Introduction
When Maria Leptin called to say that I had been awarded the 2010 EMBO Gold Medal, I was delighted and surprised. The roll call of past winners is impressive, and I had been at the Laboratory of Molecular Biology (LMB) when Jan Löwe had received the award in 2007 for his elegant work on the bacterial cytoskeleton (Michie and Löwe, 2006). I also knew that several other LMB scientists, including Barbara Pearse, Hugh Pelham and Matthew Freeman, had previously won this prestigious award for their seminal contributions to molecular biology (Pelham, 1989; Freeman, 2002).
Like many scientists that I know I invariably find the experiments we are about to do the most compelling, and our curiosity and optimism about the future, and the limitless possibilities that it offers, is perhaps part of what drives us continually forward into the unknown, but makes it difficult for us to sit down and write reviews about what we have already done. However, the recognition of our laboratory's work by this medal provides a ‘still point’ in which I can reflect, and allows me the opportunity to provide a personal account that recognizes the contributions to science made by the many talented and generous people that have educated and mentored me, and that I have had the privilege to work with. I hope that this review can honour, in a small way, the debt of gratitude I feel for the ways in which they have enriched my life in science.
I was naturally drawn to chemistry at school because it provided a systematic explanation for how all matter behaves in terms of simple sets of rules governed by invisible particles and captured in the periodic table. I had fantastic teaching in both chemistry and physics, and was taught by Mr Liasis, Mr Manthorpe, Mr Teh and Ms Pountney over the years. It was clear to me that I wanted to do chemistry at University and I was lucky enough to be accepted to Oxford. At Oxford I was tutored by Peter Atkins and Gordon Lowe, who appeared to take the view that they were teaching us to be scientists and that the examinations at the end of the degree were incidental distractions that bright people would somehow get through. Their tutorials were aimed at getting us to think, sometimes in unusual ways, and I have not yet forgotten the surreal image of Peter Atkins reclining on his office chaise longue, a glass of dry sherry in hand and his leather trousers creaking, while he shone a lamp on my face and demanded that I explain how I would establish the laws of thermodynamics on a desert island using only a coconut (of course I had a truly marvellous proof, but there is insufficient space for it here!). While tutorials were fantastic entertainment, the lecture courses were generally more prosaic, traditional and thorough. Later in the course I specialized in organic chemistry and was given tutorials with John Sutherland who, like Gordon Lowe, was continually able to relate the chemistry we were learning to biological processes. I decided to do a part II research year with John, whose laboratory then worked on both engineering penicillin biosynthesis to make new antibiotics and the chemical origins of life (Powner et al, 2009). I worked on engineering an enzyme that naturally expands the five-membered ring of penicillin to the six-membered ring in a cephalosporin, so that it would accept new substrates and make new types of cephalosporin antibiotics. This experience got me hooked on a combination of chemistry and molecular biology and the idea of doing more research.
In 1996, I moved to Yale to study for a PhD, since a US PhD allowed me to complement a thorough training in chemistry I had received at Oxford with the opportunity to take biology classes. After the first year at Yale, I had taken most of the biology courses and felt equally comfortable with both chemistry and biology. I decided to work for Alanna Schepartz at Yale, who had a great project that had been pioneered by a graduate student in the laboratory, Neal Zondlo. Neal had shown that it was possible to dissect out the DNA-binding residues of a helical protein and transfer these residues in register onto a small stable scaffold protein to generate a new functional chimeric protein with exquisite DNA-binding affinity and specificity (Zondlo and Schepartz, 1999). With Robert Grotzfeld, I developed combinatorial approaches, using phage display, to extend the approach Neal had developed. I went on to show that we could make high-affinity binders for protein and DNA targets using this approach (Chin et al, 2001; Chin and Schepartz, 2001a, 2001b). Since the approaches we developed were new to the laboratory, I learned a lot about how to do experiments and how to get things to work from scratch, which has been very valuable.
In 2001, I finished my PhD and went off to Scripps for a postdoc with Pete Schultz. From then on my research has focussed on engineering the translational machinery of cells for incorporating new amino acids. I, therefore, describe our current overall strategy for reprogramming translation first (below), and then go on to describe how my postdoctoral work fits in and provides one key part of the foundation for our current programme.
Reprogramming translation
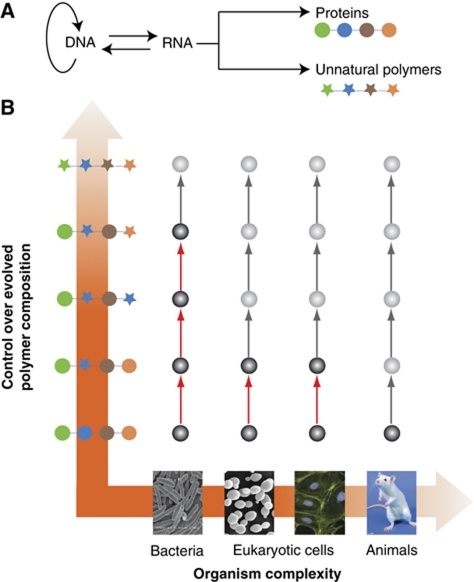
Protein translation is the process by which cells decode genetic information to build functional polymers of amino acids. While natural protein translation synthesizes proteins composed of the natural 20 amino acids, the process by which these polymers are made provides the ultimate paradigm for the synthesis of proteins containing unnatural amino acids beyond the canonical 20, and for the synthesis of entirely unnatural evolvable polymers of genetically determined length, composition and sequence (Figure 1).
Figure 1.
Reprogramming the genetic code. (A) A central paradigm in molecular biology for the synthesis of proteins containing natural amino acids (coloured circles) may be engineered for the synthesis of proteins containing unnatural amino acids (coloured stars), and by extension of the synthesis of completely unnatural polymers. (B) Progress in reprogramming the genetic code. Natural amino acids are represented by coloured circles and unnatural amino acids by coloured stars. The vertical axis shows progress in incorporating unnatural amino acids into proteins, while the horizontal axis shows progress in incorporating unnatural amino acids in increasingly complex organisms. Red arrows represent steps that have been experimentally demonstrated.
Over the last 7 years, we have engineered protein translation for several purposes. First, we have engineered protein translation to create foundational approaches for the encoded and evolvable synthesis of new polymers (Rackham and Chin, 2005a; Wang et al, 2007; Neumann et al, 2010b) (Figure 2). Second, we have developed methods for site specifically installing several key post-translational modifications into recombinant proteins, and used these methods to provide previously unattainable insight into the role of these modifications in regulating biological function (Neumann et al, 2008a, 2008b, 2009; Nguyen et al, 2009a, 2009b; Lammers et al, 2010; Virdee et al, 2010; Zhao et al, 2010; Akutsu et al, 2011; Arbely et al, 2011) (Figure 3). Third, we have developed ‘photochemical genetic’ methods to rapidly control the activity of proteins in living cells, providing insight into the dynamics of elementary steps in biological processes, as well as insight into the regulation of intracellular network connectivity in space and time (Figure 4) (Gautier et al, 2010, 2011); with these methods, we hope to understand molecular processes inside cells and organisms with the level of precision more commonly associated with in vitro biochemistry or biophysics.
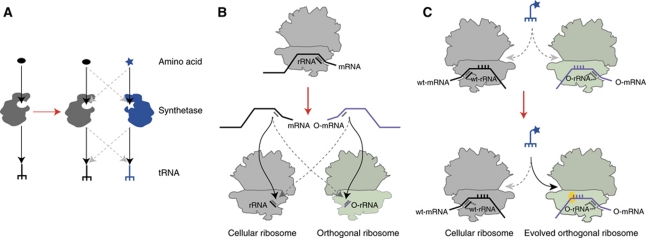
Figure 2.
Engineering the translational machinery to reprogramme the genetic code. (A) Creating orthogonal amber suppressor aminoacyl-tRNA synthetase/tRNA pairs. Cells contain natural synthetases (grey) that use natural amino acids (black oval) to aminoacylate natural tRNAs (black trident). Expanding the genetic code requires the addition of an orthogonal synthetase, tRNA, and amino acid (star) shown in blue. (B) Creation of orthogonal ribosome mRNA pairs by duplication and specialization. The natural cellular ribosome (grey) recognizes natural messages, while the new orthogonal ribosome (green) recognizes orthogonal messages (purple). (C) Evolving the orthogonal ribosome for quadruplet decoding for a parallel genetic code.
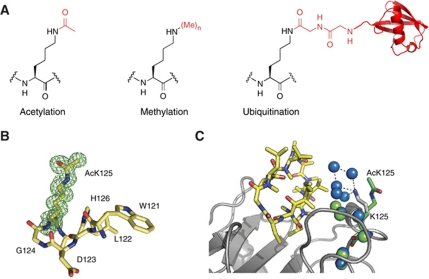
Figure 3.
Genetically encoding lysine acetylation, methylation and ubiquitination. (A) The chemical structures of the post-translationally modified amino acids. (B) Omit map of acetyl lysine density in acetylated cyclophilin A, contoured at 1σ. (C) Structural comparison of the acetylated and unacetylated cyclophilin A–cyclosporine complexes. Cyclosporine is in yellow. The backbone structure of cyclophilin and acetylated cyclophilin are very similar. The backbone of cyclophilin A from a previous structure (PBB 2CPL) is shown in grey and the acetyllysine from our structure is in green. Waters belonging to the unacetylated complex are in blue, waters belonging to the acetylated complex are in green. Acetylation leads to a reorganization of the water network at the interface. This rationalizes why acetylating cyclophilin leads to a 20-fold lower affinity for cyclosporine that may antagonize the immunosuppressive effects of cyclosporine.
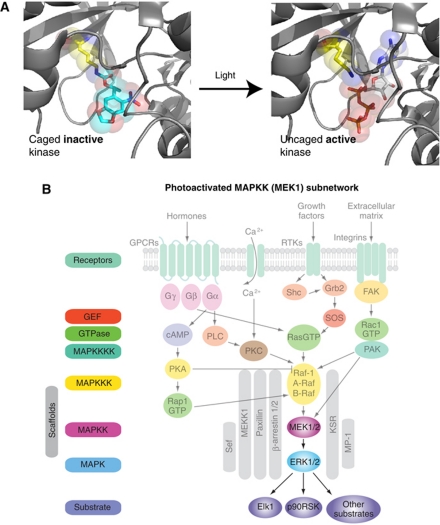
Figure 4.
Controlling specific molecular events inside living cells with light. (A) Caging a near-universally conserved lysine (K97) in the MEK1 active site inactivates the enzyme by sterically blocking ATP binding. Decaging with light rapidly removes the caging group and activates the kinase (figures created using Pymol and MEK1 structure PDB: 1S9J). (B) Isolating a synthetic photoactivateable subnetwork in MAP kinase signalling, via genetically encoding a photocaged lysine in the MEK1 active site.
The fidelity of natural translation is primarily set by two processes: (1) aminoacylation of the correct tRNA, and no other tRNA by an aminoacyl-tRNA synthetase (Ling et al, 2009) and (2) the correct decoding and translocation of each tRNA by the ribosome in response to its cognate triplet codon on the mRNA to direct peptide bond formation (Ramakrishnan, 2002). As the ribosome uses tRNA adapter molecules, the chemical identity of the monomers polymerized is chemically independent of the template; this is distinct from the case in nucleic acid-dependent nucleic acid polymerases (DNA polymerases and RNA polymerases), where the substrates for polymerization must directly pair with the template. As the ribosome uses a single set of active sites for polymerization, coupled to a translocation activity, it is—unlike NRPSs, PKSs or fatty acid synthetases—able to synthesize very long polymers of defined and arbitrarily programmed sequence.
We realized that in order to reprogramme protein translation to incorporate new amino acids into proteins, and ultimately to synthesize completely unnatural polymers, there are at least three challenges we need to address. First, we need to uniquely attach a new amino acid to a new tRNA. This requires the creation of orthogonal aminoacyl-tRNA synthetase/tRNA pairs in which the aminoacyl-tRNA synthetase is able to uniquely recognize a new amino acid that is not a substrate for endogenous synthetases in the host organism and specifically load the new amino acid onto a cognate tRNA that is not a substrate for endogenous synthetases. Next, we need a codon with which we can uniquely encode the incorporation of the new amino acid. Each of the 64 triplet codons is used in encoding the synthesis of natural proteins, but we have demonstrated that it is possible to evolve the ribosome itself to decode additional genetic information. Finally, the chemical scope of natural protein translation is limited to the synthesis of polypeptides from α-L amino acids; to synthesize a full range of new polymers will require alteration of the ribosome's peptidyl-transferase centre (Dedkova et al, 2003) as well as, potentially, alterations to other parts of the translational machinery.
Scripps, La Jolla and orthogonal aminoacyl-tRNA synthetase/tRNA pairs
While there are no blank codons in the genetic code, it is well known that the amber stop codon can be decoded, using amber suppressor tRNAs, in a variety of cells and organisms. Amber suppression is inherently inefficient because the amber stop codon is normally read as a termination signal by protein release factors that bind to the A site in the ribosome and hydrolyse the nascent polypeptide chain attached to the P-site tRNA (Capecchi, 1967; Scolnick et al, 1968; Petry et al, 2005). When both an amber suppressor tRNA and the release factor are present in cells, the tRNA and release factor compete for A-site binding. Under these conditions, 80% of protein synthesis that is initiated typically terminates in response to the amber codon, while 20% is decoded by the amber suppressor tRNA and continues to produce the full-length protein (Wang et al, 2007).
Though amber suppression is inefficient, it provides a codon that we can use as an initial insertion signal for unnatural amino-acid incorporation. This allows us to focus our attention on the first problem in reprogramming the genetic code—discovering aminoacyl-tRNA synthetase/tRNACUA pairs that are orthogonal in the host and that direct the incorporation of new amino acids. In fact, my interest in incorporating unnatural amino acids into proteins preceded any notion of wholesale genetic code reprogramming, and in 2001, I went to Pete Schultz's laboratory at Scripps to work on creating amber suppressor synthetase/tRNA pairs for incorporating unnatural amino acids into proteins in cells.
Pete's laboratory had a long-term interest in incorporating unnatural amino acids into proteins. Indeed, when I applied to work with Pete, his laboratory had already pioneered in vitro methods for incorporating unnatural amino acids into proteins (Noren et al, 1989; Mendel et al, 1995) by combining cell extracts with methods for the chemical aminoacylation of amber suppressor tRNAs (Hecht et al, 1978; Heckler et al, 1984). With Dennis Dougherty and Henry Lester at Caltech, they had extended these approaches, using microinjection of aminoacylated tRNAs, into Xenopus oocytes (Dougherty, 2008). This allowed the introduction of unnatural amino acids into channel proteins expressed in the oocyte. The Dougherty laboratory extended these unnatural amino-acid mutagenesis strategies and has performed elegant studies that probed and defined the dynamics of nicotinic receptor and the role of pi-cation interactions in protein interactions (Dougherty, 2008). Early experiments from the Schultz and Dougherty laboratories beautifully demonstrated how the ability to tailor the properties of individual amino acids atom by atom at defined sites in proteins allow new biological insights to be revealed in complicated systems, via the application of physical organic chemistry principles. However, because in vitro methods for aminoacylating tRNAs are inherently inefficient and do not allow re-acylation of the tRNA in the translation reaction, the in vitro aminoacylation and translation methods yielded small amounts of protein and were technically very challenging. The Schultz laboratory was, therefore, working hard on developing in vivo methods for incorporating unnatural amino acids by engineering aminoacyl-tRNA synthetases and tRNAs (Figure 2A).
Early indications that it might be possible to site specifically add new amino acids to proteins produced in cells came from experiments reported by Furter (1998). These experiments demonstrated that a fluorinated analogue of phenylalanine could be incorporated into a protein in Escherichia coli in response to the amber codon using the yeast phenylalanyl-tRNA synthetase tRNACUA pair. Since it is known that fluorinated analogues of phenylalanine are substrates for phenylalanine synthetases, these experiments used a strain of E. coli normally resistant to fluorinated phenylalanine, to avoid incorporation of fluorinated phenylalanine at sense codons via the endogenous E. coli PheRS/tRNAs. Since the yeast synthetase recognizes both phenylalanine and the fluorinated phenylalanine added to the cells, a mixture of fluorinated phenylalanine and natural amino acids were incorporated into the protein in response to the amber codon.
David Liu, Thomas Magliery, Miro Pasternak and Peter Schultz articulated that the discovery of aminoacyl-tRNA synthetase/tRNACUA pairs that are orthogonal in a host organism, and that direct the site specific and quantitative incorporation of new amino acids might be achieved by breaking the problem down into two sub-problems (Liu et al, 1997; Liu and Schultz, 1999): (1) discovering aminoacyl-tRNA synthetase/tRNACUA, where the synthetase uses a natural amino acid but does not aminoacylate any tRNAs in the host organism, and the tRNACUA is not a substrate for any endogenous synthetases and (2) reprogramming the synthetase enzyme so that it uniquely recognizes a new unnatural amino acid added to the cell and no natural amino acids. The first sub-problem was addressed by importing aminoacyl-tRNA synthetase/tRNA pairs from heterologous organisms, taking advantage of the evolutionary divergence of synthetase and tRNA sequence and structure between domains of life. The second sub-problem was addressed by creating large libraries (109 variants) of aminoacyl-tRNA synthetase mutants in which the mutations are targeted, using structural information, and performing a two-step genetic selection on this library to identify synthetases that specifically use an unnatural amino acid and no natural amino acids.
In Pete's laboratory, I addressed the incorporation of several of the first unnatural amino acids into proteins in response to the amber codon in E. coli using this strategy (Chin et al, 2002a, 2002b). This work took advantage of an amber suppressor derivative of the Methanococcus janaschii tyrosyl-tRNA synthetase (MjTyrRS)/tRNA pair, which is orthogonal in E. coli (Xie and Schultz, 2006). We showed that this pair could be evolved to direct the incorporation of a range of unnatural amino acids with useful properties in response to the amber codon. In particular, I demonstrated that it was possible to evolve this pair to incorporate photocrosslinking amino acids into proteins in response to the amber codon in E. coli (Chin et al, 2002a, 2002b; Chin and Schultz, 2002). This allowed the sites of protein interactions to be mapped both in vitro and in vivo by simply shining light on cells. Unlike non-covalent methods of investigating protein interactions in vivo, such as TAP tagging, this method traps the protein interaction in the cell before purification, and gives direct information about the sites within the proteins that are involved in interactions. The methods we developed have been used to obtain direct information about protein interactions in environments that are difficult to probe by other methods, for example for proteins at or in membranes. In addition, the method may be used to trap some of the most interesting weak or transient interactions that may be systematically lost in non-covalent approaches. Numerous laboratories have used the crosslinking methods we developed to provide unique insights into protein interactions in diverse systems, including the interactions of chaperones (trigger factor, ClpB and GroEL) with substrates, protein interactions important in cell-cycle regulation, conformational changes in RNAP and the topology of transcriptional initiation complexes, protein interactions at the inner and outer membrane of E. coli, protein interactions in the mitochondrial and ER membranes in yeast and protein interactions at the plasma membrane in mammalian cells (Schlieker et al, 2004; Weibezahn et al, 2004; Farrell et al, 2005; Kaiser et al, 2006; Mori and Ito, 2006; Chen et al, 2007; Haslberger et al, 2007; Lakshmipathy et al, 2007; Boos et al, 2008; Kimata et al, 2008; Mohibullah and Hahn, 2008; Panahandeh et al, 2008; Braig et al, 2009; Ieva and Bernstein, 2009; Okuda and Tokuda, 2009; Raschle et al, 2009; Tamura et al, 2009; Carvalho et al, 2010; Jensen et al, 2010; Liu et al, 2010; Tagami et al, 2010; Yamano et al, 2010).
The initial methods for incorporating unnatural amino acids into proteins could only be applied in E. coli. I was interested in incorporating unnatural amino acids into eukaryotic cells and organisms because of the enormous potential I saw in being able to make atomic perturbations at specific sites in a specific protein within complex organisms. I realized that such approaches might allow us to dissect, follow and manipulate complex biological processes in space and time directly in vivo. However, the MjTyrRS/tRNACUA pair that we had used in E. coli could not be used in eukaryotic cells because it is not orthogonal with respect to eukaryotic synthetases and tRNAs. It was clear that to expand the genetic code of eukaryotic cells, we would need (1) new synthetase tRNA pairs and (2) new methods to evolve the specificity of these pairs directly in a eukaryotic host. Schimmel's laboratory and others had shown that tyrosyl-tRNA synthetase/tRNACUA pair and the leucyl-tRNA synthetase/tRNACUA pair may be orthogonal in eukaryotes (Edwards and Schimmel, 1990), and so I created a strategy to evolve these pairs to incorporate unnatural amino acids into proteins in yeast. I not only saw yeast as both interesting in its own right for genetic code expansion but also realized that the synthetases we evolved in this system might be directly transplanted to other eukaryotic hosts, including mammalian cells, where the direct transformation with large libraries of synthetase genes and the rapid selections and deconvolution methods we developed in yeast would not have been possible. Eric Meggers, Chris Anderson and Ashton Cropp worked with me on this project, and we successfully developed a method for evolving aminoacyl-tRNA synthetase/tRNA pairs for the incorporating unnatural amino acids, including photocrosslinkers, heavy atoms, biophysical probes and bio-orthogonal labels for protein labelling, into proteins in eukaryotic cells for the first time (Chin et al, 2003a, 2003b). The synthetases we developed in this work are widely used to probe processes in yeast and mammalian cells (Hino et al, 2005, 2011; Chen et al, 2007; Huang et al, 2008; Mohibullah and Hahn, 2008; Ye et al, 2009, 2010; Carvalho et al, 2010).
Cambridge, reprogramming translation
The Cambridge laboratory began in the summer of 2003. In 2002, shortly after finishing my PhD and moving to Scripps, I had contacted Greg Winter, whose pioneering work on protein engineering I knew well. Indeed, Greg's seminal work on antibody engineering (Jones et al, 1986; Riechmann et al, 1988; Winter and Milstein, 1991) had been an inspiration for my PhD work and his work, along with Alan Fersht and others, on defining the functional centres of tyrosyl-tRNA synthetase through early site-directed mutagenesis experiments had formed a foundation for my postdoctoral work on engineering these enzymes (Winter et al, 1982; Fersht et al, 1985; Bedouelle and Winter, 1986). Greg invited me to visit the Medical Research Council LMB and this eventually led to the offer of an independent position at LMB, with the suggestion that I go away and think of something ambitious and important to do in my independent career and the promise that I would have reasonable resources to get started. I accepted with the proviso that I would stay a year and a half to finish my postdoctoral projects at Scripps. I am very fortunate to be part of a community and environment at LMB, where there are few barriers to doing science.
From my postdoctoral work with Pete, I was convinced of the enormous potential of encoding unnatural amino acids into proteins, but felt that we had only begun to scratch the surface of what might be possible. When I began to think about how we might systematically reprogramme translation in cells, I realized that we needed to take control of the engine of translation—the ribosome—and make a version of the ribosome that we could alter or evolve to do what we wanted.
The ribosome is large and complicated. But exciting progress in structural biology of the ribosome had begun to provide a detailed picture of the subunits and the functional centres of the ribosome. An electrifying talk by Venki Ramakrishnan on 9 May 2003 at the Skirball Institute at NYU convinced me that we were now entering an era in which the ribosome could be understood in molecular detail and—potentially—engineered. Indeed, the molecular insights that Venki and his group at LMB have provided, along with many insights provided by the rest of the ribosome field, have turned out to be invaluable to our work on engineering the ribosome. However, I realized that even if we understood in molecular detail how to engineer the ribosome, altering the cellular ribosome—which is the ultimate cellular hub and responsible for making every protein in the cell—would be problematic. Indeed, it is well known that many mutations in the ribosome are dominant negative or lethal, since they interfere with the synthesis of the entire proteome. I realized that if we could create a new ‘orthogonal’ ribosome that was uncoupled from the requirement to synthesize the proteome, and decoded a message that was not read by the endogenous ribosome, then this new ribosome—which would be non-essential to the cell—should, in principle, be evolvable in the laboratory. Moreover, since the genetic code is a correspondence between amino acids and codons, set by the translational machinery, I realized that the selective delivery of tRNAs aminoacylated with unnatural amino acids to the orthogonal ribosome could form the basis for a parallel and independent, or orthogonal, genetic code for the synthesis of unnatural polymers.
Oliver Rackham, who was the first postdoc in the Cambridge laboratory, began work on creating the orthogonal ribosome in E. coli. He first developed a genetic selection through which we could select for or against the expression of a single gene fusion and then showed that he could use this to select mRNA leader sequences, containing alternative Shine Dalgarno sequences (Hui and de Boer, 1987; Rackham and Chin, 2005a), that were not recognized by the endogenous ribosome, but are specifically and efficiently read by a new orthogonal ribosome (Rackham and Chin, 2005a) (Figure 2B).
Oliver Rackham began to take advantage of this new non-essential orthogonal ribosome and showed that it is possible to use different orthogonal ribosomes to produce Boolean logic in gene expression (Rackham and Chin, 2005b). More recently, Wenlin An has shown that it is possible to select genetic elements that direct orthogonal transcription by T7 RNAP and orthogonal translation by an orthogonal ribosome (An and Chin, 2009). This provides an orthogonal gene expression pathway in the cell that is entirely insulated from that of normal gene expression. We have suggested that the synthesis of orthogonal, parallel and independent systems, that are released from the constraints that are frozen in natural biology by the evolutionary process, will allow the synthetic evolution of the most fundamental systems in biology. Furthermore, the selective insulation of orthogonal systems from cellular regulation may provide foundational technologies for making biology more amenable to engineering. Orthogonal systems may, therefore, provide a key to the creation of scalable, complex dynamic synthetic biology systems constructed from a large number of biological parts (Kwok, 2010).
Wenlin demonstrated that the orthogonal gene expression pathway can be used to set up regulatory circuits that cannot be created using the endogenous, essential transcription and translation machinery (An and Chin, 2009). For example, Wenlin showed that it is possible to create a variety of transcription–translation networks, including transcription–translation feed forward loops, which would be impossible to create using the endogenous machinery. This allowed Wenlin to control the timing of gene expression in new ways and introduce information processing delays into gene expression on the order of hours. In the process of this work, Wenlin was also able to define a minimal transcript that is correctly transcribed and processed to produce a functional 16S rRNA in the ribosome small subunit. This allowed Wenlin to provide insights—into the minimal requirements for rRNA processing—that would be challenging to achieve with the natural ribosome. This work demonstrates that new dynamic properties can be accessed with orthogonal systems, and that orthogonal systems are amenable to rational manipulation and design. In the future, it may be possible to evolve orthogonal systems to provide a spectrum of tailored dynamics in gene expression. This might ultimately provide new ways to synthetically control and investigate the timing of gene expression in biological decision-making processes.
Oliver Rackham and Kaihang (Kai) Wang, the first PhD student who joined the laboratory in 2004, were interested in using the orthogonal ribosome to get functional information on the parts of the ribosome that were being structurally elucidated (Yusupov et al, 2001; Schuwirth et al, 2005). They used the orthogonal ribosome to carry out large-scale combinatorial mutagenesis and in vivo selections on 30S nucleotides that form RNA–RNA intersubunit bridges between the large and small subunit in the E. coli ribosome, as defined by structural biology approaches. They determined the co-variation and functional importance of bridge nucleotides. Comparison of the structural interface and phylogenetic data to the functional epitopes they defined with their experiments (Rackham et al, 2006) allowed Oliver and Kai to reveal how information for ribosome function is partitioned across bridges, and suggested a subset of nucleotides, at the structurally defined interface, that form ‘functional epitopes’ in the translation cycle, and may have measurable effects on individual steps of the translational cycle.
Cambridge, ribo-X
Since the orthogonal ribosome is not responsible for synthesizing the proteome, it is in principle evolvable. We realized that since the genetic code is a correspondence between amino acids and codons, and since this correspondence is set by protein translation, it should be possible to create a parallel translation pathway by selectively delivering tRNAs aminoacylated with new amino acids to the orthogonal ribosome. This would allow us to create an entirely parallel genetic code for encoding the incorporation of unnatural amino acids into proteins in cells.
To begin to exemplify this approach, we first asked whether we could evolve an orthogonal ribosome to efficiently read an amber stop codon placed within an orthogonal message as a sense codon, thereby differentiating the way the genetic code is read on an endogenous and orthogonal message (Wang et al, 2007). This is of practical importance because the truncated protein produced in an amber suppression experiment limits the yield of full-length protein and may interfere, in a dominant negative manner, with exactly the process under study in in vivo experiments; deleting release factor is lethal and interferes with the decoding of all amber stop codons in the genome. In contrast, our approach leaves the decoding of genomic amber stop codons unaltered and selectively reads the amber codons of interest, placed within the orthogonal message, as a sense codon.
To create an orthogonal ribosome that efficiently reads the amber stop codon as a sense codon, Kaihang Wang created a saturation library of mutants in the 530-loop region in the decoding centre of the orthogonal ribosome. This region of the ribosome is responsible for recognizing RF1 and correctly decoding tRNAs. Kaihang then used a selectable marker containing an amber stop codon to select for ribosomes that incorporate amino acids loaded onto amber suppressor tRNAs much more efficiently than the natural ribosome. Oliver Barrett, a PhD student in the laboratory, subsequently developed a system to purify orthogonal ribosomes (Barrett and Chin, 2010) (building on prior work from Rachel Green's laboratory on tagging endogenous ribosomes; Youngman and Green, 2005) from cells and used this to provide direct evidence that the molecular basis of this effect is a decreased affinity of the evolved orthogonal ribosome for RF1. Kaihang and Heinz Neumann demonstrated that the fidelity of the evolved orthogonal ribosome (ribo-X) was comparable to that of the natural ribosome. They also demonstrated that ribo-X allows the very efficient incorporation of single unnatural amino acids into proteins, and also allows the incorporation of multiple identical unnatural amino acids into proteins for the first time.
Cambridge, ribo-Q
While the evolution of ribo-X demonstrated for the first time that it is possible to synthetically differentiate the way genetic information is read on two distinct messages in the cell, we were interested in extending this approach to provide a whole series of additional codons that we might assign—given new orthogonal synthetases and tRNAs—to new amino acids.
We realized that if we could selectively deliver a set of tRNAs loaded with new amino acids to the orthogonal message, then we could write a new genetic code on the orthogonal message. We realized that at the molecular level, this might be achieved by creating a ‘bump’ on the tRNA and a corresponding ‘hole’ in the orthogonal ribosome (or vice versa). It is well known that tRNAs with extended anticodons are very poor substrates for natural ribosomes, and we realized that if we could evolve an orthogonal ribosome that efficiently decodes quadruplet codons on the orthogonal message using extended anticodon tRNAs then this would provide a series of additional codons on the orthogonal message that could be assigned to new amino acids (Figure 2C). Kaihang Wang designed and created 14 structurally guided libraries in the decoding centre of the orthogonal ribosome. Each library contains approximately 108 members, and together the libraries cover 144 nucleotides of ribosomal RNA. From these libraries, Kaihang selected a new orthogonal ribosome ribo-Q1 that was able to efficiently decode a series of quadruplet codons using extended anticodon tRNAs (Neumann et al, 2010b). This ribosome was actually derived from ribo-X and so was additionally able to efficiently decode the amber codon using amber suppressor tRNAs. Lloyd Davis and Kaihang Wang showed that the ribo-X had excellent fidelity in tRNA decoding. This work created a new orthogonal ribosome (ribo-Q) that provides several blank quadruplet codons that may be assigned to new amino acids on the orthogonal message. However, at this point, there was only a single aminoacyl-tRNA synthetase/tRNA pair, the MjTyrRS/tRNACUA pair, which could be evolved to incorporate unnatural amino acids in E. coli.
Heinz Neumann showed that the MjTyrRS/tRNACUA pair and the pyrrolysyl-tRNA synthetase (PylRS)/tRNA pair (which Heinz had shown was a second orthogonal synthetase/tRNA pair that could be evolved to incorporate unnatural amino acids in response to the amber codon, as discussed below) are mutually orthogonal. We then put together an orthogonal translation pathway in which two distinct unnatural amino acids are loaded onto distinct tRNAs by distinct synthetases and selectively decoded on the orthogonal message by ribo-Q. This allowed the efficient genetic incorporation of two distinct unnatural amino acids into a protein, in response to two distinct codons, for the first time (Neumann et al, 2010b).
The ability to direct two unnatural amino acids into proteins allows us to begin to programme properties into proteins that are not a property of either amino acid individually but emerge from the interaction between the two amino acids. Using the PylRS/tRNACUA pair, we incorporated an aliphatic alkyne (Nguyen et al, 2009b), and using an evolved MjTyrRS/tRNAAGGA pair, we incorporated a phenyl-azide (Chin et al, 2002b). These amino acids are ‘bio-orthogonal’ (Sletten and Bertozzi, 2009); they contain chemical functional groups (azides and alkynes) that do not react with molecules found in biology, but specifically react with each other, via a cycloaddition, to form a stable triazole linkage. Heinz demonstrated that by encoding these two amino acids at proximal sites, it was possible to genetically programme a rapid, proximity accelerated cycloaddition to form a nanoscale, redox insensitive, triazole crosslink in a protein. Extensions of this approach may allow us to rapidly explore all possible crosslinks in proteins, and this approach may find utility in trapping particular functional states of proteins or in stabilizing protein therapeutics.
Since the synthetases derived from MjTyrRS and PylRS have each been used to encode numerous unnatural amino acids, it will now be possible to encode several hundred pairwise combinations of unnatural amino acids into proteins by simple extensions of our approach. By encoding new combinations of unnatural amino acids, additional new properties, such as fluorescence, may be programmed into proteins, and this may facilitate the labelling of specific proteins in vivo.
Cambridge, de novo generation of orthogonal synthetases and tRNAs
Ribo-Q provides numerous additional codons on the orthogonal mRNA. However, since only two orthogonal synthetase/tRNA pairs exist that can be used to incorporate distinct amino acids, only two distinct unnatural amino acids can be incorporated into a protein in the cell. A clear challenge in going from incorporating two unnatural amino acids to the synthesis of completely unnatural polymers is, therefore, to discover or invent strategies for generating new orthogonal aminoacyl-tRNA synthetase/tRNA pairs that can be used to decode additional codons that may be read by ribo-Q on the orthogonal message.
The two existing orthogonal synthetase tRNA pairs in E. coli were derived by import from heterologous organisms, taking advantage of the fact that while the genetic code is near-universally conserved between known organisms, the sequences and structures of synthetases and tRNAs have diverged through evolution. Since we know much, from years of biochemistry and structural biology, about the identity elements by which synthetases and tRNAs recognize each other, it is possible to make informed guesses about which synthetases and tRNAs are likely to be orthogonal in a given heterologous host.
However, it is unclear how many mutually orthogonal synthetase/tRNA combinations can be discovered by taking advantage of natural evolutionary divergence. Moreover, since the evolutionary record suggests that the current set of synthetases and tRNAs arose by gene duplication and specialization from a simpler basis set (e.g. tyrosyl-tRNA synthetase and tryptophanyl-tRNA synthetase appear to be derived from a common ancestor), we realized that it might be possible to extend this evolutionary process in the laboratory to generate orthogonal synthetases and tRNAs de novo. Heinz Neumann demonstrated that by a series of genetic selections on structural targeted libraries in a tRNA and the synthetase, it is possible to evolve a new synthetase/tRNA pair that is orthogonal to both the synthetase from which it was evolved and every other synthetase and tRNA in the cell (Neumann et al, 2010a). This work demonstrates, for the first time, that the small number of orthogonal synthetase/tRNA pairs that have been discovered in nature does not place an intrinsic limit on the potential of genetic code expansion.
Future work will aim to couple strategies, including those we have described, for providing new codons with additional orthogonal synthetases and tRNAs to extend the orthogonal genetic code for the synthesis of completely unnatural polymers. We will also investigate further evolving the orthogonal ribosome to allow the biosynthesis of unnatural polymers composed on non-α-L amino acids. This will likely require evolution of the peptidyl-transferase centre and other parts of the ribosome, but the demonstrated evolvability of the orthogonal ribosome provides a starting point for this approach. Using cells endowed with genetically encoded heritable polymers, we may be able to explore the combinatorial biosynthesis of materials and therapeutics and investigate whether life with additional genetically encoded polymers can do things that natural biology cannot.
Cambridge, post-translational modifications
It is clear that the functions of proteins are extensively regulated by post-translational modification. The dynamically modified proteome orchestrates biological complexity and a persistent challenge in explaining the mechanistic basis of biological regulation is to define the molecular effects of post-translational modification on protein function. In order to understand the role of post-translational modifications, we require methods to synthesize proteins bearing quantitatively, and site specifically installed post-translational modifications.
While natural modifying enzymes can be used for modifying proteins in some cases, the increased power of analytical methods, in particular mass spectrometry, now means that the identification of modifications on a protein often precedes a detailed understanding of the pathways by which the modifications are installed or removed (Choudhary et al, 2009). The ability to make a natural modification by a synthetic route provides tools to study the function of a modified protein and to uncover natural regulators of the modifications discovered by the new analytical tools. Moreover, even when the natural modifying enzymes are known, they often act in large complexes that are difficult or impossible to isolate and may not modify the desired recombinant protein site specifically or completely. In the past few years, we have developed methods that allow the site-specific, quantitative installation of post-translational modifications (Figure 3), including lysine acetylation (Neumann et al, 2008b), lysine mono- and di-methylation (Nguyen et al, 2009a, 2010), and lysine ubiquitination (Virdee et al, 2010) into recombinant proteins and used the tools we have developed to provide previously unattainable new biological insight (Neumann et al, 2009; Lammers et al, 2010; Virdee et al, 2010; Zhao et al, 2010; Akutsu et al, 2011; Arbely et al, 2011). Our work installing each of these modifications takes advantage of the PylRS/tRNACUA pair.
Pyrrolysine is an unusual derivative of lysine that is incorporated into certain proteins in response to the amber codon in some methanogens. Work from several groups demonstrated that the amino acid is incorporated using an aminoacyl-tRNA synthetase/tRNACUA pair (Ambrogelly et al, 2007). This pair can recognize analogues of pyrrolysine and, unlike the pathway for incorporating selenocysteine, the synthetase/tRNA and amino acid are sufficient to direct the incorporation of the amino acid in response to the amber stop codon (Ambrogelly et al, 2007).
Cambridge, acetylation and chromatin
Heinz Neumann first showed that the PylRS/tRNA pair, which is orthogonal in E. coli, could be synthetically evolved in the laboratory to direct the quantitative, site-specific incorporation of unnatural amino acids into proteins. The first amino-acid Heinz incorporated was acetyllysine (Neumann et al, 2008b). In collaboration with Daniela Rhodes’ group at LMB, which drew heavily on Daniela's years of expertise in chromatin biology, Heinz developed methods for producing site-specifically acetylated histone proteins, including H3 acetylated on lysine 56 (Neumann et al, 2009). This modification has a demonstrated role in DNA repair, replication, regulation of transcription, chromatin assembly and defining epigenetic status (Cosgrove et al, 2004; Hyland et al, 2005; Xu et al, 2005, 2007; Celic et al, 2006; Driscoll et al, 2007; Han et al, 2007; Rufiange et al, 2007; Chen et al, 2008; Li et al, 2008; Xie et al, 2009). However, it had not been possible to synthesize H3 acetylated on K56 to quantitatively test mechanistic proposals for how this acetylation in chromatin might affect these complicated cellular phenomena. In collaboration with Daniela's group, John vanNoort's group at Leiden and Tom Owen-Hughes’ group in Dundee, we are able to measure the effect of this acetylation on chromatin compaction, remodeler activity and DNA wrapping on the histone proteins. In particular, measurements from vanNoort's group using single nucleosome FRET demonstrated that K56 acetylation increased unwrapping of the DNA around the nucleosome core providing a physical basis for numerous in vivo observations. Ongoing work aims to address the role of histone modifications and combinations of modifications on chromatin structure and function.
Cambridge, structure and function of acetylated proteins
Mass spectrometry studies have now demonstrated that thousands of proteins, beyond histones, are specifically acetylated (Choudhary et al, 2009), and we have begun to address the role of acetylation in these proteins. In a collaboration with Leo James at LMB, we have defined the effects of an identified acetylation within the active site of cyclophilin on HIV-1 capsid isomerization and cyclosporine binding (Lammers et al, 2010). In the course of this project, Michael Lammers, working with Leo James, solved the first high-resolution structures of acetylated proteins and their complexes, using proteins made using the acetyllysyl-tRNA synthetase/tRNACUA pair (Figure 3B). The structure of acetylated cyclophilin A in complex with HIV capsid, in combination with biophysical measurements allowed Michael to show that active site acetylation controls the cis/trans isomerization of HIV capsid, a step that may be important in viral capsid disassembly. The structure of acetylated cyclophilin in complex with cyclosporine (Figure 3C) reveals that acetylation reorganizes a water network at the cyclopsporine–cyclophilin interface. This decreases the affinity for cyclosporine, suggesting that acetylation may antagonize the immunosuppressive effects of cyclosporine. More recently, our laboratory has also contributed to understanding the role of acetylation in regulating metabolism as part of a large-scale study (Zhao et al, 2010).
Cambridge, chromatin and methylation
We have also used the PylRS to site specifically direct the incorporation of mono- and di-methyl lysine (Figure 3A) into recombinant proteins. We realized that it might be thermodynamically challenging to generate a synthetase that differentiates mono-methyl lysine from lysine, which is constitutively present in the cell, by a factor of 104, as required to maintain the fidelity of protein translation. However, Duy Nguyen, a PhD student in the laboratory, was able to show that we could also install mono-methyl lysine into recombinant proteins by adding a chemical protecting group to methyl lysine that increases its bulk and makes it a good substrate for the synthetase. Once the protected methyl lysine is installed in the protein at a specific site, the protecting group can be removed under mild conditions, revealing a protein with a site specifically incorporated methyl lysine (Nguyen et al, 2009a). More recently, Duy has also developed a method that also allows di-methyl lysine to be installed in recombinant proteins (Nguyen et al, 2010). We are using the methods we have developed for methylation, along with methods we have developed to install other modifications, to investigate the role of post-translational modifications in chromatin structure and function.
Cambridge, ubiquitination
Very recently, Satpal Virdee has developed methods for the creation of site-specific isopeptide bonds between a lysine in one protein and the C-terminus of another protein, as occurs in protein ubiquitination and SUMOylation (Virdee et al, 2010) (Figure 3A). These methods genetically define the site of isopeptide bond formation and are, therefore, in principle applicable to forming isopeptide bonds between proteins of any length.
Satpal has applied the method he developed, named genetically encoded orthogonal protection with activated ligation (GOPAL) to the synthesis of atypical ubiquitin chain in which one of the lysine residues, other than K63 or K48, within a ubiquitin molecule is linked to the C-terminus of another ubiquitin. These chains have been identified in cells and implicated in diverse biological processes (Peng et al, 2003; Ikeda and Dikic, 2008; Xu et al, 2009), but it has not been possible to synthesize such chains to address their function or regulation systematically.
In GOPAL, one ubiquitin molecule is expressed containing a genetically encoded version of lysine in which the amino group is blocked with a protecting group. The site of the genetically encoded protected amino acid defines the ultimate site of isopeptide bond formation. Another ubiquitin molecule is expressed containing a C-terminal thioester by intein fusion thiolysis approaches. All the lysines and other free amino groups in both ubiquitins are protected with a second chemical protecting group before the protecting group on the genetically encoded lysine derivative is removed, revealing a single free amine in the proteins. The amino group is activated and selectively couples to the thioester forming the isopeptide bond, and subsequent deprotection of all other amines reveals the native, specifically linked diubiquitin.
Using this approach, we first synthesized K6- and K29-linked diubiquitin. In collaboration with David Komander at LMB, Satpal was able to solve a crystal structure of K6-linked diubiquitin and to profile a panel of deubiquitinases, representing approximately 10% of those known in humans, on these newly synthesized linkages. These experiments revealed that atypical linkages are the preferred substrates of certain deubiquitinases, notably TRABID, which cleaves K29-linked diubiquitin 40 times more rapidly than K63-linked ubiquitin, which is a preferred substrate for TRABID with respect to K48 linkages (Tran et al, 2008). Since TRABID has been implicated as a positive regulator of Wnt signalling (Tran et al, 2008) by Mariann Bienz's laboratory at LMB, these experiments suggest that there may be a potential role for atypical chains in regulating this important signalling pathway.
Cambridge, photochemical genetics and real-time molecular cell biology
The pyrrolysyl-tRNA synthetase/tRNACUA pair is orthogonal not just in E. coli, but also in yeast and mammalian cells (Mukai et al, 2008; Gautier et al, 2010; Hancock et al, 2010). PylRS can, therefore, be evolved for new amino-acid specificity in E. coli and then used in eukaryotic cells. While the PyltRNACUA sequences used in E. coli can also be used in yeast and mammalian cells, the regulatory elements used to direct the transcription of the heterologous tRNA required extensive optimization.
Eukaryotic tRNA genes contain RNA polymerase III promoter sequences internal to the structural tRNA gene. The sequence of the promoter is, therefore, intimately associated with the structure and function of the tRNA (Galli et al, 1981). As the tRNAs we import into eukaryotic cells do not contain internal RNA polymerase III promoter sequences, they are generally not expressed in eukaryotic cells. The expression of heterologous tRNAs requires either the introduction of the sequences that direct Pol III transcription into the tRNAs or the discovery of extragenic sequences that will direct the transcription of functional tRNAs. While efforts to alter the sequences of tRNA genes to match consensus promoter sequences have not been successful, strategies for providing extragenic Pol III promoters have been successful. In yeast, Susan Hancock showed that a pyrrolysyl tRNA can be expressed as part of a di-cistronic (Schmidt et al, 1980) construct in which an arginyl-tRNA gene provides the Pol III promoter for the transcription of pyrrolysyl tRNA and, in mammalian cells, extragenic pol III promoters such as U6, widely developed for SiRNA, can be used to drive heterologous tRNAs (Mukai et al, 2008).
We have now incorporated a number of unnatural amino acids into proteins using the PylRS and its evolved variants in E. coli, yeast and mammalian cells. We are interested in building on these advances to develop methods that allow us to use the properties of genetically encoded unnatural amino acids to directly, specifically and synthetically observe and control molecular functions of user-defined proteins, with high spatial and temporal precision, in living cells and organisms. In recent work, Arnaud Gautier has developed approaches for site specifically incorporating photocaged versions of lysine, designed and synthesized by Alex Deiters’ group at NC State, with whom we have extensively collaborated, into proteins (Gautier et al, 2010). We have demonstrated that protein interactions and protein catalytic activity can be regulated by replacing crucial lysines in proteins with their photocaged version. Using this approach, an initially inactive protein, in which a key residue is photocaged, can be rapidly activated by shining light on cells to remove the photocage. Our work in this area builds on years of developments in photochemistry and previous work on in vitro and in vivo photocaging of diverse chemical functional groups in molecules (Riggsbee and Deiters, 2010).
Arnaud first demonstrated that genetically encoding a caged lysine in place of a key lysine within a nuclear localization sequence allowed us to rapidly trigger nuclear import of the protein by shining light on cells. This provides a means to measure the kinetics of nuclear import very rapidly and reproducibly in single cells, which we are currently exploiting to understand contributions to nuclear import kinetics (Gautier et al, 2010). Next, Arnaud extended the approach to controlling enzymatic activity. He showed that caging a lysine, within the active site of MEK kinase, along with introducing activating mutations into MEK, created a catalytically inactive enzyme. Kinase catalytic activity could be rapidly activated with light (Figure 4), allowing rapid, receptor independent, activation of a defined subnetwork in MAP kinase signalling in which photoactivated MEK leads to the activation of ERK, which accumulates in the nucleus and phosphorylates transcription factors (Gautier et al, 2011). This approach allowed us to directly observe the kinetics of the steps between MEK activation and ERK nuclear entry for the first time; studying the real-time adaptive response in signalling networks has not been possible using genetic or siRNA approaches because these approaches to controlling kinase levels are generally much slower than kinase network adaptation. Our data suggest that dual phosphorylation of ERK by MEK is rate determining for nuclear import, in accord with other recent work (Lidke et al, 2010). Moreover, these experiments provide a unique insight into the architecture of feedback pathways that may control network adaptation to receptor stimulation. Since the lysine caged in these experiments is near-universally conserved in kinase active sites (Manning et al, 2002), it should be possible to apply the approach we have developed to almost any protein kinase. By applying the approach to every kinase in a pathway, it should be possible to quantitatively define the contribution of every step in a signalling cascade to signal transmission and to provide further insight into adaptation and feedback mechanisms. In the future, we hope that the types of ‘photochemical genetic’ approaches we are developing will prove useful in decoding the molecular basis of many complex adaptive phenomena in organisms.
Conclusion
It has been a great pleasure to see our science grow in many new and exciting ways over the past few years. I have been fortunate to have very talented people come to my laboratory from a wide range of backgrounds, from total chemical synthesis to transgenic animals, through biochemistry, genetics, molecular evolution, structural biology and cell biology, and to be surrounded by some great collaborators. It has been a great pleasure to learn from everyone in the laboratory and to watch people in the laboratory learn from each other.
I believe that we are training a new generation of scientists who can seamlessly engineer across a range of scales from molecules to systems. They can engineer the specificity of biological networks and biological molecules as well as control the structures of small molecules atom by atom. By coupling these abilities, we have begun to provide solutions to problems previously viewed as intractable. I hope that the people that invested in training me get satisfaction from seeing the fragments of everything I learned from them fused to create something new, and I look forward to being surprised and excited by what the extraordinarily talented alumni that are beginning to emerge from my laboratory may do in the future.
Acknowledgments
I am grateful to Daniela Rhodes and Mariann Bienz for nominating me for this award, and to Venki Ramakrishnan, Kim Nasmyth and Paul Nurse for supporting the nomination. I am grateful to those people who have invested their time and effort in mentoring me over the years, including John Sutherland, Alanna Schepartz and Peter Schultz. I am grateful to the MRC, the ERC, HFSP and EMBO for their willingness to support long-term ambitious, curiosity-driven research. Finally, I am deeply indebted to all the fantastically innovative, dedicated and insightful people I have had the good fortune to work with, some of whom are mentioned in this personal account of my life in science.
Footnotes
The author declares that he has no conflict of interest.
References
- Akutsu M, Ye Y, Virdee S, Chin JW, Komander D (2011) Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci USA 108: 2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogelly A, Palioura S, Soll D (2007) Natural expansion of the genetic code. Nat Chem Biol 3: 29–35 [DOI] [PubMed] [Google Scholar]
- An W, Chin JW (2009) Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci USA 106: 8477–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E, Natan E, Brandt T, Allen MD, Veprintsev DB, Robinson CV, Chin JW, Joerger AC, Fersht AR (2011) Acetylation of Lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc Natl Acad Sci USA (advance online publication 27 April 2011; doi:10.1073/pnas.1105028108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett OP, Chin JW (2010) Evolved orthogonal ribosome purification for in vitro characterization. Nucleic Acids Res 38: 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle H, Winter G (1986) A model of synthetase/transfer RNA interaction as deduced by protein engineering. Nature 320: 371–373 [DOI] [PubMed] [Google Scholar]
- Boos D, Kuffer C, Lenobel R, Korner R, Stemmann O (2008) Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J Biol Chem 283: 816–823 [DOI] [PubMed] [Google Scholar]
- Braig D, Bar C, Thumfart JO, Koch HG (2009) Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J Mol Biol 390: 401–413 [DOI] [PubMed] [Google Scholar]
- Capecchi MR (1967) Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci USA 58: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA (2010) Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A (2006) The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16: 1280–1289 [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK (2008) Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HT, Warfield L, Hahn S (2007) The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG (2003a) An expanded eukaryotic genetic code. Science 301: 964–967 [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Chu S, Meggers E, Schultz PG (2003b) Progress toward an expanded eukaryotic genetic code. Chem Biol 10: 511–519 [DOI] [PubMed] [Google Scholar]
- Chin JW, Grotzfeld RM, Fabian MA, Schepartz A (2001) Methodology for optimizing functional miniature proteins based on avian pancreatic polypeptide using phage display. Bioorg Med Chem Lett 11: 1501–1505 [DOI] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG (2002a) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA 99: 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG (2002b) Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc 124: 9026–9027 [DOI] [PubMed] [Google Scholar]
- Chin JW, Schepartz A (2001a) Concerted evolution of structure and function in a miniature protein. J Am Chem Soc 123: 2929–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Schepartz A (2001b) Design and evolution of a miniature Bcl-2 binding protein. Angew Chem Int Ed Engl 40: 3806–3809 [DOI] [PubMed] [Google Scholar]
- Chin JW, Schultz PG (2002) In vivo photocrosslinking with unnatural amino acid mutagenesis. Chembiochem 3: 1135–1137 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Cosgrove MS, Boeke JD, Wolberger C (2004) Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol 11: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Dedkova LM, Fahmi NE, Golovine SY, Hecht SM (2003) Enhanced D-amino acid incorporation into protein by modified ribosomes. J Am Chem Soc 125: 6616–6617 [DOI] [PubMed] [Google Scholar]
- Dougherty DA (2008) Physical organic chemistry on the brain. J Org Chem 73: 3667–3673 [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards H, Schimmel P (1990) A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol Cell Biol 10: 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW (2005) Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods 2: 377–384 [DOI] [PubMed] [Google Scholar]
- Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MM, Winter G (1985) Hydrogen bonding and biological specificity analysed by protein engineering. Nature 314: 235–238 [DOI] [PubMed] [Google Scholar]
- Freeman M (2002) A fly's eye view of EGF receptor signalling. EMBO J 21: 6635–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter R (1998) Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci 7: 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G, Hofstetter H, Birnstiel ML (1981) Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294: 626–631 [DOI] [PubMed] [Google Scholar]
- Gautier A, Deiters A, Chin JW (2011) Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc 133: 2124–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW (2010) Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc 132: 4086–4088 [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Hancock SM, Uprety R, Deiters A, Chin JW (2010) Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J Am Chem Soc 132: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Hecht SM, Alford BL, Kuroda Y, Kitano S (1978) ‘Chemical aminoacylation’ of tRNA's. J Biol Chem 253: 4517–4520 [PubMed] [Google Scholar]
- Heckler TG, Chang LH, Zama Y, Naka T, Chorghade MS, Hecht SM (1984) T4 RNA ligase mediated preparation of novel ‘chemically misacylated’ tRNAPheS. Biochemistry 23: 1468–1473 [DOI] [PubMed] [Google Scholar]
- Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S (2005) Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods 2: 201–206 [DOI] [PubMed] [Google Scholar]
- Hino N, Oyama M, Sato A, Mukai T, Iraha F, Hayashi A, Kozuka-Hata H, Yamamoto T, Yokoyama S, Sakamoto K (2011) Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. J Mol Biol 406: 343–353 [DOI] [PubMed] [Google Scholar]
- Huang LY, Umanah G, Hauser M, Son C, Arshava B, Naider F, Becker JM (2008) Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry 47: 5638–5648 [DOI] [PubMed] [Google Scholar]
- Hui A, de Boer HA (1987) Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci USA 84: 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD (2005) Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA 106: 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RL, Stade LW, Wimmer R, Stensballe A, Duroux M, Larsen KL, Wingren C, Duroux L (2010) Direct site-directed photocoupling of proteins onto surfaces coated with beta-cyclodextrins. Langmuir 26: 11597–11604 [DOI] [PubMed] [Google Scholar]
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G (1986) Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321: 522–525 [DOI] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM (2006) Real-time observation of trigger factor function on translating ribosomes. Nature 444: 455–460 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Trickey M, Izawa D, Gannon J, Yamamoto M, Yamano H (2008) A mutual inhibition between APC/C and its substrate Mes1 required for meiotic progression in fission yeast. Dev Cell 14: 446–454 [DOI] [PubMed] [Google Scholar]
- Kwok R (2010) Five hard truths for synthetic biology. Nature 463: 288–290 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA (2007) Identification of nascent chain interaction sites on trigger factor. J Biol Chem 282: 12186–12193 [DOI] [PubMed] [Google Scholar]
- Lammers M, Neumann H, Chin JW, James LC (2010) Acetylation regulates cyclophilin A catalysis, immunosuppression and HIV isomerization. Nat Chem Biol 6: 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Huang F, Post JN, Rieger B, Wilsbacher J, Thomas JL, Pouyssegur J, Jovin TM, Lenormand P (2010) ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J Biol Chem 285: 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Reynolds N, Ibba M (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol 63: 61–78 [DOI] [PubMed] [Google Scholar]
- Liu C, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, Beckmann R, Hayer-Hartl M (2010) Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463: 197–202 [DOI] [PubMed] [Google Scholar]
- Liu DR, Magliery TJ, Pastrnak M, Schultz PG (1997) Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc Natl Acad Sci USA 94: 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DR, Schultz PG (1999) Progress toward the evolution of an organism with an expanded genetic code. Proc Natl Acad Sci USA 96: 4780–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Mendel D, Cornish VW, Schultz PG (1995) Site-directed mutagenesis with an expanded genetic code. Annu Rev Biophys Biomol Struct 24: 435–462 [DOI] [PubMed] [Google Scholar]
- Michie KA, Löwe J (2006) Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem 75: 467–492 [DOI] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S (2008) Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22: 2994–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K (2006) Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA 103: 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S (2008) Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun 371: 818–822 [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW (2009) A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Hazen JL, Weinstein J, Mehl RA, Chin JW (2008a) Genetically encoding protein oxidative damage. J Am Chem Soc 130: 4028–4033 [DOI] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW (2008b) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol 4: 232–234 [DOI] [PubMed] [Google Scholar]
- Neumann H, Slusarczyk AL, Chin JW (2010a) De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J Am Chem Soc 132: 2142–2144 [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW (2010b) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464: 441–444 [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW (2009a) Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J Am Chem Soc 131: 14194–14195 [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Garcia Alai MM, Virdee S, Chin JW (2010) Genetically directing epsilon-N, N-dimethyl-L-lysine in recombinant histones. Chem Biol 17: 1072–1076 [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, Chin JW (2009b) Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc 131: 8720–8721 [DOI] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244: 182–188 [DOI] [PubMed] [Google Scholar]
- Okuda S, Tokuda H (2009) Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA 106: 5877–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahandeh S, Maurer C, Moser M, DeLisa MP, Muller M (2008) Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J Biol Chem 283: 33267–33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR (1989) Heat shock and the sorting of luminal ER proteins. EMBO J 8: 3171–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FV IV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Powner MW, Gerland B, Sutherland JD (2009) Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459: 239–242 [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin JW (2005a) A network of orthogonal ribosome-mRNA pairs. Nat Chem Biol 1: 159–166 [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin JW (2005b) Cellular logic with orthogonal ribosomes. J Am Chem Soc 127: 17584–17585 [DOI] [PubMed] [Google Scholar]
- Rackham O, Wang K, Chin JW (2006) Functional epitopes at the ribosome subunit interface. Nat Chem Biol 2: 254–258 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Raschle T, Speziga D, Kress W, Moccand C, Gehrig P, Amrhein N, Weber-Ban E, Fitzpatrick TB (2009) Intersubunit cross-talk in pyridoxal 5′-phosphate synthase, coordinated by the C terminus of the synthase subunit. J Biol Chem 284: 7706–7718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann L, Clark M, Waldmann H, Winter G (1988) Reshaping human antibodies for therapy. Nature 332: 323–327 [DOI] [PubMed] [Google Scholar]
- Riggsbee CW, Deiters A (2010) Recent advances in the photochemical control of protein function. Trends Biotechnol 28: 468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A (2007) Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615 [DOI] [PubMed] [Google Scholar]
- Schmidt O, Mao J, Ogden R, Beckmann J, Sakano H, Abelson J, Soll D (1980) Dimeric tRNA precursors in yeast. Nature 287: 750–752 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 A resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M (1968) Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA 61: 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten EM, Bertozzi CR (2009) Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl 48: 6974–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S (2010) Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468: 978–982 [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T (2009) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol 184: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Hamada F, Schwarz-Romond T, Bienz M (2008) Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev 22: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW (2010) Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol 6: 750–757 [DOI] [PubMed] [Google Scholar]
- Wang K, Neumann H, Peak-Chew SY, Chin JW (2007) Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol 25: 770–777 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Winter G, Fersht AR, Wilkinson AJ, Zoller M, Smith M (1982) Redesigning enzyme structure by site-directed mutagenesis: tyrosyl tRNA synthetase and ATP binding. Nature 299: 756–758 [DOI] [PubMed] [Google Scholar]
- Winter G, Milstein C (1991) Man-made antibodies. Nature 349: 293–299 [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG (2006) A chemical toolkit for proteins—an expanded genetic code. Nat Rev Mol Cell Biol 7: 775–782 [DOI] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M (2009) Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M (2007) Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell 27: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T (2010) Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem 285: 41222–41231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Huber T, Vogel R, Sakmar TP (2009) FTIR analysis of GPCR activation using azido probes. Nat Chem Biol 5: 397–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Zaitseva E, Caltabiano G, Schertler GF, Sakmar TP, Deupi X, Vogel R (2010) Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature 464: 1386–1389 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Green R (2005) Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods 36: 305–312 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondlo NJ, Schepartz A (1999) Highly specific DNA recognition by a designed DNA binding protein. J Am Chem Soc 121: 6938–6939 [Google Scholar]