES at the same time.
Figure 8.
Schematic diagrams depicting the delivery of an initiator tRNA to the P site of a HCV IRES-associated 40S ribosomal subunit under normal and stress conditions. (A) eIF2 delivery of initiator tRNA to the 40S ribosomal subunit under normal conditions. (B) eIF2A delivery of initiator tRNA to the 40S ribosomal subunit under stress conditions. The anticodon of the initiator tRNA is associated with the initiation codon in the mRNA. eIF5B, which may facilitate 60S ribosome joining under normal and stress conditions, is also depicted. eIF5B and the direct interaction between eIF2A and the IIId domain of the HCV IRES may allow proper positioning of the eIF2A-associated initiator tRNA at the P site of the ribosome.
Because HCV IRES-dependent translation was not affected by knockdown of eIF2A when cells were cultivated under normal conditions, we speculate that eIF2, not eIF2A, has the major role in delivering Met-tRNAi to the 40S ribosomal subunit on the HCV IRES under normal conditions. Thus, when functional eIF2–GTP–Met-tRNAi ternary complexes are plentiful, the translation mechanism of HCV IRES mRNA is similar to that of cap-dependent and other IRES-dependent mRNAs (Figure 2). Moreover, eIF2 has been shown to outcompete eIF2A in the methionyl puromycin synthesis assay when both proteins are present (Adams et al, 1975), indicating the preferential utilization of eIF2 rather than eIF2A in translation under normal conditions. In addition, the majority of eIF2A protein is localized in the nucleus under normal conditions, which limits the availability of eIF2A for translation (Figure 7). Whether eIF2A is involved in the translation of HCV mRNA under normal conditions is not entirely clear. However, the fact that the binding affinity of the HCV IRES for eIF2A protein was indistinguishable between normal and stressed cell extracts (data not shown) suggests that eIF2A may also participate in the translation of HCV mRNA under normal conditions.
A recent study that included in vitro reconstitution experiments showed that eIF5B promotes binding of Met-tRNAi to 40S–eIF3–HCV IRES complexes without eIF2 (Terenin et al, 2008). However, no mechanism was suggested in an attempt to explain how Met-tRNAi was delivered to the HCV IRES-associated complex, without a carrier. Intriguingly, eIF2A and eIF5B mutations result in synthetic growth defects in yeast cells (Zoll et al, 2002), suggesting that eIF2A and eIF5B function in related pathways. These observations are consistent with the idea that both eIF2A and eIF5B participate in the translation of HCV mRNA under stress conditions, but the detailed relationship between eIF2A and eIF5B remains to be elucidated.
On the basis of data presented here and in previous reports, we propose a model for the continuous translation of HCV mRNA under stress conditions that centres on the role of the Met-tRNAi carriers eIF2 and eIF2A in translation (Figure 8). According to this model, the HCV IRES element binds to the solvent side of the 40S ribosomal subunit along with eIF3 (Siridechadilok et al, 2005). Domain II of the HCV IRES is exposed to the inter-subunit side of the 40S ribosomal subunit (Boehringer et al, 2005). Under normal conditions, Met-tRNAi is loaded to the P site of the 40S ribosomal subunit as a ternary complex with eIF2 and GTP. Under stress conditions, eIF2A bound to the IIId domain of the HCV IRES participates in the recruitment of Met-tRNAi to the P site of the ribosome because eIF2A possess an intrinsic initiator tRNA-binding property (Figure 3). eIF5B may assist in this initiator tRNA recruitment process and/or the subsequent 60S ribosome-joining step.
In conclusion, we describe a novel mechanism whereby a specific mRNA is translated under stress conditions. This mode of translation is most likely to be employed by a subset of cellular mRNAs that are typically translated under stress conditions. It is intriguing that yeast eIF2A has translational inhibitory function for a yeast cellular mRNA URE2 (Komar et al, 2005; Reineke and Merrick, 2009). Identification of cellular mRNAs that utilize eIF2A, which is conserved from yeast to humans, and investigations into the physiological roles of eIF2A in translation, will add a new dimension to our understanding of translational regulation under different physiological conditions.
Materials and methods
Cell culture and preparation of 293T cell lysates for in vitro translation
293T, Huh-7, and Huh-7.5.1 cells were cultivated in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal bovine serum (JRH). 293T cell lysates were generated by treating forty 100-mm dishes of 293T cells with control siRNA or siRNA against eIF2A (600 pmole/dish). After incubating with siRNA for 48 h, cells were harvested and translation-competent cell lysates were prepared as follows: cell pellets were washed three times with ice-cold phosphate-buffered saline (PBS) and then incubated with 1.5 volumes of cold hypotonic buffer on ice for 10 min. Cells were lysed by homogenization in a small Dounce homogenizer (40 strokes) and then centrifuged at 12 000 g for 15 min. The supernatant was dialysed against buffer containing 10 mM HEPES-KOH (pH 7.4), 90 mM K(CH3CO2), 1.5 mM Mg(CH3CO2)2, and 2.5 mM dithiothreitol. Translation reactions with the 293T lysate were performed as described previously (Paek et al, 2008) using 40 nM reporter RNA and 180 mM K(CH3CO2). Translation reactions in micrococcal nuclease-treated rabbit reticulocyte lysates were performed as recommended by the supplier (Promega).
Plasmid construction and siRNA
Dual reporters harbouring the HCV IRES, EMCV IRES, and poliovirus IRES were constructed as described previously (Kim et al, 2004). Monocistronic reporters containing a given HCV IRES variant followed by firefly luciferase, kindly provided by Dr Takashi Shimoike, (National Institute of Infectious Diseases, Japan), were prepared as described previously (Shimoike et al, 2006). For construction of plasmid pQE31/His–eIF2A, eIF2A gene was amplified by PCR from pSK(−)/human eIF2A (Korea Unigene) using the following primers: 5′-AGCGGATCCAGCGCCGTCCACGCCG-3′ and 5′-AGCGTCGACTTAAATACCCAATTCCAAATC-3′. The PCR product was treated BamHI-SalI and cloned into BamHI-SalI-treated pQE31 (Qiagen). SMART pool siRNA against eIF2A was purchased from Dharmacon.
Transfections and luciferase assays
Cell were transfected with monocistronic firefly and Renilla luciferase reporters or bicistronic Renilla and firefly luciferase RNA reporter constructs using Lipofectamine, as described by the manufacturer (Invitrogen). Cells were harvested 2 h after transfection. For experiments employing reporters, RLuc and FLuc activities in cell lysates were measured using a dual luciferase assay system, as described previously (Grentzmann et al, 1998).
Filter-binding assays
The reaction mixture (25 μl each) containing indicated components (Figure 3) in binding buffer (20 mM Tris–HCl (pH 7.4), 100 mM KCl, 5 mM Mg(CH3CO2)2, and 1 mM DTT) was incubated at 25°C for 20 min. The amounts of components are as follows: 2.5 pmole of 40S ribosomal subunit, 1 pmole of [35S]Met-tRNAi (10 000 c.p.m./pmole), 0.5 pmole of HCV IRES, and 3 pmole of eIF2A. In all, 175 μl of dilution buffer (20 mM Tris–HCl (pH 7.4), 100 mM KCl, 5 mM Mg(CH3CO2)2, 1 mM DL-methionine) was added to the reaction mixture, and the sample was filtered through a membrane (HVPP membrane, Millipore MAHVN45, 0.45 μm), which has low binding affinity to proteins and nucleic acids, under negative pressure. The filter was washed five times with 1 ml of binding buffer, and [35S]Met-tRNAi bound to the filter was monitored by a FLA-5100 (Fujifilm).
In vitro transcription and pull-down with biotinylated RNAs
Reporter RNAs used to assess translation in intact cells and cell-free systems were generated with T7 RNA polymerase from HpaI-treated dual reporters and XhoI-treated monocistronic reporters. HCV IRES RNAs used in RNA pull-down assays were generated with T7 RNA polymerase in the presence of biotinylated UTP from AccI-treated monocistronic reporters. RNA-affinity chromatography was performed using purified His–eIF2A or Huh-7 cell lysates, as described elsewhere (Kim et al, 2004).
Protein expression and purification
Escherichia coli strain BL21 was used to produce recombinant His–eIF2A from plasmid pQE31/His–eIF2A. Isopropyl-β-D-thiogalactopyranoside (final concentration, 1 mM) was added to induce eIF2A protein expression at an optical density at 600 nm of 0.7. After incubation at 16°C for 18 h, the cells (culture volume: 6 l) were harvested, resuspended in lysis buffer (20 mM sodium phosphate (pH 7.6), 300 mM NaCl, 0.5 mM phenylmethylsulphonyl fluoride, 1 mM β-mercaptoethanol, and 10% glycerol), and sonicated. The resulting cell extracts were loaded onto a Ni-nitrilotriacetic acid-agarose column (Qiagen) equilibrated with lysis buffer containing 10 mM imidazole. The columns were washed with lysis buffer containing 30 mM imidazole, and bound eIF2A was eluted with 80 mM imidazole. The same column purification step was repeated three times for higher purity. About 10 μg of recombinant eIF2A proteins was obtained from 6 l of E. coli culture.
Fluorescence microscopy
After JFH1 infection, Huh-7.5.1 cells were grown for 48 h on coverslips coated with 0.2% gelatin and then washed three times with PBS. The cells were fixed with 3.5% paraformaldehyde (Sigma) at room temperature (RT) for 12 min. After washing three times with PBS, cells were permeabilized with 0.1% Triton X-100 at RT for 2 min, and then washed three times with PBS. The samples were soaked in blocking solution (PBS containing 1% bovine serum albumin) for 30 min at RT and then incubated with primary antibodies for 1 h at RT. After washing three times with PBS, the samples were treated with rhodamine-conjugated and/or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at RT. Finally, the coverslips were washed three times with PBS, placed on a glass slide, and then sealed with transparent nail polish. The fluorescent images were captured with a cooled charge-coupled device camera mounted on a Zeiss Axioplan microscope (Jena, Germany).
Antibodies
The primary antibodies used were anti-eIF2A (ProteinTech Group), anti-eIF3b (Santa Cruz), anti-RpS6 (Cell Signaling), anti-phospho eIF2α (Cell Signaling), anti-eIF2α (Novous), anti-His (R&D Systems), anti-GAPDH (AbD Serotec), and anti-HCV core protein (Thermo Scientific).
Supplementary Material
Acknowledgments
We thank Dr Sunghoon Kim for providing Gemt-Easy vectors containing initiator tRNAi, elongator tRNAm, and tRNALeu. We thank Dr Takashi Shimoike for providing monocistronic reporters containing a given HCV IRES variant followed by firefly luciferase. This work was supported in part by a grant FPR08B1-220 of the 21C Frontier Functional Proteomics Project from MEST, NRF NCRC grant (no. 2010-0028447), NRF BRL grant (no. 2010-0019706), Bio R&D program through the NRF grant (no. 2010-0018167) and WCU program through the NRF grant (no. R31-2009-000-10105-0) funded by the Korea government (MEST).
Author contributions: JHK and SKJ designed the experiments; JHK, SMP, JHP and SJK performed the experiments; JHK and SKJ analysed data; JHK and SKJ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams SL, Safer B, Anderson WF, Merrick WC (1975) Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem 250: 9083–9089 [PubMed] [Google Scholar]
- Ali N, Siddiqui A (1995) Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol 69: 6367–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, Paterlini-Brechot P (2005) Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24: 4921–4933 [DOI] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H (2005) Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE (1998) Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN (2010) GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ, Balkow K, Hunt T, Jackson RJ, Trachsel H (1977) Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell 11: 187–200 [DOI] [PubMed] [Google Scholar]
- Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG (1997) Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230: 217–227 [DOI] [PubMed] [Google Scholar]
- Garaigorta U, Chisari FV (2009) Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70: 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF (1998) A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Hahm B, Kim YK, Kim JH, Kim TY, Jang SK (1998) Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol 72: 8782–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N (2005) Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM (2000) Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol 74: 10430–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, Ahn BY (2009) PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res 142: 51–56 [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA (2001) Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Jung JH, Wakita T, Yoon SK, Jang SK (2007) Monitoring the antiviral effect of alpha interferon on individual cells. J Virol 81: 8814–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK (2004) A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol 24: 7878–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU (2000) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC (2005) Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 280: 15601–15611 [DOI] [PubMed] [Google Scholar]
- Lancaster AM, Jan E, Sarnow P (2006) Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12: 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ (2009) Structure and function of HCV IRES domains. Virus Res 139: 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Merrick WC, Anderson WF (1975) Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 250: 1197–1206 [PubMed] [Google Scholar]
- Paek KY, Kim CS, Park SM, Kim JH, Jang SK (2008) RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82: 12082–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU (2008) eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (2005) Reconstitution of eukaryotic translation elongation in vitro following initiation by internal ribosomal entry. Methods 36: 261–269 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Shirokikh NE, Hellen CU (2005) Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol 328: 589–605 [DOI] [PubMed] [Google Scholar]
- Reineke LC, Merrick WC (2009) Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA 15: 2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, Pelletier J (2006) Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell 17: 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Cao C, Dever TE, Burley SK (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103: 781–792 [DOI] [PubMed] [Google Scholar]
- Samuel CE (2001) Antiviral actions of interferons. Clin Microbiol Rev 14: 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Naveau M, Mechulam Y (2010) Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett 584: 405–412 [DOI] [PubMed] [Google Scholar]
- Shimoike T, Koyama C, Murakami K, Suzuki R, Matsuura Y, Miyamura T, Suzuki T (2006) Down-regulation of the internal ribosome entry site (IRES)-mediated translation of the hepatitis C virus: critical role of binding of the stem-loop IIId domain of IRES and the viral core protein. Virology 345: 434–445 [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU (1998) Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV (2010) Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Mellor J, Jarvis LM, Davidson F, Kolberg J, Urdea M, Yap PL, Simmonds P (1995) Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. The International HCV Collaborative Study Group. J Gen Virol 76(Part 7): 1749–1761 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Tardif KD, Mori K, Siddiqui A (2002) Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol 76: 7453–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM (1999) Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285: 107–110 [DOI] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN (2008) Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 15: 836–841 [DOI] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M (2006) Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KL, Jang SI, You JC (2009) Identification of in vivo interaction between Hepatitis C Virus core protein and 5′ and 3′ UTR RNA. Virus Res 145: 285–292 [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV (2005) Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA 102: 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC (2002) Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem 277: 37079–37087 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.