Abstract
2D and 3D cryo-electron microscopy, together with adsorption kinetics assays of ϕCb13 and ϕCbK phage-infected Caulobacter crescentus, provides insight into the mechanisms of infection. ϕCb13 and ϕCbK actively interact with the flagellum and subsequently attach to receptors on the cell pole. We present evidence that the first interaction of the phage with the bacterial flagellum takes place through a filament on the phage head. This contact with the flagellum facilitates concentration of phage particles around the receptor (i.e., the pilus portals) on the bacterial cell surface, thereby increasing the likelihood of infection. Phage head filaments have not been well characterized and their function is described here. Phage head filaments may systematically underlie the initial interactions of phages with their hosts in other systems and possibly represent a widespread mechanism of efficient phage propagation.
Keywords: cryo-electron tomography, alpha-proteobacteria
Many bacteriophages target host appendages, such as flagella and pili, as initial points of adsorption (1–4) and have been shown to preferentially attach to host receptors localized at cell poles (5, 6). By using the flagella and/or pili to initiate contact with the host cell, phages may improve the likelihood of attachment and successful infection. Many of the phages that are flagellotropic include members of the Siphoviridae or Myoviridae families, including Bacillus subtilis phage PBS1 (3), Escherichia coli phage Chi (2), Proteus vulgaris phage PV22 (7), and Asticcacaulis biprosthecum phage AcM2 (8). A number of flagellotropic phages that infect members of Caulobacteraceae (α-proteobacteria) have been described (1, 9–11). Caulobacter crescentus, while in its swarmer state or flagellated form (Fig. S1), has been shown to host several flagellotropic phages, including ϕCb13 and ϕCbK (11).
ϕCb13 and ϕCbK are noncontractile-tailed, dsDNA Siphoviridae phages (12). They have an elongated head and a large genome (>200 kb) that are characteristics of the relatively uncommon B3 morphotype. ϕCb13 and ϕCbK have been essential in studies of bacterial development (13), cell cycle-dependent gene expression (14–16), and determination of polarity in C. crescentus (17, 18). Most importantly, ϕCbK has been a model system for cryo-electron microscopy and image reconstruction (19, 20). In several landmark studies, it was the first bacteriophage in which the five- to threefold symmetry mismatch between the head and tail was identified (20), a possible model for the distribution of the capsid proteins was produced (21), and the structure of the tail was assessed (20, 22, 23). Initial reports also revealed that both ϕCb13 and ϕCbK exploit the flagellum and/or the pili for adsorption and attachment to the host cell (6, 24, 25). However, the actual mechanisms involved remain undefined.
In this paper, we identify the contacts made between ϕCb13 or ϕCbK and the bacterium C. crescentus during the initial stages of infection by determining their 3D structures using cryo-electron tomography (cryo-ET). Bacterial motility assays, phage one-step growth curves and adsorption assays were also used to determine how the flagellum, pili, and cellular motility affect phage infection efficiency. The 3D structures of the interface between the ϕCb13 and ϕCbK head-associated filament and the cell's flagellum illustrate a unique mechanism of infection. In brief, the head filament loosely wraps around the flagellum, allowing flagellar rotation to draw phages toward the cell pole, and host receptors. The assays revealed that motility, directionality of flagellum rotation, and the presence of the pili are critical determinants of infection. Therefore, our structural studies and microbiological assays of ϕCb13- and ϕCbK-infected C. crescentus cells reveal a previously undescribed mechanism that these phages use for initial adsorption to the cell's flagellum and the subsequent irreversible attachment of the phage tail to the host receptors.
Results and Discussion
Ultrastructure of C. crescentus Cells by Cryo-Electron Microscopy.
The interactions of motility mutants of C. crescentus with ϕCb13 and ϕCbK were examined to determine the relationship between flagellar assembly and function in the initial stages of phage infection. 2D images and 3D reconstructions of C. crescentus revealed characteristic cellular ultrastructure. Unique specific densities corresponding to subunits of potential phage receptors were not evident at the resolution levels of the 2D images and 3D reconstructions presented. However, common attributes of these cells are evident and include the S layer, the cell inner and outer membranes and peptidoglycan layer, and the macromolecule-dense cytoplasm (26). Other pole-localized appendages or complexes, previously found to be specific to either swarmer (flagella, pili, and chemoreceptor arrays; refs. 27 and 28) or divisional cells (stalks) were also encountered during 2D and 3D EM data surveys (Fig. S2).
Ultrastructure of ϕCb13 and ϕCbK by Cryo-EM and Cryo-ET Reveals Distinctive Head Filaments.
To confirm the results of previous structural studies of ϕCb13 and ϕCbK particles (21, 23), we generated 2D cryo-EM images and 3D cryo-ET reconstructions of isolated phages and of phages infecting C. crescentus cells. In addition to confirming that the prolate heads are 60 nm in diameter and 200 nm in length and their noncontractile tails are 290 nm long, we showed that a phage head appendage (head filament) was present (Fig. 1 and Fig. S3). Furthermore, our analyses also enabled us to optimize procedures for the 3D segmentation of the cells and phages by using the 3D EM Amira toolbox (29) so that we were able to accurately distinguish the head filament from the bacterial flagellum (Fig. 2). These results suggested that the head filament is responsible for the initial attachment of the phage to the flagellum.
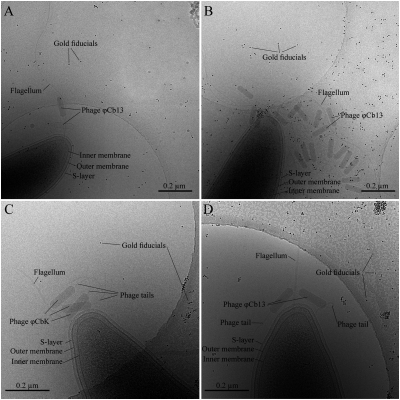
Fig. 1.
Cryo-ET image, 3D reconstruction, and image analysis of ϕCb13 phages. (A) An image at 0° tilt of C. crescentus pole with two ϕCb13 phages attached. Fiducial gold particles are 10 nm in diameter. (B) A tomographic slice from Amira of the same region of the sample after tomographic reconstruction. (C) Same image after contrast inversion using the toolbox for 3D electron microscopy (EM) in Amira. (D) After thresholding with the island label module of the 3D EM toolbox the different regions are visualized. (E) Labels identify specific regions independently (i.e., each phage and the S-layer). Here, regions are visualized with different colors, yellow corresponding to the phages and green to the cell's S-layer. Arrowheads in E point to phage head filaments. The mouse cursor is used to select the labels to be included in the segmentation. Object selection is done through several slices and several times until all regions of interest are selected.
Fig. 2.
Automatically generated segmentations of ϕCb13-infected NA1000 C. crescentus cells. Notice the head filaments that extend from the phage and wrap around the flagellum. Orange-labeled densities inside yellow empty phages correspond to dislodged tail-head connectors. Full phages are depicted in blue and bacterial cell components in green. In A and B, the segmentation is superimposed over a Voltex (direct volume rendering where each point in the volume is assumed to emit and absorb light). Automatic segmentations were generated by using the IslandLabel module from the 3D-EM toolbox developed by Pruggnaller et al. (29).
Role of Flagellar Machinery in Phage Infection.
ϕCb13 and ϕCbK preferentially infect swarmer cells (6, 30), where a correlation exists between flagellar rotation and the ability of ϕCbK to successfully infect cells and generate progeny (6). It has been established that WT C. crescentus cells have a swimming reversal frequency of 50% (31). To swim forward, the cell's flagellum rotates clockwise (CW), although when the cell swims in reverse, the flagellum turns counterclockwise (CCW). To examine the effect of cell motility on phage infection, we first established baseline motility data for each C. crescentus strain (Table 1). Motility assays confirmed that strain ΔpilA, containing an in-frame deletion that results in cells without pili (25), is able to swarm and exhibits similar motility patterns to those of WT C. crescentus (Table 2). Conversely, the strains motA:Tn (a transposon mutant with a normal flagellum and a nonfunctioning flagellar motor; refs. 32 and 33); cheR::Tn (NS209) and cheR::Tn (NS338) (strains exhibiting CW flagellar rotation; ref. 31); cheB144::Tn and cheB148::Tn (strains exhibiting CCW flagellar rotation; ref. 31); and ΔtipF (flagellum-lacking strain; ref. 32) were unable to swarm efficiently.
Table 1.
Bacterial strains used during the course of this study
| Strain name | Phenotype | Ref. |
| C. crescentus NA1000 | Synchronizable derivative of CB15 | 34 |
| C. crescentus motA:Tn | Incomplete stator and deficient motility | 32, 33 |
| C. crescentus cheB144::Tn | Counterclockwise flagellar rotation (CCW) | 31 |
| C. crescentus cheB148::Tn | Counterclockwise flagellar rotation (CCW) | 31 |
| C. crescentus cheR::Tn (NS209) | Clockwise flagellar rotation (CW) | 32 |
| C. crescentus cheR::Tn (NS338) | Clockwise flagellar rotation (CW) | 32 |
| C. crescentus ΔtipF | Flagellum not present | 32 |
| C. crescentus ΔpilA | Pili not present | 25 |
Table 2.
Motility assays and rate of phage adsorption to wild-type and motility mutants
| ϕCb13 adsorption (K values = × 10−11 mL/min) |
ϕCbK adsorption (K values = × 10−11 mL/min) |
||||||
| Strain name | Motility zone diameter (48 h), mm | Slope | r value | k value | Slope | r value | k value |
| C. crescentus NA1000 | 6 | −0.0150 | 0.9888 | 3.71 | −0.0157 | 0.9955 | 3.49 |
| C. crescentus motA:Tn | 0 | −0.0036 | 0.9073 | 0.60 | −0.0073 | 0.9975 | 1.62 |
| C. crescentus cheB144::Tn | 2 | −0.0090 | 0.9960 | 2.40 | −0.0092 | 0.9672 | 2.04 |
| C. crescentus cheB148::Tn | 2 | −0.0011 | 0.9753 | 2.78 | −0.0105 | 0.9678 | 2.33 |
| C. crescentus cheR::Tn (NS209) | 4 | −0.0043 | 0.9127 | 0.62 | −0.0058 | 0.9947 | 1.29 |
| C. crescentus cheR::Tn (NS338) | 4 | −0.0047 | 0.9385 | 0.71 | −0.0079 | 0.9965 | 1.76 |
| C. crescentus ΔtipF | 0 | −0.0023 | 0.9256 | 0.67 | −0.0083 | 0.9938 | 1.84 |
| C. crescentus ΔpilA | 7 | −0.0007 | 0.6481 | 0.13 | −0.0027 | 0.9521 | 0.60 |
Upon confirmation of the motility phenotype, experiments were performed to assess the rate of adsorption of ϕCb13 and ϕCbK to host cell strains (Table 2 and Fig. S4). The adsorption efficiency of ϕCb13 to the wild-type strain NA1000 (34) is represented as a k value (adsorption constant) of 3.71 × 10−11 mL/min, whereas ϕCbK adsorbs at a rate of 3.49 × 10−11 mL/min. There was a dramatic sixfold reduction in the adsorption rate of ϕCb13 to the flagellar motor defective cells. A similar reduction was observed in the k values of the flagellum-lacking ΔtipF mutant, compared with NA1000 (Table 2). This similarity in the reduction rate of adsorption suggests that efficient binding of ϕCb13 requires not only the presence of the flagellum, but an actively rotating flagellum. In contrast, there was only a twofold reduction in the ϕCbK adsorption rate to the cells, indicating that ϕCbK is more efficient than ϕCb13 in adsorbing to both flagellum defective and deficient strains.
When cells exhibiting exclusively CW (cheR mutants) or CCW (cheB mutants) flagellar rotation were exposed to ϕCbK, it was apparent that phages adsorbed less efficiently, with cheB mutations inhibiting phage adsorption less than cheR (Table 2). Interestingly, the reduction of ϕCb13 or ϕCbK phage adsorption to the cheR mutants was comparable to that of the flagellar motor defective or flagellum-lacking mutants, suggesting that the direction of flagellar rotation is important for phage adsorption. The adsorption of both phages is significantly more efficient in the cheB mutants that exhibit CCW rotation, suggesting that CCW rotation may facilitate the wrapping of the phage head filament around the flagellum.
Model for Phage Adsorption.
As indicated above, our results support the hypothesis that the motion of the C. crescentus flagellum facilitates the positioning of phages in proximity to the cell pole. The translocation of the phages along the flagellum increases the likelihood of interactions between the phage tail and polar receptors necessary for irreversible attachment.
ϕCb13 and ϕCbK elicit a two- to threefold reduction in their rates of adsorption to nonflagellated and flagellar rotation mutants and both phages still formed plaques on nonmotile C. crescentus lawns. This reduced adsorption rate indicates that the presence of an active flagellum is important for efficient adsorption, but the absence of a flagellum does not confer phage resistance. Because phage adsorption is more efficient in CCW (cheB) mutants than in the CW (cheR) mutants (Table 2), the direction of rotation might be important in this process. A possible explanation for these results is that the natural rotation of the flagellum, which alternates between CW and CCW directions, favors the accumulation of phages around the cell pole (Movie S1). Therefore, the normal mode of rotation may increase the likelihood of phage–receptor contact and make the direction of flagellar rotation a limiting factor in adsorption kinetics. The higher likelihood of infection with a CCW rotational bias is consistent with the nut and bolt model proposed by Berg and Anderson (35). It has been demonstrated with other phages, such as Chi, that the time it takes for the phage to reach the cell is less than the flagellum rotation reversal frequency (36). Simply, the speed of the phage along the flagellum favors its approach to the cell body for irreversible attachment. Unlike Chi and other phages, such as PBS1 (3), where the phage tail and tail fiber are responsible for both the initial adsorption to the flagellum and the irreversible attachment to the cell, ϕCbK and ϕCb13 initially adsorb to the flagellum via the head filament followed by irreversible attachment to the appropriate receptor via the phage tail. Thus, the interaction of the ϕCbK and ϕCb13 head filament with the flagellum is likely to differ from that of the Chi phage tail fiber with the flagellum.
We hypothesize that, in cells with flagellar rotation biased in the CW direction, this directionality causes phage particles (with their head filaments wrapped around the flagellum) to migrate away from the cell pole (Movie S2). In addition, CW rotation causes the bacterium to move forward in the aquatic environment, which results in phages being dragged by the cell. Under these conditions, the phage tails are oriented away from the cell pole, resulting in a reduction in the likelihood of phage tail interaction with its receptor. In contrast, phages adsorbed to the flagellum of cells with a CCW flagellar rotation migrate toward the cell pole (Movie S3). In addition, the ensuing backward motion of the cell orients the phage tails toward the cell pole, favoring attachment to a receptor.
Adsorption of Phage Heads to Bacterial Flagellum.
The use of cryo-EM and cryo-ET to study host–pathogen interactions allows us to directly observe structures and processes that would otherwise be damaged by heavy metals after negative staining or dehydration and staining procedures during sample embedding for sectioning. This analysis uncovered the unusual mechanism that phages ϕCbK and ϕCb13 use to initially adsorb to their host. They use an apparently flexible and rather variable length head filament that is used to wrap around the flagellum, likely as a means for positioning themselves close to the cell pole and the final sites of irreversible attachment to the receptor (herein identified as the pilus portals) (Fig. 2 A and B and Movie S4, Movie S5, and Movie S6). This type of association was consistently observed throughout our transmission electron microscopy survey of both negatively stained and plunge frozen cells when phages were found in close proximity to the flagellum. In addition, the head filament is found on phage particles when cells lack a flagellum, indicating that the filament is of phage origin (Fig. 1 and Fig. S3).
In the course of this study, we were able to capture specific events that take place during phage infection of C. crescentus cells. These images illustrate that ≈15 min after infection, phages ϕCbK and ϕCb13 were localized along the bacterial flagellum by means of the head filament (Figs. 2 A and B and 3 A and B). One-step growth curves indicate that 15 min still corresponds to initial steps of infection because phage progeny are not detectable before 60 min of infection (Fig. S5). Initial reports of head filaments in C. crescentus phages were published decades ago by Leonard et al. (21, 23) and confirmed by Papadopoulos and Smith (22). However, no insight into their function was offered at the time. It has also been shown that other bacteriophages, such as the cyanophage Syn5 of a marine Synechococcus species, express a hornlike or filamentous structure on their capsid (37). As was the case for ϕCbK, the function of the Syn5 horn has not yet been determined. Our proposed mechanism for ϕCbK and ϕCb13 head filament adsorption to the flagellum of C. crescentus is consistent with the suggestion of Bender et al. (15) that a weak affinity between the flagellum and a phage structure (unspecified in their model but identified herein as the head filament) guides and concentrates phages near the cell receptor.
Fig. 3.
Cryo-electron micrographs of plunge-frozen C. crescentus strains infected with either phage ϕCb13 or ϕCbK. (A) A NA1000 cell infected with phage ϕCb13. (B) A ΔmotA mutant cell infected with ϕCb13. Notice that flagellum-associated phage particles are oriented with their heads toward the flagellum. Phages attached to the cell pole are also observed. C and D correspond to ΔpilA mutants depicting successful adsorption of phage particles ϕCbK (C) and ϕCb13 (D) to the bacterial flagellum. Phage tails in C and D are not attached to the cell pole despite their close proximity. Scale bars 0.2 μm.
Initial flagellar adsorption of phages by their tails or tail fibers has been described in several systems as a necessary step leading to irreversible attachment to their host (4, 38). Successful contact of the phage tail with the receptor, located on the cell body, then requires the temporary separation of the phage from the cell or the participation of auxiliary fibers of filaments in phage tails to reestablish a connection between the phage tail and the cell body. The mechanism of flagellar adsorption by ϕCbK and ϕCb13 may increase their chance of irreversible attachment by separating the two processes. In this case, initial contact is mediated by a separate appendage (i.e., the head-filament), which allows their tails to remain free for attachment to the cell receptor and subsequent injection of their genetic material.
Role of Pilus Portals in Phage Attachment.
We studied the pilin-deficient deletion mutant ΔpilA (25, 39) to determine how the absence of pili would affect phage infection. As mentioned above, flagellar motion facilitates ϕCb13 and ϕCbK adsorption. Nonetheless, we have established for ϕCb13 and confirmed for ϕCbK that this behavior is not considered a requirement for successful infection of C. crescentus. Previous research indicates that a specific receptor for ϕCbK is present in nonswarming strains of C. crescentus (15). In addition, Scholl and Jollick (40) determined that the C. crescentus dsDNA phage ϕ6 uses components of the pilus apparatus for infection. In our study, adsorption assays of both ϕCbK and ϕCb13 to the ΔpilA mutant elicited the lowest k values for each phage (Table 2 and Fig. S4). In the case of ϕCbK, we confirmed (Table 2, Fig. S4, and Fig. S6) what has been suggested for this phage: Pili are necessary for successful infection of C. crescentus (6, 14). Similarly, our ϕCb13 data show that adsorption is hindered when the cells lack pili, indicating that this phage also uses pili portals as the site for attachment (Table 2, Fig. S4, and Fig. S6). Extensive microscopy data screening of EM grids prepared from ≈50 individual cultures of either ϕCb13- or ϕCbK-infected ΔpilA mutants (two representative images are shown in Fig. 3 C and D) reveals that phage do not attach to the bacterial cell surface when pili are not present. Our tomography data further indicate that irreversible attachment of ϕCb13/ϕCbK tails occurs in areas of the cell pole where pili portals are located (Figs. 4 and 5 and Movie S7). There are several reports of pilus-dependent phage infection in the literature but, to our knowledge, they mostly conclude that phage tails initially attach to bacterial pili and subsequent pili retraction facilitates genome injection after successful, irreversible attachment to the host. This mechanism of infection is found in phages C22, M6, PE69, C5 (41), and KMV (42) of Pseudomonas aeruginosa, phages ϕCb5 and ϕCbK of C. crescentus (9, 11, 25), and phage AcM2 of Asticcacaulis biprosthecum (8). For ϕ6, it has been observed that final attachment occurs in areas adjacent to pilus insertion sites on the cell surface (40). Therefore, we conclude that both phages attach to pili via their tails and then attach to the pili portals after pilus retraction.
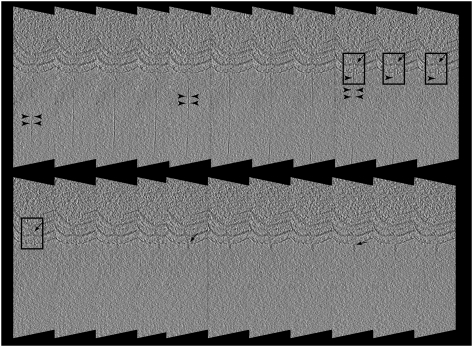
Fig. 4.
Image collage of the interaction between C. crescentus pilus and the tail of ϕCb13. Arrowheads point to the pilus filament in all images. (A) Slice through a tomogram of an infected NA1000 cell. The black rectangle highlights the area of pilus-phage interaction. (B) Segmented volume imposed in 3D over the same slice in A. Phages are depicted in blue, bacteria surface layer in green and pilus in orange. (C) Rotation of B to position the site of pilus-phage interaction in the front of the view. (D) Enlarged segmented volume.
Fig. 5.
Averaged 13-nm tomography slices of NA1000 C. crescentus cell infected with ϕCb13. Images are restricted to the cell pole and highlighted areas (rectangles) are shown where contact between the phage tail and cell pilus is evident. Arrowheads indicate pilus and arrows indicate portions of the phage tail.
Conclusions
We have shown that the first interactions of ϕCb13 and ϕCbK phages with the bacterial flagellum take place through a filament located on the phage head. This contact facilitates the concentration of phage particles around the phage receptor (i.e., the pilus portals) on the bacterial cell surface, thereby increasing the likelihood of successful adsorption. There is a relatively low likelihood of encounters between phages and their hosts in aquatic environments, especially for bacteria such as the Caulobacters that live at low densities in environments with limiting nutrients. Therefore, this mechanism of flagellar attachment may have evolved in C. crescentus phages to increase the probability of successful infection of the bacterial host. Additionally, by targeting C. crescentus flagellated cells, the phage genome is injected into a bacterium that recently arose by cell division, which is the ideal environment to support the production of new phage progeny. Further studies should focus on the head filament to reveal its genetic origin, composition, and contribution to phage adaptation. In addition, an examination of the particular contacts of the head filament with C. crescentus flagellin subunits should provide additional insight into this unique mechanism of reversible adsorption.
Materials and Methods
Bacterial Cultures, Motility Assay, and Phage Propagation.
Fresh colonies of C. crescentus strains differing in flagellar assembly or rotation (Table 1) were inoculated and grown overnight at 30 °C with aeration, in peptone yeast extract (PYE; 0.2% peptone, 0.1% yeast extract, 1 mM MgSO4, 0.5 mM CaCl2) modified from Schmidt and Stanier (1, 43). For plating medium, either 1.5% (regular growth plates) or 0.3% (motility plates) agar was added. For adsorption assays, PYE medium contained 4 mM MgSO4 (1). When necessary, swarmer cells were separated by using a 75% solution of Percoll (Sigma-Aldrich) from a heterogeneous cell culture by means of their higher buoyant density (1.07 g/mL) compared with the lower density (1.01 g/mL) of stalked cells (44).
Motility assays used freshly prepared PYE motility plates air-dried under a UV-illuminated, biosafety cabinet for at least 15 min. Using sterile plastic needles, fresh colonies from overnight cultures were stabbed into the agar and incubated for several days at 30 °C. The diameter of motility zones was measured every 24 h to establish motility phenotype.
Phages were propagated by using modified double agar overlay plate assay (45). Briefly, 400 μL of a wild-type C. crescentus overnight culture were infected with phage ϕCb13 or ϕCbK and incubated without shaking at 30 °C for 15 min to allow adsorption. Infected cells were added to 4.5 mL of molten 0.5% PYE agar and immediately overlaid on 1.5% PYE agar. Plates were incubated at 30 °C for 24 h, when small plaque-forming units (pfu) were visible. PYE medium (2 mL) was added to allow phages to diffuse into it overnight at 4 °C. Phage-containing PYE was recovered and treated with chloroform to inactivate remaining bacterial cells.
Determination of Phage Infection Efficiency.
To establish whether either flagellar rotation or integrity had an effect on the efficiency of phage infection, adsorption assays were performed to determine the change in abundance of unadsorbed phage over time (1, 15). Nine milliliters of overnight cultures of NA1000 or mutant C. crescentus at an OD600 of 0.6 (3–5 × 108 cells per mL) in PYE adsorption media (4 mM MgSO4) were mixed with either ϕCb13 or ϕCbK at a multiplicity of infection of 0.002 in 125-mL flasks. Adsorption was for 60 min at 30 °C with slow shaking. one hundred-microliter samples were taken every 5 min, mixed with 900 μL of ice-cold PYE medium, and then 30 μL of chloroform was added. Bacteria and bacterial debris were separated from unadsorbed phages by centrifugation for 5 min at 12,000 × g. Subsequently, 100 μL of phage-containing supernatant were mixed with 400 μL of a phage infection indicator strain (bNY30A; OD600 = 0.3), mixed with molten 0.5% PYE agar and overlaid on 1.5% agar. Plates were incubated at 30 °C overnight before counting pfu. A control flask containing only phage was maintained in parallel for the duration of the experiment. The adsorption rate constant (k) was calculated by plotting relative pfu over time and dividing the slope of the regression line by the viable bacteria cell number. Table 2 depicts the k values for ϕCb13 and ϕCbK.
cryo-ET Preparation and Data Collection.
C. crescentus strains (Table 1) grown in PYE medium were flash-frozen (plunge frozen) onto glow-discharged, 200 mesh, copper Quantifoil grids in liquid ethane by using either a Vitrobot Mark II or a Vitrobot Mark III system (FEI). BSA-treated colloidal gold with a particle size of 10 nm was applied onto the grids and let air dry before sample application and freezing. To characterize C. crescentus phage infection by cryo-ET, overnight cultures were infected with either ϕCb13 or ϕCbK phages and incubated for 15 min at 30 °C without shaking. Four-microliter aliquots were immediately applied onto grids and plunge frozen as mentioned above.
Cryo-EM and cryo-ET data collection was performed by using two microscopes: an FEI Polara 300-kV FEG-TEM equipped with a Gatan energy filter (slit width 20 eV) coupled to a lens-coupled 4k × 4k Gatan UltraCam CCD camera at the California Institute of Technology or an FEI Tecnai G2 F30 300-kV FEG-TEM using a Gatan 4k × 4k Ultrascan CCD camera at the University of Minnesota Characterization Facility. Images were acquired with pixel size ranging from 0.372 nm to 0.648 nm on the specimen. The magnifications used allowed us to resolve bacterial components including the outer membrane, inner membrane, and flagellar basal body (Fig. S1). A total electron dose between 150 and 200 electrons per squared Angstrom (e−/Å2) was sufficient to allow collection of tilt series ranging from −65° to +65° (131 images) without sample destruction (bubbling). Data were collected at a defocus of 8.0 μm under focus (first CTF zero: 1/4 nm−1) with the purpose of enhancing the contrast of various cell and phage components. Tilt series images were taken automatically with 1° tilt increments by using the predictive UCSF tomography package (46).
Image Processing and Analysis and Movie Generation.
Tomographic reconstructions were generated by using IMOD (47), and the data were binned twofold during this process. 3D volumes were rendered by using Amira (Visage Imaging Systems). A series of supporting tools in a visualization and segmentation toolbox, developed specifically for 3D EM data analysis in Amira (Amira toolbox; ref. 29), were used to achieve a more objective segmentation of the data and to avoid manual contouring (entirely subjective tracing of edges or features; Fig. 1). This method of segmentation automatically assigns labels to specific densities in tomogram slices and connects these regions in three dimensions, thereby removing user bias and making interpretation of tomographic data more reliable. The user defines the threshold for selecting regions of density; however, selection is purely derived from the histogram values present within the tomogram. For the most accurate representation of the filamentous structures of the phage head filament and the bacterial flagellum, we found the 3D EM Amira toolbox to be the most effective method available.
Reconstructions were loaded into Amira and cropped to focus on the areas of interest. Slices were filtered after contrast inversion by using 3D Gaussian smoothing with a kernel size of 3 × 3 × 3. Subsequently, the IslandLabel module was applied, which bins the data by using a user-defined threshold and then tags connected regions of the binary image with a label.
Direct volume rendering and z-slice progressions were done in Amira. Animated sequences of operations were created by using the DemoMaker module and converted into image sequences with the MovieMaker module. Individual images were labeled with Adobe Illustrator CS5 (Adobe Systems). Animations were generated by using Autodesk Maya 2011 (Autodesk).
Supplementary Material
Acknowledgments
We thank Dr. Jens M. Holl and Mr. Grant M. Williams for valuable help with manuscript preparation, graphic arts, and helpful discussions; Dr. Ryland Young and Dr. Ian Molineux for advice and mentoring on phage biology; Ms. Jeannette Taylor, Ms. Hong Yi, Dr. Wei Zhang, Dr. Ozan Ugurlu, and Mr. Chris Frethem for assistance during cryo-EM data collection; and Ms. Sabine Pruggnaller for providing the Amira toolbox for 3D segmentation. This work was supported in part by Emory University, Children's Healthcare of Atlanta, and the Georgia Research Alliance (to E.R.W.); and Human Frontier Science Program Grant RGP0051 (to P.H.V. and E.R.W.). The work at California Institute of Technology was supported by National Institutes of Health Grant P01 GM066521 (to G.J.J.) and a gift to Caltech from the Gordon and Betty Moore Foundation. Parts of this work were carried out in the Institute of Technology Characterization Facility, University of Minnesota, which receives partial support from National Science Foundation through the Materials Research Science & Engineering Centers (MRSEC) program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012388108/-/DCSupplemental.
References
- 1.Schmidt JM, Stanier RY. Isolation and characterization of bacteriophages active against stalked bacteria. J Gen Microbiol. 1965;39:95–107. doi: 10.1099/00221287-39-1-95. [DOI] [PubMed] [Google Scholar]
- 2.Schade SZ, Adler J, Ris H. How bacteriophage chi attacks motile bacteria. J Virol. 1967;1:599–609. doi: 10.1128/jvi.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raimondo LM, Lundh NP, Martinez RJ. Primary adsorption site of phage PBS1: The flagellum of Bacillus subtilis. J Virol. 1968;2:256–264. doi: 10.1128/jvi.2.3.256-264.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovett PS. PBPI: A flagella specific bacteriophage mediating transduction in Bacillus pumilus. Virology. 1972;47:743–752. doi: 10.1016/0042-6822(72)90564-8. [DOI] [PubMed] [Google Scholar]
- 5.Edgar R, et al. Bacteriophage infection is targeted to cellular poles. Mol Microbiol. 2008;68:1107–1116. doi: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagenaur C, Farmer S, Agabian N. Adsorption properties of stage-specific Caulobacter phage phiCbK. Virology. 1977;77:401–407. doi: 10.1016/0042-6822(77)90436-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhilenkov EL, et al. The ability of flagellum-specific Proteus vulgaris bacteriophage PV22 to interact with Campylobacter jejuni flagella in culture. Virol J. 2006;3:50. doi: 10.1186/1743-422X-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pate JL, Petzold SJ, Umbreit TH. Two flagellotropic phages and one pilus-specific phage active against Asticcacaulis biprosthecum. Virology. 1979;94:24–37. doi: 10.1016/0042-6822(79)90435-5. [DOI] [PubMed] [Google Scholar]
- 9.Bendis I, Shapiro L. Properties of Caulobacter ribonucleic acid bacteriophage phi Cb5. J Virol. 1970;6:847–854. doi: 10.1128/jvi.6.6.847-854.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agabian-Keshishian N, Shapiro L. Stalked bacteria: Properties of deoxriybonucleic acid bacteriophage phiCbK. J Virol. 1970;5:795–800. doi: 10.1128/jvi.5.6.795-800.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt JM. Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J Gen Microbiol. 1966;45:347–353. doi: 10.1099/00221287-45-2-347. [DOI] [PubMed] [Google Scholar]
- 12.Ackermann HW. Frequency of morphological phage descriptions in the year 2000. Brief review. Arch Virol. 2001;146:843–857. doi: 10.1007/s007050170120. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda A, Iba H, Okada Y. Stalkless mutants of Caulobacter crescentus. J Bacteriol. 1977;131:280–287. doi: 10.1128/jb.131.1.280-287.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagenaur C, Agabian N. Caulobacter crescentus pili: Structure and stage-specific expression. J Bacteriol. 1977;131:340–346. doi: 10.1128/jb.131.1.340-346.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bender RA, Refson CM, O'Neill EA. Role of the flagellum in cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J Bacteriol. 1989;171:1035–1040. doi: 10.1128/jb.171.2.1035-1040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer JM, Newton A. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J Bacteriol. 1989;171:392–401. doi: 10.1128/jb.171.1.392-401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda A, Okada Y. Effect of macromolecular synthesis on the coordinate morphogenesis of polar surface structures in Caulobacter crescentus. J Bacteriol. 1977;130:1199–1205. doi: 10.1128/jb.130.3.1199-1205.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda A, et al. Regulation of polar morphogenesis in Caulobacter crescentus. J Bacteriol. 1981;145:559–572. doi: 10.1128/jb.145.1.559-572.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 20.Lake JA, Leonard KR. Bacteriophage structure: Determination of head-tail symmetry mismatch for Caulobacter crescentus phage phiCbK. Science. 1974;183:744–747. doi: 10.1126/science.183.4126.744. [DOI] [PubMed] [Google Scholar]
- 21.Leonard KR, Kleinschmidt AK, Agabian-Keshishian N, Shapiro L, Maizel JV., Jr Structural studies on the capsid of Caulobacter crescentus bacteriophage phiCbK. J Mol Biol. 1972;71:201–216. doi: 10.1016/0022-2836(72)90346-4. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulos S, Smith PR. The structure of the tail of the bacteriophage phi CbK. J Ultrastruct Res. 1982;80:62–70. doi: 10.1016/s0022-5320(82)80032-4. [DOI] [PubMed] [Google Scholar]
- 23.Leonard KR, Kleinschmidt AK, Lake JA. Caulobacter crescentus bacteriophage phiCbK: Structure and in vitro self-assembly of the tail. J Mol Biol. 1973;81:349–365. doi: 10.1016/0022-2836(73)90146-0. [DOI] [PubMed] [Google Scholar]
- 24.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 25.Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briegel A, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruggnaller S, Mayr M, Frangakis AS. A visualization and segmentation toolbox for electron microscopy. J Struct Biol. 2008;164:161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Poindexter JS, Hornack PR, Armstrong PA. Intracellular development of a large DNA bacteriophage lytic for Caulobacter crescentus. Arch Mikrobiol. 1967;59:237–246. doi: 10.1007/BF00406337. [DOI] [PubMed] [Google Scholar]
- 31.Ely B, et al. General nonchemotactic mutants of Caulobacter crescentus. Genetics. 1986;114:717–730. doi: 10.1093/genetics/114.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks ME, et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol. 2010;192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 36.Samuel AD, et al. Flagellar determinants of bacterial sensitivity to chi-phage. Proc Natl Acad Sci USA. 1999;96:9863–9866. doi: 10.1073/pnas.96.17.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pope WH, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: A “horned” bacteriophage of marine synechococcus. J Mol Biol. 2007;368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jollick JD, Wright BL. A flagella specific bacteriophage for caulobacter. J Gen Virol. 1974;22:197–205. doi: 10.1099/0022-1317-22-2-197. [DOI] [PubMed] [Google Scholar]
- 39.Brinton CC., Jr The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 40.Scholl DR, Jollick JD. Pilus-dependent, double-stranded DNA bacteriophage for Caulobacter. J Virol. 1980;35:949–954. doi: 10.1128/jvi.35.3.949-954.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley DE, Pitt TL. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J Gen Virol. 1974;24:1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- 42.Chibeu A, et al. The adsorption of Pseudomonas aeruginosa bacteriophage phiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol Lett. 2009;296:210–218. doi: 10.1111/j.1574-6968.2009.01640.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kropinski AM, et al. Enumeration of bacteriophages by double agar overlay plaque assay. In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols. Vol. 1. New York: Humana; 2009. p. 307. [DOI] [PubMed] [Google Scholar]
- 46.Zheng SQ, et al. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.