Abstract
The anaphase-promoting complex/cyclosome (APC/C) is a cell cycle-regulated E3 ubiquitin ligase that controls the degradation of substrate proteins at mitotic exit and throughout the G1 phase. We have identified an APC/C substrate and cell cycle-regulated protein, KIAA0101/PAF15. PAF15 protein levels peak in the G2/M phase of the cell cycle and drop rapidly at mitotic exit in an APC/C- and KEN-box–dependent fashion. PAF15 associates with proliferating cell nuclear antigen (PCNA), and depletion of PAF15 decreases the number of cells in S phase, suggesting a role for it in cell cycle regulation. Following irradiation, PAF15 colocalized with γH2AX foci at sites of DNA damage through its interaction with PCNA. Finally, PAF15 depletion led to an increase in homologous recombination-mediated DNA repair, and overexpression caused sensitivity to UV-induced DNA damage. We conclude that PAF15 is an APC/C-regulated protein involved in both cell cycle progression and the DNA damage response.
Keywords: DNA replication, proteolysis
Remodeling of the cellular proteome occurs throughout the cell cycle. This remodeling both drives and responds to the changing cell cycle state and is dictated largely by transcriptional alteration of gene expression and ubiquitin-mediated proteolysis. For example, entry into S phase requires transcription of numerous cell cycle genes, as well as ubiquitin-mediated destruction of cyclin-dependent kinase (CDK) inhibitors. Together, this creates the burst of CDK activity needed to enter S phase and provides the machinery required for its progression. Following S-phase entry, the proteins involved in licensing replication origins are degraded, limiting replication to once per cell cycle. Thus, in addition to transcriptional controls, ubiquitin-mediated proteolysis remodels the cellular proteome and provides directionality to the eukaryotic cell cycle.
The anaphase-promoting complex or cyclosome (APC/C) is a multisubunit E3 ubiquitin ligase that acts as a major regulator of proteome remodeling during the cell cycle, specifically controlling mitotic exit and G1 progression (1). Cdc20 and Cdh1/Fzr1 are the only two specificity factors that function with the APC/C during somatic cell growth and are conserved from yeast to humans (reviewed in 2). In mitosis, the metaphase-to-anaphase transition is driven by the activation of APC/C-Cdc20–dependent ubquitylation of cyclin B and securin/PTTG1. Following mitotic exit, APC/C-Cdh1 becomes active in G1 phase (2). APC/C-Cdh1 targets a number of proteins for destruction, including Cdc20, the mitotic cyclins, the replication inhibitor geminin, and various spindle assembly factors (3, 4). APC/C-Cdh1 recognizes substrates through a lysine–glutamic acid–asparagine tripeptide motif, termed the KEN box (5).
Once replication begins, DNA polymerases are tethered to DNA through their association with proliferating cell nuclear antigen (PCNA), a homo-trimeric complex that acts as a polymerase processivity factor (reviewed in 6). A growing number of proteins are associated with PCNA through one of two known motifs. The PCNA interacting motif, or PIP box, was first identified in the CDK inhibitor p21/CIP1 (7). More recently, the APIM motif has been identified as an alternative PCNA-binding sequence (8). In addition to its role in DNA replication, PCNA plays a role in both DNA repair and translesion synthesis across sites of damage. In response to DNA damage, PCNA is mono- and poly-ubiquitylated. These modifications provide a platform for the recruitment of repair factors as well as specialized translesion polymerases that can bypass bulky adducts in the DNA (reviewed in 9 and 10). A large, and growing, number of proteins that localize to sites of damage have been discovered, but only a small number are dependent on PCNA for that localization.
The ongoing identification of functional relationships between proteins, and the classification of their biological roles, is critical to achieving a deeper understanding of cellular biology. With respect to the DNA damage response and cell cycle control, this includes the identification of proteins that localize to breaks and those that are regulated transcriptionally and posttranslationally throughout the cell cycle. We show here that the PCNA-interacting protein KIAA0101/p15PAF/PAF15 (11–14) is a cell cycle-regulated phospho-protein targeted by APC-Cdh1 through a conserved KEN box motif and that PAF15 localizes to sites of DNA damage. We conclude that PAF15 plays a role in both DNA replication and the response to DNA damage.
Results
PAF15 Protein Levels Are Cell Cycle-Regulated.
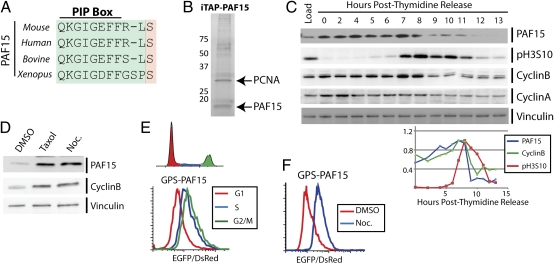
Using an ongoing screen to identify stability regulated proteins, we recovered the PCNA-associated protein PAF15. PAF15 contains a highly conserved PCNA-interacting motif (PIP box: Fig. 1A). To examine the function of PAF15, HA-tagged PAF15 was affinity-purified from HEK-293T cells (15). Tandem mass spectroscopy (MS/MS) identified copurifying proteins. Accordingly, we identified PCNA as having a strong interaction with PAF15, as has been reported by others (11–14). Silver staining of the immunoprecipitates together with MS/MS data suggested that PAF15 is stoichiometrically associated with PCNA in a 1:1 complex (Fig. 1B). Additional interacting proteins were also identified (Table S1). In addition, MS/MS of PAF15 purifications identified phosphorylation sites on PAF15 at serine 31 and 72 (Table S2). We had previously identified serine 72 phosphorylation in a MS/MS-based screen for mitotic phospho-proteins (16). Interestingly, serine 72 resides in an SP motif like that found in the CDK consensus, is highly conserved throughout evolution, and is located within the PAF15 PIP-box motif (Fig. 1A, orange shading).
Fig. 1.
PAF15 protein levels are regulated throughout the cell cycle. (A) PAF15 contains a PIP box that is highly conserved between vertebrate homologs. (B) HA-PAF15 was affinity-purified from G2 synchronized 293T cells and analyzed by silver stain and MS/MS. (C) HeLa cells were synchronized at the G1/S boundary using a double thymidine block and were collected for Western blot following release into the cell cycle. Vinculin was blotted for a loading control. Semiquantitative analysis of Western blots for cyclin B, PAF15, and phosho-serine 10 on histone 3 are graphed (Lower). Load refers to asynchronous cells. (D) HeLa cells were treated with either taxol or nocodazole to induce mitotic arrest or with DMSO alone. Lysates were blotted for PAF15, cyclin B, and for the loading control vinculin. (E) 293T cells expressing GPS-PAF15 were treated with the DNA dye Hoechst and analyzed by flow cytometry. Cells were gaited on the basis of DNA content (in G1, S, and G2/M phases). The EGFP/DsRed ratios for cells in G1, S, and G2/M phases are plotted as a histogram, showing an increase in PAF15 stability throughout the cell cycle. (F) 293T cells expressing GPS-PAF15 were arrested in mitosis using nocodazole. Histograms of asynchronous (DMSO-treated) and nocodazole-arrested cells are overlaid.
Given the tight interaction between PCNA and PAF15, the identification of mitotic phosphorylation on a conserved PAF15 CDK consensus site, and our discovery that PAF15 is ubquitylated, we tested whether PAF15 protein levels might be regulated throughout the cell cycle. To address this, we synchronized cells at the G1–S boundary using a double thymidine block. Cells were collected for Western blot analysis following release. Cyclin B steadily accumulates until ∼9 h post release (Fig. 1C). Similarly, PAF15 levels plateau at ∼8–9 h following release from thymidine. Semiquantitative Western blotting showed that cyclin B and PAF15 undergo similar decreases in protein levels at the exit from mitosis, which is marked by the decrease in phosphorylated histone H3 on serine 10 (Fig. 1C) (17). Similar to cyclin B, PAF15 levels are increased in mitotic cells arrested with taxol and nocodazole, compared with an asynchronous control population (Fig. 1D).
The precipitous drop in PAF15 at mitotic exit, coincident with cyclin B, suggests that it is controlled at the protein level. To examine this possibility, we used our dual-fluorescent reporter system, termed global protein stability (GPS) (18), to examine PAF15 levels at different points in the cell cycle under the control of an exogenous promoter. Enhanced green fluorescent protein (EGFP)-tagged PAF15 was expressed in the context of the GPS retroviral reporter, which produces a single transcript encoding both DsRed and EGFP-PAF15, separated by an internal ribosome entry site (18, 19). Unperturbed, asynchronous 293T and HeLa cells stably expressing this construct were treated with the DNA dye Hoechst and analyzed, without fixation, by flow cytometry. This system allows us to assess cell cycle-dependent changes in protein stability. Cells were gaited on the basis of their DNA content (in the G1, S, and G2/M phases), and the EGFP/DsRed ratio was compared for cells at different points in the cell cycle (Fig. 1E, Upper). Overlaid histograms demonstrate that, as cells progress from G1 to S to G2/M, there is a measurable increase in the EGFP/DsRed ratio, indicative of an increase in the protein levels of EGFP-PAF15 (Fig. 1E, Lower). Furthermore, mitotic arrest using nocodazole caused a large increase in the EGFP/DsRed ratio compared with asynchronous cells (Fig. 1F). Because expression in the GPS system is driven by an exogenous promoter, these data indicate that PAF15 protein levels are controlled posttranscriptionally throughout the cell cycle.
PAF15 Is an APC/C-Cdh1 Substrate.
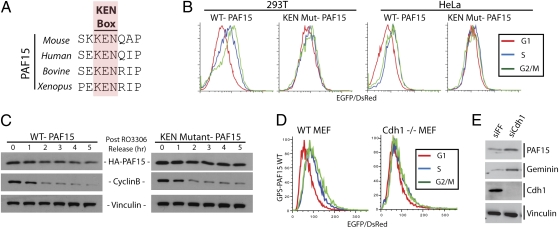
APC/C ubiquitylates substrates, targeting them for degradation at mitotic exit and throughout the G1 phase. Because PAF15 protein levels were regulated posttranscriptionally, we scanned its sequence for a known APC/C degron. Sequence analysis of vertebrate PAF15 homologs revealed a highly conserved KEN box, the recognition motif for APC/C-Cdh1 (Fig. 2A). In addition, analysis using a protein disorder prediction algorithm suggests that this region of the protein is intrinsically disordered, a common feature of KEN box motifs (20). If PAF15 protein stability is controlled by APC/C-mediated ubiquitylation, mutation of its KEN box should block proteolysis during G1. The essential KEN box lysine was mutated to alanine (K78A), and cells expressing the PAF15 GPS reporter were examined by flow cytometry for cell cycle-dependent differences. In both HeLa and 293T cells, wild-type PAF15 protein increases as cells progress from G1 to S to G2/M, as measured by flow cytometry (Fig. 2B). Importantly, mutation of the KEN box abrogated the cell cycle-dependent changes in PAF15 protein in both cell lines (Fig. 2B). To further confirm that a KEN box mutant of PAF15 is stabilized in G1, HeLa cells expressing an HA-tagged PAF15 were synchronized at the G2/M boundary using the Cdk1 inhibitor RO3006. After release from the inhibitor, cells progressed through M phase and back into G1 phase. Wild-type PAF15 is degraded in G1 as expected, whereas the KEN box mutant remained more stable (Fig. 2C). Cyclin B is readily degraded in cells expressing either wild-type or the KEN mutant PAF15, demonstrating that expression of the KEN-mutated PAF15 does not affect mitotic exit or APC/C activity.
Fig. 2.
PAF15 is degraded in G1 by the anaphase-promoting complex. (A) PAF15 contains a KEN box that is highly conserved among its vertebrate homologs. (B) Wild-type and KEN mutant (K78A) PAF15 was expressed using the GPS vector in both 293T and HeLa cells. Cells were gaited on the basis of their cell cycle state after staining with Hoechst. Overlaid histograms of the EGFP/DsRed ratio for cells in G1, S, and G2/M phases are shown. (C) HeLa cells were arrested at the G2/M boundary using the CDK1 inhibitor RO3306. Following release, HA-tagged WT and KEN mutant PAF15 were followed by Western blot. Immunoblotting for cyclin B and vinculin provides a marker for cell cycle status and a loading control, respectively. (D) GPS-PAF15 was expressed in WT and Cdh1 null (−/−) MEFs. Cells were gaited on the basis of cell cycle state after staining with Hoechst. Overlaid histograms of the EGFP/DsRed ratio for cells in G1, S, and G2/M phases are shown. (E) HeLa cells were treated with siRNA targeting firefly luciferase (FF) as a negative control or Cdh1. Seventy-two hours after transfection cells were collected for Western blot and analyzed with antibodies to PAF15, geminin, Cdh1, and vinculin (loading control).
To confirm that APC/C-Cdh1 is in fact responsible for PAF15 degradation, we used Cdh1 null mouse embryonic fibroblasts (MEFs). PAF15 was expressed from the GPS construct in both Cdh1 null (−/−) or wild-type (WT) MEFs. WT MEFs expressing GPS-PAF15 show a cell cycle-dependent distribution similar to that found in human 293T and HeLa cells. The Cdh1−/− MEFs show almost no change in the EGFP/DsRed levels throughout the cell cycle, indicating little change in PAF15 stability (Fig. 2D). To further confirm the role of APC/C-Cdh1 in PAF15 stability, Cdh1 was depleted from HeLa cells using siRNA. Immunoblotting for PAF15, 72 h after siRNA transfection, showed an increase in its protein level similar to that of the known APC/C substrate geminin (Fig. 2E). Immunoblotting for Cdh1 demonstrates that it is effectively depleted by siRNA. These results demonstrate that PAF15 is targeted for destruction by the APC/C-Cdh1 ubiquitin ligase.
PAF15 Plays a Role in the DNA Damage Response.
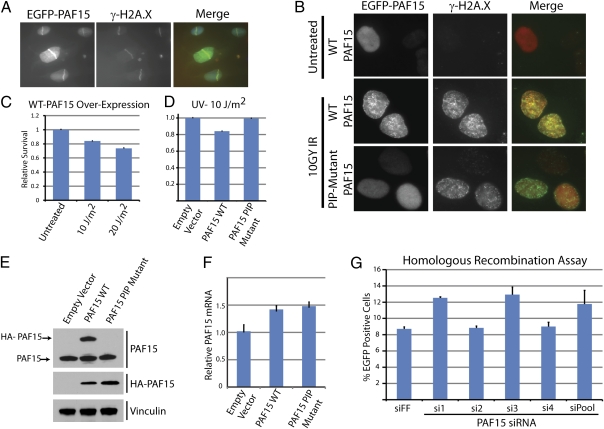
A subset of DNA damage response proteins access sites of damage through PCNA. For example, the translesion synthesis polymerase, Pol-η, binds to ubiquitylated PCNA. Because mass spectrometry and silver staining of PAF15 pull-downs suggested that PCNA and PAF15 are in 1:1 complex (Fig. 1B), we asked whether PAF15 could also localize to sites of DNA damage. Cells expressing EGFP-tagged PAF15 were analyzed by fluorescence microscopy following DNA damage induced by laser microirradiation and ionizing γ-radiation (IR). Following laser-induced microirradiation, EGFP-PAF15 localized to sites of damage and colocalized with phosphorylated H2A.X (γH2A.X: Fig. 3A). Similarly, after 10 Gy of IR, EGFP-PAF15 localized to damage-induced foci with γH2A.X (Fig. 3B). We tested whether this localization was dependent on the PAF15 PIP-box motif. The two consecutive phenylalanine residues within the PIP box were mutated to alanine (PIP mutant). Following 10 Gy of IR, PAF15 harboring a PIP mutant was unable to localize to damage foci (Fig. 3B), demonstrating that its recruitment to sites of damage is dependent on its interaction with PCNA.
Fig. 3.
PAF15 is involved in the cellular response to DNA damage. (A) U2OS cells stably expressing EGFP-PAF15 were laser-microirradiated and analyzed by immunofluorescence imaging after staining for EGFP-PAF15 and γH2A.X. (B) U2OS cells expressing EGFP-PAF15 wild-type or a PIP-box mutant were irradiated with 10 Gy of ionizing radiation. Irradiated cells were analyzed by immunofluorescence imaging after staining for EGFP-PAF15 and γH2A.X. (C) Cells stably overexpressing PAF15 wild type were irradiated with 10 or 20 J/m2 of UV and examined for survival using cell titer glo 4 d later. (D) Cells expressing PAF15 wild type or a PIP-box mutant were irradiated with 10 J/m2 of UV and examined for survival using cell titer glo 4 d later. (E and F) U2OS cells overexpressing HA-tagged PAF15 WT and PIP-box mutants were analyzed by immunoblot with PAF15 and HA antibodies (E) and using quantitative RT-PCR (F). Note that the PAF15 antibody does not recognize the PIP-box mutant protein (PAF15 immunoblot, Top). (G) DR-U2OS cells, which express a GFP reporter for homologous recombination, were depleted of PAF15 using siRNA, subsequently infected with adenovirus expressing I-SceI, and assayed for homologous recombination by measuring the reconstitution of GFP. Error bars represent SD; P value for siRNA 1 = 0.003 and for siRNA 3 = 0.01.
Given the localization of PAF15 to sites of damage, we next examined whether overexpression of PAF15 had an effect on cell survival following damage. U2OS cells were transduced with either wild-type or the PIP-box mutant PAF15 and subjected to both IR and UV damage. Four days after damage, cell survival was assessed using cell titer glo, a surrogate for cell number and viability. Cells overexpressing PAF15 by approximately twofold (Fig. 3E, Top) were mildly sensitive to increasing doses of UV, but not IR, relative to undamaged controls (Fig. 3C). Mutation of the PAF15 PIP box relieved this damage sensitivity (Fig. 3D). Because the antibody that we used does not recognize the PIP-box mutant of PAF15 (Fig. 3E, Top, PAF15 immunoblot), we used quantitative RT-PCR and HA immunoblots to determine the degree to which HA-PAF15 was overexpressed. We found that both WT and PIP-box mutant PAF15 were expressed at similar levels (Fig. 3E, Middle), and their mRNAs were each expressed at ∼1.5-fold relative to empty vector-expressing cells (Fig. 3F), consistent with the immunoblot analysis for WT PAF15 overexpression (Fig. 3E). This demonstrates that a when PAF15 is present in twofold excess, it impairs cell growth or survival following UV-induced DNA damage through its interaction with PCNA.
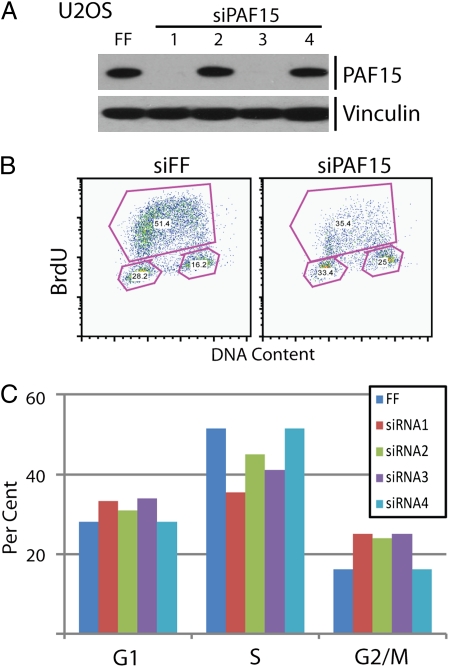
PCNA modification has been implicated in homologous recombination (HR)-mediated repair. To examine whether PAF15 plays a role in HR, we used a well-established fluorescent GFP reporter assay (21). Cells were subjected to a single double-strand break within the sequence of a truncated GFP using the restriction endonuclease I-SceI. Following double-strand break formation, repair of the GFP sequence by homologous recombination, using an alternative, truncated GFP sequence located downstream, leads to expression of a functional GFP that is assessed by flow cytometry. PAF15 was depleted from cells using four independent siRNAs. Only two of the four siRNAs depleted PAF15, as judged by immunoblot for the endogenous protein (Fig. 4A). Interestingly, the two siRNAs that deplete PAF15 (siRNA 1 and 3; see Fig. 4A) induce a hyper-recombination phenotype (Fig. 3G). Depletion with PAF15 siRNAs 1 and 3 leads to a 44% and 48% increase in HR, respectively. These data suggest a role for PAF15 in restricting HR during the S and G2 phases of the cell cycle.
Fig. 4.
PAF15 is involved in cell cycle progression. Depletion of PAF15 from U2OS cells caused a decrease in BrdU-labeled S-phase cells. (A) Western blot for PAF15 following depletion with four independent siRNA. (B) Flow cytometric analysis of BrdU-labeled cells treated with siRNA targeting firefly luciferase or PAF15. (C) Quantification of BrdU-labeling experiment for cells in G1, S, and G2/M phases.
PAF15 Controls Cell Cycle Progression.
To determine whether PAF15 affects cell cycle progression, we depleted it from cells using siRNA and examined cell cycle state following bromodeoxuridine (BrdU) labeling. Four independent siRNA were tested for their ability to deplete PAF15 and affect cell cycle progression in two independent cell lines. Consistently, siRNA 1 and 3 were able to deplete PAF15 protein to undetectable levels in 293T and U2OS cells (Fig. 4A and Fig. S1). Forty-eight hours after treatment with siRNA, cells were pulsed with BrdU for 20 min. Staining for BrdU and DNA content revealed a reproducible decrease in BrdU-positive cells, suggesting a decrease in the number of cells in S phase. Data from a representative experiment are shown in Fig. 4. In 293T cells this coincides with an increase in the G1 population (Fig. S1). In U2OS, there was a relative increase in both G1 and G2 cells (Fig. 4 B and C). The decrease in S-phase cells in both cell lines, together with PAF15’s tight association with PCNA, suggests that PAF15 plays a role in DNA replication.
Discussion
PAF15 depletion has been reported to sensitize cells to DNA cross-linking agents (13). We were unable to recapitulate these results using U2OS, HeLa, HCT116, DLD1, and 293T cells with UV, IR, hydroxyurea, and mitomycin c-mediated DNA damage, even though we can deplete PAF15 below detectable levels. The reason for this discrepancy is currently unclear. Instead, we found that overexpression of PAF15 caused a defect in survival/growth following UV damage. Importantly, sensitivity is dependent on the PAF15–PCNA interaction because overexpression of PAF15 harboring PIP-box mutations does not produce sensitivity to UV. The molecular mechanism underpinning this sensitivity remains unclear. It is possible that PAF15 restricts the localization of repair factors or translesion synthesis polymerases to sites of damage through a stoichiometric interaction with PCNA. A similar mechanism has been proposed for the role of the PCNA-associated CDK inhibitor p21CIP1. A p21CIP1 mutant that cannot be degraded following damage remains associated with PCNA and blocks docking of the translesion synthesis polymerase Pol-η (22). Unlike p21CIP1, which is degraded by the ubiquitin E3 ligase CRL4-Cdt2 following DNA damage (23, 24), we did not detect a change in PAF15 levels following treatment with UV, IR, mitomycin C, and hydroxyurea, suggesting that PAF15 differs from p21/CIP1 in response to DNA damage (23). Additionally, although PAF15 and PCNA appear to interact in a 1:1 complex, because PCNA forms a trimer we cannot formally exclude the possibility that a fraction of PAF15 interacts with intact PCNA trimers.
PAF15 depletion led to a hyper-recombination phenotype using two independent siRNA oligonucleotides, and the degree of depletion correlated precisely with phenotype, suggesting that this was not due to off-target effects associated with siRNA. Homologous recombination occurs primarily in the S and G2 phases of the cell cycle. Because PAF15 depletion decreases the number of cells in S phase, it is unlikely that the hyper-recombination phenotype is a consequence of cell cycle perturbation. In U2OS cells, siRNA 2, which does not deplete PAF15, caused a modest increase in G2/M phase cells, similar to siRNAs 1 and 3, which produce a strong PAF15 depletion. Although we suspect that the cell cycle perturbation of siRNA 2 is due to an off-target effect, this result suggests that simply stalling cells in the G2 phase of the cell cycle is insufficient to cause a 50% increase in homologous recombination. Furthermore, we observed an increase in HR after only 30 h of PAF15 depletion, a time at which no cell cycle perturbation is apparent. Because all of the PAF15 appears to be in complex with PCNA, its depletion could provide PCNA-interacting proteins access to PCNA that were otherwise restricted from binding, leading to an alteration in the balance between repair choices and tolerance mechanisms that occur at the site of break.
Double-strand breaks are repaired by either nonhomologous end joining or HR. By providing HR factors premature or inappropriate access to PCNA, the scales could be tipped in favor of HR and the chance of repair consequently reduced through nonhomologous end joining. PAF15 is present only in cells during S and G2/M phases of the cell cycle. It may seem counterintuitive to express an HR restriction factor during the only time in the cell cycle that HR occurs. However, it has become increasingly clear that the choice between different repair pathways is an essential component in mediating the overall DNA damage response. Expression of PAF15 in S-G2/M could buffer repair pathway initiation by restricting access to PCNA at the site of a break or by allowing the recruitment of distinct accessory factors while the decision of which pathway to execute is being biochemically initiated. Alternatively, PAF15 could play a role in the recruitment of nonhomologous end joining components to the break site.
The coding sequence of PAF15 contains three known single-nucleotide polymorphisms (SNPs). One of these polymorphisms is a mis-sense mutation that corresponds to the KEN box glutamic acid, mutating it to serine (SNP ID rs11554313). Although this SNP has not been implicated in any disease state, our functional analysis suggests that mutation of the PAF15 KEN box leads to G1 stabilization and that overexpression of PAF15 could impair the cellular response to DNA damage. The other two SNPs are located within the PAF15 PIP box (SNP IDs rs11554309 and rs11554320). Interestingly, one of these mutations is located in one of the two critical phenylalanine residues that mediate interaction with PCNA.
The high conservation of PAF15 among vertebrates, its strong interaction with PCNA, and its ubiquitin-dependent regulation throughout the cell cycle suggest that it plays an important role in cell cycle regulation and DNA replication. Its degradation in G1, through APC/C-Cdh1–dependent ubiquitylation, and its modest phenotype in some tissue culture cell lines are reminiscent of the protein geminin. Geminin binds to and sequesters the replication licensing factor Cdt1. Surprisingly, geminin is essential only for restraining replication licensing in certain human tissue culture cell lines. Importantly, geminin-dependent sequestration is not the only mechanism by which Cdt1 is restrained. There are at least two E3 ubiquitin ligases that target Cdt1 for destruction. Presumably, the particular balance of specific proteins that exists in different cell lines is responsible for these phenotypic differences. In light of these differences, in vitro examination of PAF15’s role in promoting DNA replication would likely shed significant light on its function. Similarly, testing its function in a biochemically tractable system that mirrors physiological protein levels and that is well suited for studying replication, such as the Xenopus egg extract system, could provide us with key insights into the function of PAF15 in the cell cycle and DNA replication.
Materials and Methods
Tissue Culture, Reagents, and Procedures.
HeLa, U2OS, MEF, and 293T cells were maintained in Dulbecco's modified Eagle media supplemented with penicillin/streptomycin and 10% FBS. All media and supplements are from Invitrogen.
293T cells were transfected using TransIT transfection reagent (Mirus) when packaging retroviruses, as described (25). Retroviral infections were supplemented with hexadimethrine bromide (polybrene) at 8 μg/mL. The RNAiMAX (Invitrogen) lipid transfection reagent was used for transfection of siRNA oligonucleotides (used at a final concentration of 20 nM) according to the manufacturer's protocols. siRNA were obtained from Dharmacon as a “set of four siGenome upgrades.” Cdh1 siRNA were used together as a smart pool. A complete list of the siRNA sequences used in this study is listed in Table S3.
The CDK inhibitor RO3306 (Calbiochem) was used at a final concentration of 10 μM. Taxol and nocodazole were used at a concentration of 100 nM and 100 μg/mL, respectively (Sigma). Thymidine was used at a final concentration of 2.5 mM. Cell titer glo was used to assess cell viability according to the manufacturer's protocol (Promega). Homologous recombination was assayed using adenovirus expressing I-SceI and DR-U2OS cells containing an integrated GFP HR reporter (21).
Flow cytometry was performed on a BD-LSRII Flow Cytometer (Becton Dickinson). Data were collected using BD FACS Diva software (Becton Dickinson), and analysis was performed on FloJo software. BrdU labeling was done using the APC-BrdU flow kit (BD Pharmingen). Cells were laser-microirradiated and processed for immunofluorescence as described (26).
Immunological reagents used are listed in Table S3. Semiquantitative immunoblot analysis was performed on images collected with an air-cooled CCD camera using ImageJ (National Institutes of Health). Silver staining was performed using a SilverQuest Silver Staining kit (Invitrogen). Protein disorder prediction was performed using DisEMBL. Sequence alignments were performed using ClustalW.
Immunoprecipitation and Mass Spectrometry.
PAF15 interactors were identified as described previously (15, 27). TCA-precipitated proteins were dissolved in 100 mM ammonium bicarbonate (pH 8.0) with 10% acetonitrile and 10 ng/μL trypsin (Promega). They were subsequently incubated at 37 °C for 5 h and desalted using C18 stage tips as described previously (28). The peptide mixture was then shot twice in succession on an LTQ Orbitrap Discovery (Thermo Scientific). For each run, the peptide mixture, dissolved in 5% formic acid and 5% acetonitrile, was loaded onto a 15-cm C18 column, and peptides were eluted using a 60-min 5–25% acetonitrile gradient. Tandem MS spectra were searched using Sequest with a 25-ppm mass window and dynamic modification of 79.96633 Da on serine, threonine, and tyrosine. Searching occurred against a target-decoy database of tryptic human peptides (29), and identified peptides were filtered to a false discovery rate of less than 1%. The Ascore algorithm (30) was applied to assess the probability of correct phosphorylation site assignment.
Supplementary Material
Acknowledgments
We thank the members of the S.J.E. laboratory for helpful discussions and insights. Special thanks to Britt Adamson for help with the homologous recombination reporter assay and Anna Burrows for help with quantitative RT-PCR. We thank Steve Gygi for access to MS/MS analysis software and Stephen Taylor for providing the Cdh1 null MEFs. M.J.E. is the Philip O'Bryan Montgomery, Jr., MD fellow of the Damon Runyon Cancer Research Foundation (DRG-1996-08). A.E.H.E. is supported by fellowships from the Jane Coffin Childs Foundation and the American Society for Radiation Oncology. A.C. is a recipient of a European Molecular Biology Organization long-term fellowship. This work was supported by a National Institutes of Health grant (to S.J.E.). S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106136108/-/DCSupplemental.
References
- 1.Peters JM. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 2.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 5.Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 6.Bruck I, O'Donnell M. The ring-type polymerase sliding clamp family. Genome Biol. 2001;2(1) doi: 10.1186/gb-2001-2-1-reviews3001. REVIEWS3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 8.Gilljam KM, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich HD. Conservation of DNA damage tolerance pathways from yeast to humans. Biochem Soc Trans. 2007;35:1334–1337. doi: 10.1042/BST0351334. [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa M, et al. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res. 2007;67:2568–2576. doi: 10.1158/0008-5472.CAN-06-4356. [DOI] [PubMed] [Google Scholar]
- 12.van Bueren KL, et al. Murine embryonic expression of the gene for the UV-responsive protein p15(PAF) Gene Expr Patterns. 2007;7:47–50. doi: 10.1016/j.modgep.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Turchi L, et al. ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 2009;16:728–737. doi: 10.1038/cdd.2009.2. [DOI] [PubMed] [Google Scholar]
- 14.Yu P, et al. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene. 2001;20:484–489. doi: 10.1038/sj.onc.1204113. [DOI] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 18.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 19.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 20.Michael S, Travé G, Ramu C, Chica C, Gibson TJ. Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- 21.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J Cell Sci. 2008;121:3271–3282. doi: 10.1242/jcs.027730. [DOI] [PubMed] [Google Scholar]
- 23.Abbas T, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciccia A, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 29.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 30.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.