Abstract
Glycosaminoglycan polysaccharides play critical roles in many cellular processes, ranging from viral invasion and angiogenesis to spinal cord injury. Their diverse biological activities are derived from an ability to regulate a remarkable number of proteins. However, few methods exist for the rapid identification of glycosaminoglycan–protein interactions and for studying the potential of glycosaminoglycans to assemble multimeric protein complexes. Here, we report a multidisciplinary approach that combines new carbohydrate microarray and computational modeling methodologies to elucidate glycosaminoglycan–protein interactions. The approach was validated through the study of known protein partners for heparan and chondroitin sulfate, including fibroblast growth factor 2 (FGF2) and its receptor FGFR1, the malarial protein VAR2CSA, and tumor necrosis factor-α (TNF-α). We also applied the approach to identify previously undescribed interactions between a specific sulfated epitope on chondroitin sulfate, CS-E, and the neurotrophins, a critical family of growth factors involved in the development, maintenance, and survival of the vertebrate nervous system. Our studies show for the first time that CS is capable of assembling multimeric signaling complexes and modulating neurotrophin signaling pathways. In addition, we identify a contiguous CS-E-binding site by computational modeling that suggests a potential mechanism to explain how CS may promote neurotrophin-tyrosine receptor kinase (Trk) complex formation and neurotrophin signaling. Together, our combined microarray and computational modeling methodologies provide a general, facile means to identify new glycosaminoglycan–protein–protein interactions, as well as a molecular-level understanding of those complexes.
Glycosaminoglycans (GAGs) regulate a wide range of physiological processes, including viral invasion, blood coagulation, cell growth, and spinal cord injury (1–4). Assembled from repeating disaccharide units, GAGs display diverse patterns of sulfation (SI Appendix, Fig. S1). These sulfation patterns are believed to have important functional consequences, enabling the polysaccharides to interact with a wide variety of proteins (1, 2). However, the precise sulfation motifs involved in protein recognition are understood in only a few cases (1, 4, 5). Moreover, studies of heparan sulfate (HS) interactions with fibroblast growth factors suggest that GAGs can assist in the assembly of multimeric protein complexes, thereby modulating signal transduction pathways (6–10). Yet, only a few such examples have been elucidated, and the extent to which other GAGs such as chondroitin sulfate (CS) engage in the formation of multimeric protein complexes remains unknown. Elucidating the interactions of specific GAG substructures with proteins and large protein–protein complexes will be critical for understanding the structure-activity relationships of GAGs and the mechanisms underlying important biological processes.
Several methods have been developed to study GAG–protein interactions, including affinity chromatography, analytical ultracentrifugation, electrophoretic mobility shift assays, competition experiments, mass spectrometry-based approaches, isothermal titration calorimetry, and surface plasmon resonance (9–16). Although powerful, these approaches are low throughput, often labor intensive, and require significant quantities of carbohydrate and/or protein. Notably, no methods are available to rapidly screen various GAGs for their ability to assemble multimeric protein complexes. In addition, existing methods often require oligosaccharides or polysaccharides that are relatively homogeneous in chain length and charge density, such as fractionated heparin or chemically modified HS (11, 12, 14). As such, it has been difficult to study the interactions of proteins with other GAG classes and physiologically relevant GAG preparations, which are more heterogeneous and structurally diverse.
Similarly, structural studies of GAG–protein interactions have been limited by the complexity and heterogeneity of naturally occurring GAGs. For example, the majority of crystal structures contain fully sulfated heparin oligosaccharides instead of the physiological HS ligands of lower charge density (17). As an alternative strategy, recent advances in molecular modeling have provided several approaches for understanding the interaction of heparin or HS with proteins, including IL-8, TSG-6, and PECAM-1 (18–20). However, GAG–protein interactions pose a unique set of challenges for computational modeling, such as highly flexible sugar ligands with many rotatable bonds, interaction energies dominated by electrostatics, and shallow, solvent-accessible binding pockets. Importantly, computational approaches have not yet been applied to other GAGs, such as the less highly charged CS class, nor have they been developed to investigate large GAG–protein–protein complexes.
Here, we describe an integrated approach that combines carbohydrate microarray methodologies with computational modeling to provide unique insights into GAG interactions with proteins and multimeric protein complexes. We demonstrate that carbohydrate microarrays can be used to rapidly screen proteins and protein–protein complexes for binding to specific sulfation motifs and GAG classes. Such information can then be used in conjunction with previously undescribed computational modeling approaches to predict GAG-binding sites within proteins and to determine the potential for GAGs to assemble multimeric protein complexes. Using this combined approach, we identify a specific interaction between the sulfated chondroitin sulfate-E (CS-E, SI Appendix, Fig. S1) epitope and the neurotrophin (NT) family of growth factors and receptors. Our computational modeling results suggest a contiguous CS-E-binding site that spans the NT–tyrosine receptor kinase (Trk) complex, providing a potential mechanism to explain how CS modulates complex formation and NT signaling pathways. Together with cellular data, we provide evidence that CS plays an active role in cellular signaling by regulating the interactions between growth factors and their receptors.
Results and Discussion
General Microarray Approach.
The binding of proteins to GAGs was examined using carbohydrate microarrays containing either synthetic tetrasaccharides of defined sulfation sequence (21, 22) or naturally occurring polysaccharides representing various GAG classes (23). Microarrays of synthetic tetrasaccharides displaying the CS-A, CS-C, or CS-E sulfation motifs (SI Appendix, Fig. S1) were robotically printed on aldehyde-coated glass surfaces at varying concentrations (1–300 μM). Polysaccharide microarrays were printed on poly-dl-lysine-coated glass surfaces and contained varying concentrations (0.25–25 μM) of chondroitin sulfate enriched in the CS-A, CS-C, CS-D, CS-E motifs, dermatan sulfate (CS-B), hyaluronic acid (HA), heparin (Hep), heparan sulfate (HS), or keratan sulfate (KS). In all cases, the microarrays were incubated with the protein or protein–protein complex of interest, and binding was detected using primary antibodies against the protein(s) followed by secondary Cy3- and/or Cy5-labeled antibodies. Notably, this miniature array format permitted the rapid detection of multiple binding events simultaneously and required minimal amounts (1–100 μg) of carbohydrate and protein. As described below, the arrays allowed for comparisons of the binding of large families or functional classes of proteins to various GAG subtypes to provide a more comprehensive understanding of the specificity of proteins for different GAGs classes and sulfation sequences. We also applied the microarray technology toward the discovery of previously undescribed GAG–protein interactions and toward an understanding of the assembly of multimeric protein complexes.
General Computational Approach.
Once GAG–protein interactions were identified using carbohydrate microarrays, we predicted the GAG binding sites on proteins using computational methods. First, rigid-body docking of one oligosaccharide conformation to the entire molecular surface of the protein was performed to locate the most favorable binding sites (24). Next, rigid-body docking of the oligosaccharide conformation into those binding sites was continued until a diverse set of ligand orientations with respect to the protein was obtained (25). The protein side chains in the binding sites were then optimized (26), and the energy of the entire oligosaccharide–protein complex was minimized. Residues within 5 Å of the oligosaccharide in more than one of the five minimum energy structures were considered part of the GAG binding site.
Approach Validation.
VAR2CSA.
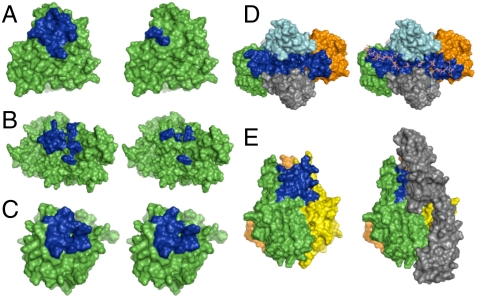
To test the computational methods, we first examined the protein VAR2CSA, a CS-A-binding protein involved in placental malaria pathogenesis (27). CS-A binds to the DBL3x and DBL6 (Duffy binding-like 3x and 6) domains of VAR2CSA with micromolar affinity, and basic residues important for the interaction have been identified by site-directed mutagenesis (28). In addition, 1.8- and 3.0-Å crystal structures of the DBL3x and DBL6 domains of VAR2CSA, respectively, have been solved (28, 29). We predicted the lowest energy conformation of a CS-A tetrasaccharide by performing molecular dynamics simulations in explicit water (22) and used ScanBindSite and GenMSCDock to determine the CS-A binding site on DBL3x or DBL6. CS-A was found to interact with both of the lysine residues predicted by mutagenesis to comprise the primary CS-A binding site on DBL6 (Fig. 1A and SI Appendix, Table S1). Moreover, five of the seven residues determined to be important for CS-A binding to DBL3x were successfully identified, further validating the computational approach (Fig. 1B and SI Appendix, Table S2). The side chain of Lys1515, one of the residues not identified by computational modeling, is buried in the crystal structure and makes electrostatic contacts with two internal Glu residues (Glu1464 and Glu1518), suggesting that this residue may not engage directly in interactions with CS-A. Together, these results show that CS binding sites on proteins can be correctly identified using our computational approach. Furthermore, we identify additional residues within DBL6 and DBL3x that may participate in close van der Waals and other interactions with the sugar.
Fig. 1.
(A and B) CS-A binding site (blue) on the DBL6 (A) and DBL3x (B) domains of VAR2CSA, as predicted by computational modeling (Left) and mutagenesis (Right). (C) Hep binding site (blue) on FGF2 as predicted computationally (Left) and determined crystallographically (Right). (D) Hep binding site (blue) in the FGF2-FGFR1 complex as predicted computationally (Left) and determined crystallographically (Right). The two FGF2 subunits are shown in green and orange, and the two FGFR1 subunits are shown in light blue and gray. (E) Predicted CS-E binding site (blue) in the trimeric structure of TNF-α (Left). Homology model of the TNF-α-TNFR1 complex (Right) shows that the CS-E binding site overlaps with the TNFR1 binding site. TNFR1 is depicted in gray.
Fibroblast growth factor 2 (FGF2) and its complex with FGF receptor 1 (FGFR1).
To further test our computational methods, we modeled the heparin binding site on FGF2 and extended our approach to the larger FGF2-FGFR1 complex. Using the solution structure of FGF2 and the crystal structure of a heparin tetrasaccharide, we identified all of the charged residues, as well as six of the seven other residues, located within 5 Å of heparin in the cocrystal structure of FGF2 complexed to a tetrasaccharide (Fig. 1C and SI Appendix, Table S3) (30). Two additional contacts were predicted using our computational approach, one of which (Lys26) is found in the cocrystal structure of FGF2 bound to a heparin hexaccharide (30).
Having correctly predicted the heparin binding site, we next tested whether our computational approach could be used to provide insight into the interaction of GAGs with large, multimeric protein complexes. Biochemical, structural, and cellular studies have established that heparin forms a ternary complex with FGF2 and the FGFR1 receptor and makes multiple contacts with both proteins (7, 31). Initial attempts to dock a heparin octasaccharide to the FGF2-FGFR1 complex identified only the heparin binding site on FGFR1. To overcome this problem, we determined each of the heparin binding sites on FGF2 and FGFR1 individually and superimposed those binding sites onto the structure of the FGF2-FGFR1 complex. The majority of the residues found in the sugar binding site of the heparin-FGF2-FGFR1 crystal structure were identified (Fig. 1D and SI Appendix, Table S4). Most importantly, we observed a contiguous binding site that spanned the FGF2-FGFR1 complex, consistent with the crystal structure. Thus, our computational methods can be used to predict GAG binding sites and to provide insights into the potential for GAGs to assemble multimeric protein complexes.
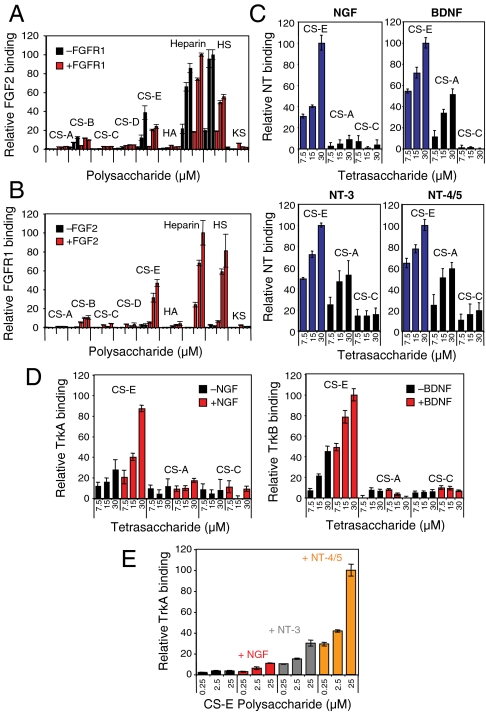
The ability of HS to mediate the formation of protein complexes is critical for its biological functions, enabling it to regulate growth factor, chemokine, and other signal transduction pathways (8, 32). As experimental methods for studying carbohydrate-mediated protein–protein interactions require considerable material and are low throughput, we sought to expand carbohydrate microarray methodologies to rapidly screen for carbohydrate–protein–protein complexes. We chose the well-established heparin-FGF2-FGFR1 interaction as our first test case. FGF2, FGFR1-Fc fusion protein, or a 1∶1 mixture of FGF2:FGFR1-Fc was incubated with the polysaccharide microarrays, and after treatment with a primary antibody against FGF2, growth factor or receptor binding was detected using orthogonal secondary antibodies conjugated to Cy3 or Cy5 dyes. We found that FGF2 bound strongly to heparin and HS polysaccharides in the absence of FGFR1 (Fig. 2A), whereas FGFR1 alone showed minimal binding to the array (Fig. 2B). Notably, FGFR1 binding increased significantly in the presence of FGF2, suggesting that binding of the growth factor to heparin or HS enhances binding of the receptor. Moreover, colocalization of both proteins was detected on the arrays (SI Appendix, Fig. S2), indicating the formation of carbohydrate–protein–protein complexes. Complex formation was observed with heparin, HS, and, to a lesser extent, with CS-E-enriched polysaccharides, consistent with the demonstrated selectivity of FGFs for these GAG subclasses (12, 23).
Fig. 2.
(A) Relative binding of FGF2 to the indicated glycosaminoglycan (0.5-, 5-, and 10-μM concentration) on polysaccharide microarrays in the presence (red) or absence (black) of FGFR1. (B) Relative binding of FGFR1 to the indicated glycosaminoglycan (0.5-, 5-, and 10-μM concentration) on polysaccharide microarrays in the presence (red) or absence (black) of FGF2. (C) Relative binding of NGF, BDNF, NT-3, and NT-4/5 to the indicated CS tetrasaccharide on carbohydrate microarrays. (D) Relative binding of the Trk receptors to CS tetrasaccharides on carbohydrate microarrays in the presence (red) or absence (black) of the indicated NTs. (E) Comparison of the relative binding of TrkA to CS-E-enriched polysaccharides in the presence of NGF, NT-3, or NT-4/5. Binding relative to the maximum signal for each plot is shown in A–C and E; binding relative to the maximum signaling for the series of two plots is shown in D. Each protein was analyzed in triplicate, and the data represent an average of 8–10 spots for a given carbohydrate concentration.
Tumor necrosis factor-α (TNF-α) and its complex with TNF receptor 1 (TNFR1).
As final validation of our computational and carbohydrate microarray approaches, we examined the interaction of CS with TNF-α, a proinflammatory cytokine involved in autoimmune diseases such as rheumatoid arthritis, Crohn’s disease, and psoriasis (33, 34). Previous studies in our laboratory have demonstrated that a tetrasaccharide displaying the CS-E sulfation motif binds to TNF-α and antagonizes its interaction with TNFR1, thereby inhibiting TNF-α-induced cell death (21). To test our microarray approach, we incubated TNF-α, TNFR1-Fc, or a 1∶1 mixture of TNF-α:TNFR1-Fc with CS tetrasaccharide microarrays. Both TNF-α and TNFR1-Fc selectively bound to CS-E tetrasaccharides when incubated alone with the microarrays (SI Appendix, Fig. S3). However, binding of these proteins to CS-E was abolished when the proteins were incubated together. These data indicate that once formed, the TNF-α-TNFR1 complex cannot bind to CS-E tetrasaccharides on the array. Thus, CS-E, TNF-α, and TNFR1 do not appear to form a ternary complex, consistent with previous studies (21) and further validating the use of microarrays to rapidly probe the interactions of GAGs with multimeric protein complexes.
We next applied our computational approach to gain insight into the CS-E binding site on TNF-α. We found that CS-E binds predominantly to two loops between antiparallel β-strands c–d of monomer A and β-strands e–f of monomer B in the TNF-α trimer structure (Fig. 1E and SI Appendix, Table S5). As the structure displays 3-fold symmetry, CS-E binding sites are also predicted between monomers B and C, and C and A. Additionally, in support of our model, we found that the average binding energies of CS-A and CS-C at this site were significantly worse than the average binding energy of CS-E (SI Appendix, Table S6). We constructed a homology model of the TNF-α-TNFR1 complex based on the known crystal structure of the TNF-β-TNFR1 complex). Notably, the CS-E binding site on TNF-α overlaps with that of TNFR1, as determined by site-directed mutagenesis (35) and homology modeling. These findings are consistent with the carbohydrate microarray results above and with previous ELISA and cellular studies (21), further validating the computational methods. Collectively, we have shown that the integration of computational modeling and microarray approaches can be used to gain important insights into GAG–protein interactions and to rapidly establish whether specific GAG subclasses or sulfation motifs interact with multimeric protein complexes.
Identification of New Glycosaminoglycan–Protein Interactions: The NTs and Their Receptors.
The NTs have critical functions in many aspects of neuronal development, including neurite outgrowth, cell survival, differentiation, and proliferation (36, 37). They also play important roles in synaptic plasticity and maintenance of the adult nervous system (36–38) and have been implicated in neurodegenerative diseases (36). Previously, we found that a tetrasaccharide containing a specific sulfation motif, CS-E, stimulates the outgrowth of developing hippocampal neurons (22, 39). Our studies implicated brain-derived neurotrophic factor (BDNF) as one of the proteins responsible for mediating the effects of CS-E (22). In addition to BDNF, the NT family includes nerve growth factor (NGF), neurotrophin-3 (NT-3), and neurotrophin-4/5 (NT-4/5), which share approximately 50% sequence homology to BDNF and strikingly similar structures, with rmsd of less than 2 Å between any two NTs. We used our microarray approach to rapidly compare the binding specificity across this protein family. Notably, all of the NTs showed concentration-dependent binding to CS-E tetrasaccharides, with NGF displaying the greatest specificity (Fig. 2C). However, the ability of BDNF and other NTs to bind weakly to other sulfation motifs was unexpected, given that only the CS-E motif stimulated neurite outgrowth (22, 39). Thus, we postulated that CS-E might interact with additional proteins, possibly forming protein–protein complexes between NTs and their receptors and that the formation of such complexes might impart greater selectivity for the CS-E motif.
To test this hypothesis, we examined the binding of various NT-receptor pairs to CS tetrasaccharide and polysaccharide microarrays. The NTs activate signal transduction pathways by binding to the Trk receptors A, B, and C (36, 37, 40, 41). In particular, TrkA binds to NGF, TrkB binds to BDNF and NT-4/5, and TrkC binds to NT-3. However, “cross-talk” among the NT family has also been observed, whereby certain NTs bind to additional Trk receptors with lower affinity (e.g., NT-3 and NT-4/5 to TrkA) (36, 42). This cross-talk raises the interesting question of how specific NT signaling pathways are differentially activated in vivo.
We first probed the ability of CS-E to assemble NT-Trk complexes, starting with the primary-binding partners NGF-TrkA, BDNF-TrkB, NT-3-TrkC, and NT-4/5-TrkB. In the absence of NT, TrkA and TrkB bound weakly to the CS-E tetrasaccharide, whereas TrkC showed no apparent binding to the microarray (Fig. 2D and SI Appendix, Fig. S4). Notably, the presence of NGF and BDNF significantly enhanced the binding of TrkA and TrkB, respectively, to CS-E, and colocalization of the proteins was observed on the array, suggesting the formation of CS-E-NGF-TrkA and CS-E-BDNF-TrkB complexes. Complex assembly was highly selective for the CS-E sulfation motif and did not occur in the presence of CS-A or CS-C tetrasaccharides. Similar results were obtained using polysaccharide microarrays (SI Appendix, Fig. S5). Thus, the complex of NT and Trk showed greater selectivity for CS-E than each NT alone, reinforcing the notion that formation of GAG–protein–protein complexes can impart greater selectivity for specific sulfation motifs. Interestingly, NT-3 and NT-4/5 did not increase the binding of TrkC and TrkB to CS-E tetra- or polysaccharides, respectively (SI Appendix, Figs. S4 and S5), suggesting that CS-E forms complexes only with certain NT-receptor pairs and raising the possibility that the spatiotemporal expression of CS-E in vivo may differentially regulate specific NT signaling pathways.
We next investigated the secondary cross-talk between the NTs and their receptors. Specifically, TrkA binding to the arrays was evaluated in the presence or absence of NGF, NT-3, or NT-4/5. Selective, but weak, binding of TrkA to CS-E-enriched polysaccharides was observed in the absence of NT (Fig. 2E and SI Appendix, Fig. S5). Addition of NGF, NT-3, or NT-4/5 significantly increased the binding of TrkA to the array (Fig. 2E), suggesting that CS-E is capable of forming complexes with NGF-TrkA, NT-3-TrkA, and NT-4/5-TrkA. Interestingly, TrkA binding to CS-E was enhanced the most by the presence of NT-4/5, followed by NT-3. In the absence of CS-E, however, TrkA showed the greatest binding affinity for NGF, followed by NT-3, and NT-4/5, as measured by ELISA (SI Appendix, Fig. S6) and consistent with previous reports (40). Thus, the ability of CS-E to enhance receptor binding was inversely related to the relative affinity of the NT-TrkA interaction. These results suggest that CS-E may help to stabilize weaker NT-receptor interactions and may enable activation of secondary NT signaling pathways.
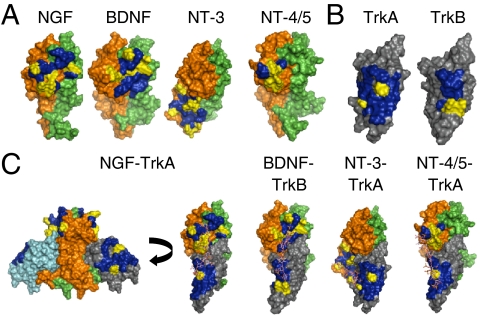
To gain molecular-level insights, we computationally modeled the CS-E binding sites in the NT and NT-receptor complexes. The CS-E tetrasaccharide structure (22) was first docked to known crystal structures of human NGF, NT-3, or NT-4/5 dimers. For the BDNF dimer structure, we built a homology model by replacing NT-3 with BDNF in the BDNF-NT-3 dimer structure. We found that the predicted CS-E binding sites share several common features (Fig. 3A and SI Appendix, Figs. S7 and S8 and Table S7). First, each site contains a high density of basic amino acids, ranging from four in the case of the NT-4/5 dimer to seven in the case of the NT-3 dimer. Although these basic residues are highly conserved across many species for a given NT, they are not entirely conserved among different NT family members (SI Appendix, Fig. S7), which may explain in part the observed differences in selectivity and affinity for CS-E. Second, each site contains lysine and arginine residues separated by distances that would allow them to interact with multiple sulfate groups on the CS-E tetrasaccharide. For example, the average distance between the sulfur atoms of the sulfate groups in the same disaccharide of CS-E is 5.5 Å and in adjacent disaccharides is 12.9 Å. In the CS-E binding site on NGF, the ϵ-amino groups of Lys32 and Lys34 are 5.6 Å apart and an average distance of 12.7 Å away from the ϵ-amino group of Lys95. Notably, we found that in each case, the average binding energy of CS-E was much lower than that of either CS-A or CS-C (SI Appendix, Table S6), although we could not resolve the relative binding energies of CS-A and CS-C due to their weaker binding interactions with the NT.
Fig. 3.
(A) Predicted CS-E binding site [blue (nonbasic residues) and yellow (basic residues)] in the BDNF, NGF, NT-4/5, and NT-3 dimer. (B) Predicted CS-E binding site [blue (nonbasic residues) and yellow (basic residues)] in the NT binding domain (domain 5) of TrkA and TrkB. (C) Predicted CS-E binding sites [blue (nonbasic residues) and yellow (basic residues)] in the BDNF-TrkB, NGF-TrkA, NT-4/5-TrkA, and NT-3-TrkA complexes. The CS octasaccharides were manually docked.
We also observed some differences in the CS-E binding sites among NT family members. Overall, CS-E bound to the BDNF, NGF, and NT-4/5 dimers in a similar manner, interacting with residues within loop 1, loop 4, and β-strand 8 of monomer A (SI Appendix, Fig. S7). However, CS-E also made contacts with residues in loop 2 and β-strands 5 and 6 of monomer B in BDNF and NGF. The predicted CS-E binding site on the NT-3 dimer was the most distinct. Many of the basic residues found in the binding sites of BDNF, NGF, and NT-4/5 were absent in NT-3 and vice versa, and CS-E interacted primarily with residues in loop 3 and β-strands 5 and 6 of monomer A (corresponding to β-strands 7 and 8 of the other NTs). Although residues in loop 3 were not well resolved in the human NGF and NT-4/5 crystal structures used for our modeling studies, we confirmed that the CS-E interaction with loop 3 was unique to NT-3 by modeling the CS-E binding site in the mouse NGF crystal structure and a different human NT-4/5 structure, both of which contain a highly resolved structure for loop 3. In both cases, the CS-E binding site was unchanged by the presence of loop 3, suggesting a distinct mode of binding to NT-3.
We next modeled the CS-E binding sites in the Trk receptors by docking the CS-E tetrasaccharide structure to known crystal structures of the ligand binding domains of TrkA and TrkB. In contrast to the CS-E binding sites on the NTs, the binding sites on TrkA and TrkB comprised primarily β-strands (specifically β-strand C, F, and G) rather than loops, and they contained only two basic residues (Fig. 3B and SI Appendix, Fig. S8 and Table S7). The presence of fewer basic residues in the binding site may account for the weaker binding affinity of CS-E for the Trks compared to the NTs. Importantly, the CS-E binding sites on TrkA and TrkB showed no overlap with the NT interaction surface, suggesting that the sugar binds to a distinct site on the receptor. Indeed, superimposing the CS-E binding sites for each protein onto the NGF-TrkA crystal structure or the BDNF-TrkB, NT-3-TrkA, and NT-4/5-TrkA homology models revealed a contiguous sugar binding site that spanned a single face of the complex (Fig. 3C and SI Appendix, Fig. S9). As the structures of the NT-Trk complex have C2 symmetry, a second CS-E binding site is predicted that would enable formation of a 2∶2∶2 complex. Each sugar binding site readily accommodates a single octasaccharide, suggesting a molecular mechanism by which CS polysaccharides might assist in the assembly of NT-Trk receptor complexes and promote NT signaling.
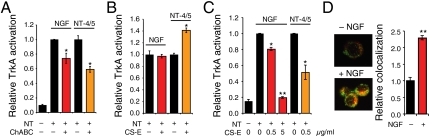
As independent confirmation of our microarray and computational results, we performed several cellular studies. Pheochromocytoma 12 (PC12) cells express high levels of TrkA and have been used extensively to study NGF-TrkA signaling pathways (43, 44). We first examined whether the CS-E motif was expressed on PC12 cells using a CS-E-specific monoclonal antibody developed by our laboratory (22, 39). We observed strong CS-E-positive staining on the cell surface, which could be removed using chondroitinase ABC (ChABC), an enzyme that hydrolyzes CS chains (SI Appendix, Fig. S10). Notably, removal of endogenous CS-E polysaccharides on PC12 cells significantly attenuated TrkA activation by NGF or NT-4/5 by 24 ± 7% and 37 ± 3%, respectively, as measured using a phospho-TrkA antibody (Fig. 4A). The greater effect of CS on NT-4/5-induced activation of TrkA compared to NGF is consistent with our microarray data (Fig. 2E) indicating that CS-E enhances the NT-4/5-TrkA interaction more than that of NGF-TrkA. These results further support the notion that CS-E promotes the formation of specific NT-Trk complexes and the activation of NT signaling pathways.
Fig. 4.
CS-E modulates NGF- or NT-4/5-mediated TrkA activation in cells. (A) Removal of endogenous CS-E on PC12 cells using ChABC reduced NGF- and NT-4/5-mediated TrkA phosphorylation. (B) CS-E-enriched polysaccharides enhanced TrkA phosphorylation by NT-4/5, but not NGF, when coated on a substratum at 500 ng·mL-1. (C) Addition of exogenous CS-E (500 ng·mL-1) to the media reduced NGF-induced and NT-4/5-induced TrkA phosphorylation. NGF-induced activation was further inhibited by higher concentrations (5,000 ng·mL-1) of CS-E. For A–C, relative TrkA activation was calculated with respect to total TrkA levels for each condition and was plotted relative to that of untreated cells in the presence of the indicated NT. n = 4, *P < 0.05, **P < 0.005. (D) Prolonged NGF treatment (60 min), increases the colocalization of CS-E and TrkA. Representative images show minimal colocalization in untreated cells and increased colocalization (yellow) after treatment with NGF. The extent of colocalization was quantified as described in SI Methods and plotted relative to that of untreated cells. n = 24 cells. **P < 5 × 10-6.
Similarly, we found that CS-E-enriched polysaccharides adsorbed onto a substratum activated NT-4/5-mediated TrkA signaling by 42 ± 6%, but had no appreciable effect on NGF-mediated TrkA signaling at the CS-E concentration tested (Fig. 4B). Furthermore, the addition of exogenous CS-E-enriched polysaccharides to the medium interfered with NT signaling, reducing NGF- and NT-4/5-mediated TrkA activation by 19 ± 2% and 49 ± 11%, respectively (Fig. 4C). A greater reduction (81 ± 1%) in NGF-induced TrkA activation was achieved by using a tenfold higher concentration of polysaccharide, indicating that CS-E can modulate NGF-TrkA interactions, albeit less effectively compared to NT-4/5-TrkA interactions. These findings are consistent with previous studies that have shown stimulatory effects on neurite outgrowth for adsorbed CS (22, 39, 45) and inhibitory effects for CS in solution (46–48). Our data support the model that CS-E polysaccharides on cell-surface proteoglycans or coated on a substratum recruit NTs to the cell surface, thereby promoting complex formation and stimulating NT signaling pathways. By adding exogenous CS-E in solution, the polysaccharide acts as a competitive inhibitor, sequestering NTs away from the cell surface and thereby disrupting NT-mediated signaling (47). Finally, we found that prolonged exposure of PC12 cells to NGF increased the colocalization of TrkA and CS-E by 2.3 ± 0.1-fold (Fig. 4D). As TrkA is known to form signaling endosomes in PC12 cells after prolonged exposure to NGF (49), these findings lend further support to the notion that CS-E is a key component of the NGF-TrkA signaling complex.
Collectively, our microarray, computational and cellular studies demonstrate that NT-Trk interactions and signaling pathways are modulated by CS-E polysaccharides. Furthermore, we suggest that NT-4/5-TrkA pathways should be more sensitive than NGF-TrkA pathways to CS-E levels. More broadly, these results provide evidence that CS GAGs regulate this important family of growth factors and function in the assembly of multimeric signaling complexes.
Conclusion
We have developed carbohydrate microarray and computational modeling approaches for the rapid screening and understanding of GAG interactions with proteins and multimeric protein complexes. Using these methods, we identify previously undescribed interactions between a specific sulfated epitope, CS-E, and the neurotrophin family of growth factors. Moreover, we show that CS is capable of assembling multimeric signaling complexes and modulating interactions between specific NTs and their receptors. Our computational modeling studies identify potential CS binding sites on NTs and other proteins. We also discover a contiguous CS-E-binding site within the NT-Trk receptor complex, which suggests a potential mechanism for how CS promotes complex formation and modulates NT signaling. Taken together, we have developed a general method for studying GAG–protein–protein interactions that can be applied to screen various GAG subclasses (HS, DS, CS, etc.) and particular sulfation motifs (CS-A, CS-E, etc.) for the ability to assemble specific multimeric complexes. The approach permits the identification of potentially low affinity carbohydrate–protein–protein interactions that would be difficult to study using existing methods. When combined with the computational methods demonstrated herein, this strategy provides unique molecular-level insights into the diverse biological functions of GAGs.
Materials and Methods
Microarray analyses, computational methods, cellular assays, immunochemistry, and ELISA methods are described in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Jose Luis Riechmann, Dr. Igor Antoshechkin, and the Caltech Millard and Muriel Jacobs Genetics and Genomics Laboratory for assistance and instrumentation for the microarray studies. We also thank Dr. Nagarajan Vaidehi, Adam Griffith, and other members of the Goddard group for their assistance. This work was supported by National Institutes of Health Grants R01 GM093627 (to L.H.W.), Training Grant 5T32 GM07616 (to C.J.R.), Training Grant 5T32 GM07737 (to P.M.C.), R01 NS071112 and 1R01 NS073115 (to W.A.G.), and by a National Science Foundation Predoctoral Fellowship (to S.E.T.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102962108/-/DCSupplemental.
References
- 1.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Raman R, Sasisekharan V, Sasisekharan R. Structural insights into biological roles of protein-glycosaminoglycan interactions. Chem Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Sugahara K, et al. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr Opin Struct Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Gama CI, Hsieh-Wilson LC. Chemical approaches to deciphering the glycosaminoglycan code. Curr Opin Chem Biol. 2005;9:609–619. doi: 10.1016/j.cbpa.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Petitou M, van Boeckel CA. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew Chem Int Ed Engl. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 7.Schlessinger J, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 8.Spivak-Kroizman T, et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahimi OA, Zhang F, Hrstka SC, Mohammadi M, Linhardt RJ. Kinetic model for FGF, FGFR, and proteoglycan signal transduction complex assembly. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- 10.Pantoliano MW, et al. Multivalent ligand-receptor binding interactions in the fibroblast growth factor system produce a cooperative growth factor and heparin mechanism for receptor dimerization. Biochemistry. 1994;33:10229–10248. doi: 10.1021/bi00200a003. [DOI] [PubMed] [Google Scholar]
- 11.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: Appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 12.Wu ZL, et al. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J Biol Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 13.Martin L, et al. Structural and functional analysis of the RANTES-glycosaminoglycans interactions. Biochemistry. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
- 14.Sweeney MD, Yu Y, Leary JA. Effects of sulfate position on heparin octasaccharide binding to CCL2 examined by tandem mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1114–1119. doi: 10.1016/j.jasms.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Keiser N, Venkataraman G, Shriver Z, Sasisekharan R. Direct isolation and sequencing of specific protein-binding glycosaminoglycans. Nat Med. 2001;7:123–128. doi: 10.1038/83263. [DOI] [PubMed] [Google Scholar]
- 16.de Paz JL, Noti C, Bohm F, Werner S, Seeberger PH. Potentiation of fibroblast growth factor activity by synthetic heparin oligosaccharide glycodendrimers. Chem Biol. 2007;14:879–887. doi: 10.1016/j.chembiol.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Mulloy B, Linhardt RJ. Order out of complexity—Protein structures that interact with heparin. Curr Opin Struct Biol. 2001;11:623–628. doi: 10.1016/s0959-440x(00)00257-8. [DOI] [PubMed] [Google Scholar]
- 18.Bitomsky W, Wade RC. Docking of glycosaminoglycans to heparin-binding proteins: Validation for aFGF, bFGF, and antithrombin and application to IL-8. J Am Chem Soc. 1999;121:3004–3013. [Google Scholar]
- 19.Mahoney DJ, et al. Characterization of the interaction between tumor necrosis factor-stimulated gene-6 and heparin: Implications for the inhibition of plasmin in extracellular matrix microenvironments. J Biol Chem. 2005;280:27044–27055. doi: 10.1074/jbc.M502068200. [DOI] [PubMed] [Google Scholar]
- 20.Gandhi NS, Coombe DR, Mancera RL. Platelet endothelial cell adhesion molecule 1 (PECAM-1) and its interactions with glycosaminoglycans: 1. Molecular modeling studies. Biochemistry. 2008;47:4851–4862. doi: 10.1021/bi702455e. [DOI] [PubMed] [Google Scholar]
- 21.Tully SE, Rawat M, Hsieh-Wilson LC. Discovery of a TNF-alpha antagonist using chondroitin sulfate microarrays. J Am Chem Soc. 2006;128:7740–7741. doi: 10.1021/ja061906t. [DOI] [PubMed] [Google Scholar]
- 22.Gama CI, et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 23.Shipp EL, Hsieh-Wilson LC. Profiling the sulfation specificities of glycosaminoglycan interactions with growth factors and chemotactic proteins using microarrays. Chem Biol. 2007;14:195–208. doi: 10.1016/j.chembiol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Vaidehi N, et al. Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SK, Li Y, Park C, Abrol R, Goddard WA., 3rd Prediction of the three-dimensional structure for the rat urotensin II receptor, and comparison of the antagonist binding sites and binding selectivity between human and rat receptors from atomistic simulations. ChemMedChem. 2010;5:1594–1608. doi: 10.1002/cmdc.201000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kam VWT, Goddard WA., 3rd Flat-bottom strategy for improved accuracy in protein side-chain placements. J Chem Theory Comput. 2008;4:2160–2169. doi: 10.1021/ct800196k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamain B, et al. Identification of multiple chondroitin sulfate A (CSA)-binding domains in the var2CSA gene transcribed in CSA-binding parasites. J Infect Dis. 2005;191:1010–1013. doi: 10.1086/428137. [DOI] [PubMed] [Google Scholar]
- 28.Khunrae P, Philip JM, Bull DR, Higgins MK. Structural comparison of two CSPG-binding DBL domains from the VAR2CSA protein important in malaria during pregnancy. J Mol Biol. 2009;393:202–213. doi: 10.1016/j.jmb.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins MK. The structure of a chondroitin sulfate-binding domain important in placental malaria. J Biol Chem. 2008;283:21842–21846. doi: 10.1074/jbc.C800086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, et al. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor. Fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–8386. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proudfoot AE, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raza A. Anti-TNF therapies in rheumatoid arthritis, Crohn’s disease, sepsis, and myelodysplastic syndromes. Microsc Res Tech. 2000;50:229–235. doi: 10.1002/1097-0029(20000801)50:3<229::AID-JEMT6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 34.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 35.Mukai Y, et al. Structure-function relationship of tumor necrosis factor (TNF) and its receptor interaction based on 3D structural analysis of a fully active TNFR1-selective TNF mutant. J Mol Biol. 2009;385:1221–1229. doi: 10.1016/j.jmb.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 36.Chao MV. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 37.Huang EJ, Reichardt LF. Neurotrophins: Roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dechant G, Barde YA. The neurotrophin receptor p75(NTR): Novel functions and implications for diseases of the nervous system. Nat Neurosci. 2002;5:1131–1136. doi: 10.1038/nn1102-1131. [DOI] [PubMed] [Google Scholar]
- 39.Tully SE, et al. A chondroitin sulfate small molecule that stimulates neuronal growth. J Am Chem Soc. 2004;126:7736–7737. doi: 10.1021/ja0484045. [DOI] [PubMed] [Google Scholar]
- 40.Bothwell M. Functional interactions of neurotrophins and neurotrophin receptors. Annu Rev Neurosci. 1995;18:223–253. doi: 10.1146/annurev.ne.18.030195.001255. [DOI] [PubMed] [Google Scholar]
- 41.Roux PP, Barker PA. Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol. 2002;67:203–233. doi: 10.1016/s0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 42.Shelton DL, et al. Human trks: Molecular cloning, tissue distribution, and expression of extracellular domain immunoadhesins. J Neurosci. 1995;15:477–491. doi: 10.1523/JNEUROSCI.15-01-00477.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao MV. Neurotrophin receptors: A window into neuronal differentiation. Neuron. 1992;9:583–593. doi: 10.1016/0896-6273(92)90023-7. [DOI] [PubMed] [Google Scholar]
- 44.Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: Making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 45.Clement AM, Sugahara K, Faissner A. Chondroitin sulfate E promotes neurite outgrowth of rat embryonic day 18 hippocampal neurons. Neurosci Lett. 1999;269:125–128. doi: 10.1016/s0304-3940(99)00432-2. [DOI] [PubMed] [Google Scholar]
- 46.Rawat M, Gama CI, Matson JB, Hsieh-Wilson LC. Neuroactive chondroitin sulfate glycomimetics. J Am Chem Soc. 2008;130:2959–2961. doi: 10.1021/ja709993p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oohira A, Matsui F, Katoh-Semba R. Inhibitory effects of brain chondroitin sulfate proteoglycans on neurite outgrowth from PC12D cells. J Neurosci. 1991;11:822–827. doi: 10.1523/JNEUROSCI.11-03-00822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapp A, Brandl N, Volpi N, Huettinger M. Evaluation of chondroitin sulfate bioactivity in hippocampal neurones and the astrocyte cell line U373: Influence of position of sulfate groups and charge density. Basic Clin Pharmacol Toxicol. 2005;96:37–43. doi: 10.1111/j.1742-7843.2005.pto960106.x. [DOI] [PubMed] [Google Scholar]
- 49.Valdez G, et al. Trk-signaling endosomes are generated by Rac-dependent macroendocytosis. Proc Natl Acad Sci USA. 2007;104:12270–12275. doi: 10.1073/pnas.0702819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.