Abstract
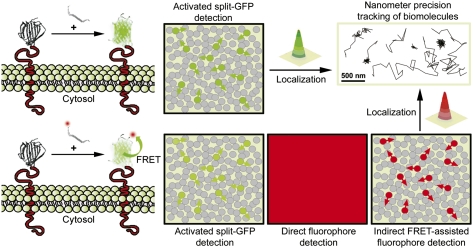
Single-molecule (SM) microscopy allows outstanding insight into biomolecular mechanisms in cells. However, selective detection of single biomolecules in their native environment remains particularly challenging. Here, we introduce an easy methodology that combines specific targeting and nanometer accuracy imaging of individual biomolecules in living cells. In this method, named complementation-activated light microscopy (CALM), proteins are fused to dark split-fluorescent proteins (split-FPs), which are activated into bright FPs by complementation with synthetic peptides. Using CALM, the diffusion dynamics of a controlled subset of extracellular and intracellular proteins are imaged with nanometer precision, and SM tracking can additionally be performed with fluorophores and quantum dots. In cells, site-specific labeling of these probes is verified by coincidence SM detection with the complemented split-FP fusion proteins or intramolecular single-pair Förster resonance energy transfer. CALM is simple and combines advantages from genetically encoded and synthetic fluorescent probes to allow high-accuracy imaging of single biomolecules in living cells, independently of their expression level and at very high probe concentrations.
Keywords: biomolecular imaging, membrane biophysics, single-molecule fluorescence, single-particle tracking, high-resolution microscopy
In recent years, parallel developments in imaging technologies, optical probes, and genetic engineering have contributed to the fast emergence of single molecule (SM) fluorescence techniques. These techniques now permit the imaging of subcellular structures with nanometer resolution and tracking of individual proteins as well as stochiometric analysis of molecular complexes in living cells (1, 2). A key requirement for SM microscopy is to limit the number of biomolecules that are simultaneously imaged to maintain an SM detection regimen while recording a statistically representative number of events. Hence, advanced labeling and targeting strategies are needed to ensure specific and sensitive detection of individual biomolecules in complex cellular environments.
In situ, protein labeling can be achieved by incorporating molecular tags that are posttranslationally coupled to exogenous fluorophores (3) or fusion to fluorescent proteins (FPs) (4). Posttranslational labeling of molecular tags necessitates probe concentrations in the micromolar range (3), well above the typical subnanomolar range required for SM imaging. Such high concentrations increase the risks of nonspecific binding in cells, impose extensive washing of the nontargeted probes, and complicate SM detection. When using FPs, low expression levels required for SM imaging are often difficult to achieve, and high expressions demand potentially toxic photobleaching. Some of these issues can be alleviated by using switchable fluorescent probes that permit low background and controlled imaging of only a fraction of tagged biomolecules in live cells (5–7). The growing panel of photoswitchable FPs is particularly useful, because it does not require advanced cellular targeting chemistries (8). However, genetically encoded FPs are neither as bright nor as photostable as the best organic and inorganic fluorophores.
Split-FPs are another type of genetically encoded fluorescent probes that switch on when reconstituted. They have been extensively used to study protein interactions in living cells by bimolecular fluorescence complementation (BiFC) (9). In BiFC assays, two proteins of interest are fused to two nonfluorescent FP fragments of roughly equal size, and protein–protein interaction drives the complementation of the fragments into a fluorescent FP. Recently, a highly asymmetric split-GFP based on a super-folder GFP was engineered in a large GFP 1–10 fragment (amino acids 1–214) and a small GFP 11 peptide fragment corresponding to the 11th β-strand of the super-folder GFP β-barrel (amino acids 214–230) (10). Both fragments spontaneously self-complement in solution, and the GFP 11 fragment can be mutated for fast complementation (11) or produced synthetically for in vitro complementation with GFP 1–10 (12, 13). When used as protein tags, GFP 11 and GFP 1–10 helped visualize synaptic contacts and distribution of bacterial effectors in cells (14, 15).
Here, we have exploited several aspects of this asymmetric split-GFP system to target, image, and track individual proteins in living cells. We have developed complementation-activated light microscopy (CALM), an SM imaging methodology that relies on the irreversible and stochastic complementation of GFP 1–10 fusion proteins by synthetic versions of the GFP 11 peptide fragment. In living cells, a controlled subset of proteins fused to GFP 1–10 is activated into bright GFPs and continuously imaged by simply adjusting the concentrations and incubation times with engineered peptides. Using CALM, low-background and ultra-specific tracking of individual transmembrane, GPI-anchored, or caveolae-associated proteins was achieved in various cell lines within minutes of peptide addition. We also used CALM for site-directed posttranslational targeting of synthetic and fluorescent moieties to individual proteins. In living cells, specific and stochiometric labeling using single fluorophores and quantum dots (qdots) was verified by coincident SM detection with the complemented split-FP fusion proteins and allowed for advanced bioimaging modalities. For instance, when combining CALM and single-pair Förster resonance energy transfer (spFRET) from a complemented GFP to an acceptor peptide conjugate, single proteins could be tracked independently of their expression level and at probe concentrations in the micromolar range.
Results
In Vitro Bulk Complementation of Split-GFP with Small Synthetic Peptides.
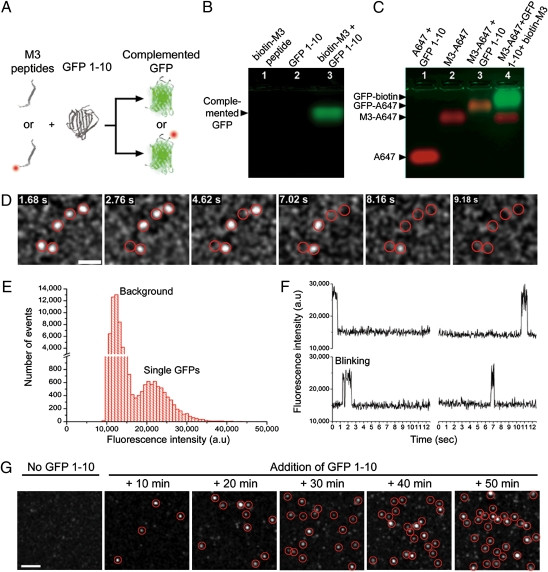
We first tested the complementation of GFP 1–10 with a variety of small synthetic peptides containing a mutated version of the GFP 11 fragment referred to as M3 peptides (Fig. 1A). We designed a 24-aa complementary peptide with an N-terminal biotin and a flexible linker to the C-terminal M3 sequence (biotin-M3) and verified its in vitro complementation with soluble GFP 1–10. Neither GFP 1–10 nor the synthetic biotin-M3 peptide is fluorescent but they self-assemble into a complemented, bright, and monomeric GFP-biotin with an apparent molecular weight (MW) of 25.0 kDa (Fig. 1B and SI Appendix, Fig. S1). Binding of M3 peptides to GFP 1–10 and GFP complementation was also tested with fluorescently labeled M3 peptides. On incubation with GFP 1–10, M3 peptides conjugated to the Alexa 647 fluorophore (M3-A647; >97% purity) (SI Appendix, Fig. S2) were shifted on native gels compared with Alexa 647 alone or unreacted M3-A647 (Fig. 1C). Specific binding of M3-A647 to GFP 1–10 and GFP complementation was confirmed by the appearance of a colocalizing GFP fluorescence band and competition with excess nonfluorescent biotin-M3 peptides (Fig. 1C). Similar results were obtained by size-exclusion chromatography (SI Appendix, Fig. S1).
Fig. 1.
Complementation of GFP 1–10 with synthetic M3 peptides and in vitro SM imaging. (A) Schematic of self-complementation between nonfluorescent (Upper) and fluorescent (Lower) synthetic M3 peptides and GFP 1–10. (B) Native gel electrophoresis of biotin-M3 peptide (lane 1), GFP 1–10 (lane 2), and complementation reaction of biotin-M3 with GFP 1–10 (lane 3). (C) Native gel shift of free Alexa 647 (A647; lane 1), unreacted M3-Alexa 647 conjugate (M3-A647; lane 2), and M3-A647 reaction with GFP 1–10 in the absence (lane 3) or presence (lane 4) of a competing excess of biotin-M3 peptides. (D) Sequential photobleaching of complemented and purified individual GFP-biotin nonspecifically bound to a glass coverslip and imaged by TIRF (Movie S1). GFP diffraction-limited spots are intentionally expanded to facilitate visualization. (Scale bar: 2 μm.) (E) Fluorescence intensity distribution for 152 single-complemented GFP-biotin molecules and background fluorescence from coverslips. (F) Fluorescence time traces of four single split-GFPs complemented on M3 peptide-coated coverslips. Single-step photobleaching and blinking events are observed. (G) Representative TIRF fields of view taken at different incubation times before and after addition of GFP 1–10 to M3 peptide-coated coverslips. The number of individual-complemented GFP-biotin per fields of view increases with increasing incubation times. GFP diffraction-limited spots are intentionally expanded to facilitate visualization. (Scale bar: 2 μm.)
In solution, the complementation kinetics of GFP 1–10 with synthetic M3 peptides were fast and spread over a 2-h period before reaching saturation (SI Appendix, Fig. S3). Consistent with previous results (10), GFP fluorescence resulting from the complementation scaled linearly with the concentration of peptides under nonsaturating conditions.
To further assess the flexibility of our designed peptides and evaluate the complementation efficiency under restricted conformational freedom, two biotinylated M3 sequences were affixed to streptavidin-coated agarose beads. As for solution-based assays, the beads rapidly became fluorescent when incubated with GFP 1–10 (SI Appendix, Fig. S4), indicating that surface attachment did not sterically hinder the complementation.
In Vitro SM Imaging of Split-GFP Complementation.
Next, we tested our ability to detect single GFP copies complemented by M3 peptides. GFP 1–10 in bacterial extract was incubated with biotin-M3, purified by high-pressure liquid chromatography (HPLC), and diluted solutions of complemented GFP-biotin deposited on glass coverslips were imaged by total internal reflection fluorescence (TIRF) microscopy. Diffraction-limited fluorescence spots corresponding to single complemented GFP-biotin were detected (Fig. 1D). The single emitter nature of GFP-biotin molecules was confirmed by their unimodal fluorescence intensity distribution (Fig. 1E), their single-step photobleaching, and blinking events (Fig. 1F and Movie S1).
Single GFPs could also be detected during the complementation process. M3-biotin peptides were affixed to avidin-functionalized coverslips and imaged by TIRF while incubating coverslips with GFP 1–10. In the absence of GFP 1–10, no fluorescence was detected, but single diffraction-limited spots of complemented GFP-biotin appeared at the coverslip surface within 10 min of incubation (Fig. 1G). The number of single GFPs per field of view increased over time, which is in agreement with the solution-based complementation kinetic assays. Sudden appearances of single GFPs during imaging were also observed (Fig. 1F), clearly indicating that the complementation of single split-GFPs could be directly imaged.
Live Cell CALM Imaging of Split-GFP Fusion Proteins.
To evaluate if CALM would be similarly efficient in living cells, we expressed GFP 1–10 as an N- or C-terminal fusion to extracellular and intracellular membrane proteins in different mammalian cell lines (Fig. 2A). As extracellular proteins, we used two membrane raft-associated proteins, a transmembrane CD4 split-GFP fusion (14) (GFP 1–10-CD4), and a glycosylphosphatidyl inositol (GPI)-anchored humanized split-GFP fusion (GFP 1–10(h)-GPI) attached to the upper leaflet of the plasma membrane. As an intracellular protein, we fused GFP 1–10(h) to caveolin-1 (cav1-GFP 1–10(h)), an integral membrane protein participating in the scaffolding of 50- to 100-nm caveolae invaginations at the plasma membrane (16).
Fig. 2.
CALM imaging in living cells. (A) Schematic representation of plasma membrane split-GFP fusions used in this work. (B) Wide-field fluorescence imaging of GFP 1–10-CD4 expression and complementation in U2OS, COS-7, and HEK cells. Expressing cells (+) are detected with a fluorescent anti-GFP antibody. When incubated with M3 peptides (+biotin-M3 or +FCC-M3), GFP 1–10-CD4 proteins are activated into bright GFP-CD4 proteins, and expressing cells become fluorescent (overlay). No GFP signal is seen in the absence of peptides (−biotin-M3) or for nonexpressing cells (−). Binding of the complementary biotin-M3 peptides on the cell surface is verified by staining with fluorescent streptavidin (SAV-A647). (Scale bar: 20 μm.) (C) Wide-field fluorescence imaging of GFP 1–10(h)-GPI expression and complementation in U2OS and COS-7 cells. (Scale bar: 10 μm.) (D) Fluorescence confocal, wide-field, and TIRF imaging of cav1-GFP 1–10(h) expression and intracellular complementation in U2OS cells. (Upper) Ventral plasma membrane confocal images of fixed cells immunolabeled for endogenous cav1 (anti-cav1) and cav1-GFP 1–10(h) (anti-GFP) showing cav1-GFP 1–10(h) colocalization with endogenous cav1 (overlays and Insets). 3D reconstructions of cells are available in Movie S4. (Scale bar: 20 μm.) (Lower) Fluorescence wide-field imaging of live U2OS cells coexpressing cav1-GFP 1–10(h) and ABP-mCherry (+). A cell microinjected with M3 peptides (star; ∼25 μM final intracellular M3 peptide concentration) and imaged after 45 min incubation at 37 °C shows a perinuclear pool of complemented cav1-GFP(h) (arrows) and a plasma membrane pool of caveolae-associated cav1-GFP(h) (arrowheads). The typical punctuated pattern of complemented caveolae is better seen by TIRF imaging of the plasma membrane (white square). The TIRF image is a pixel-based maximum intensity projection (ΣImax) overlay image for all frames of the dual-color Movie S5. (Scale bar: wide-field, 10 μm; TIRF, 5 μm.)
When expressed in U2OS, COS-7, and HEK cells, GFP 1–10-CD4 and GFP 1–10(h)-GPI were properly directed to the plasma membrane, which was verified by live cell labeling with anti-GFP antibodies recognizing GFP 1–10 (Fig. 2 B and C). Their membrane distribution was homogenous, with no aggregation or mislocalization. On incubation with high concentrations of M3 peptides (50 μM) for 45–60 min, GFP fluorescence was specifically detected at the membrane of transfected cells (Fig. 2 B and C), which was confirmed by confocal and TIRF imaging (SI Appendix, Fig. S5 and Movie S2). The appearance of the GFP signal was solely triggered by the complementation reaction, because no GFP fluorescence was observed for nontransfected cells or when M3 peptides were omitted (Fig. 2B and SI Appendix, Fig. S5). Specific binding of M3 peptides to GFP 1–10 fusion proteins was further established by incubating cells with biotin-M3 and observing colocalized membrane signals from GFP and a fluorescently labeled streptavidin (SAV-A647) (Fig. 2B). At the membrane, GFP signal was uniform and proportional to the membrane expression level of GFP 1–10-CD4 and GFP 1–10(h)-GPI as implied by the equivalent intensity ratios between anti-GFP and GFP fluorescence in different cells (Fig. 2B). Consistent with in vitro complementation kinetics assays, long incubations of M3 peptides with expressing cells resulted in increased total GFP signal, and intense fluorescence was detected after 48 h incubation (Movie S3).
Intracellular expression and association of cav1-GFP 1–10(h) with caveolae were verified by double immunolabeling of U2OS cells with anti-cav1 and anti-GFP antibodies. The typical punctuated membrane pattern of caveolae-associated endogenous cav1 (17, 18) colocalized with that of cav1-GFP 1–10(h) (Fig. 2D and Movie S4) for transfected cells, indicating that functional cav1-GFP 1–10(h) fusion proteins contribute together with endogenous cav1 to the formation of membrane caveolea in U2OS cells.
Intracellular complementation of cav1-GFP 1–10(h) was then tested by direct microinjection of M3 peptides in live and expressing U2OS cells identified with the coexpression marker actin-binding peptide mCherry-LifeAct (ABP-mCherry). Cells became GFP-fluorescent when microinjected with a high concentrations of M3 peptides (∼25 μM final intracellular concentration) followed by 45 min incubation (Fig. 2D). In wide-field fluorescence images, a perinuclear pool of cav1-GFP(h) was detected together with the typical punctuated pattern of caveolae-associated cav1-GFP(h) at the plasma membrane (Fig. 2D). Complemented and GFP-fluorescent caveolae were clearly visible when the cell membrane was imaged by TIRF microscopy (Fig. 2D and Movie S5). No GFP fluorescence was observed for nonexpressing or noninjected cells (Movie S5).
Single Biomolecule Imaging in Living Cells by CALM.
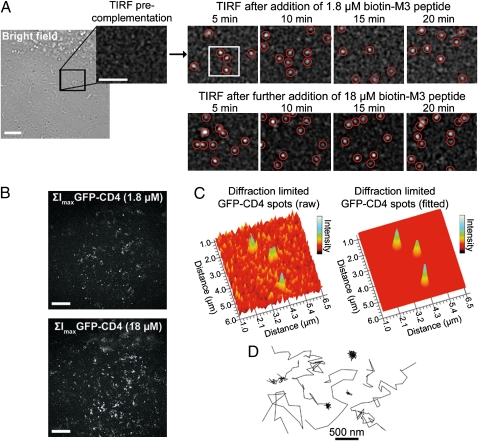
Making use of the concentration and time-dependant complementation kinetics of GFP 1–10, an SM detection regimen where individual split-GFP fusion proteins were continuously activated, imaged, and photobleached could easily be attained using low concentrations of M3 peptides. For this, the ventral plasma membrane of cells stably expressing GFP 1–10-CD4 or GFP 1–10(h)-GPI proteins was repeatedly imaged by TIRF for 1 min in 5-min intervals before and during the addition of 1.8 μM M3 peptides. Before addition of the peptides, rare spurious SM events corresponding to cellular or imaging media materials emitting fluorescence at 520 nm were detected (typically less than 3% of all detected events). A few diffraction-limited GFP spots were immediately detected on addition of biotin-M3 to cells, and lateral membrane diffusion of individual complemented GFP-CD4 proteins was clearly observed within 5 min of incubation (Fig. 3A and Movie S6) as confirmed by single-step photobleaching and blinking events. The number of single GFP-CD4 detected steadily increased during the first 10–15 min and remained relatively constant at longer incubation times (Fig. 3A and Movie S6) in line with in vitro complementation kinetic assays. On addition of 10-fold more biotin-M3 (18 μM), a higher number of GFP-CD4 appeared at the cell membrane (Fig. 3B and Movie S6), but GFP spots were still sufficiently separated to clearly identify individual biomolecules (Fig. 3A). No GFP signal was detected for nonexpressing cells imaged with similar peptide concentrations and incubation times (Movie S6). Thus, regardless of the protein expression levels, single biomolecules could easily be imaged in living cells by relying on the stochastic binding of M3 peptides and activation of split-GFP fusion proteins.
Fig. 3.
SM imaging and tracking of extracellular GFP 1–10-CD4 proteins by CALM. (A) A region of interest (black square) in the plasma membrane of a U2OS cell stably expressing GFP 1–10-CD4 is imaged by TIRF before and after complementation with biotin-M3 peptides at different incubation times. Diffraction-limited single GFP-CD4 spots appear and diffuse in the plasma membrane within minutes of M3 peptide addition. Single GFP-CD4 spots are intentionally expanded to facilitate visualization. (Scale bar: 5 μm.) (B) Pixel-based maximum intensity projections (ΣImax) TIRF images of all complemented GFP-CD4 detected during 20-min complementation with 1.8 or 18 μM biotin-M3 peptides (Movie S6). The field of view corresponds to the bright field image in A. (Scale bar: 5 μm.) (C) 3D rendering of raw and Gaussian-fitted diffraction-limited spots corresponding to individual complemented GFP-CD4 in the cell plasma membrane (white square in A). (D) Representative trajectories from single GFP-CD4 diffusing in the plasma membrane of U2OS cells during CALM imaging.
The high signal to background obtained by lighting up single GFP-CD4 in the otherwise dark cell membrane enabled the precise localization and tracking of individual CD4 with a mean uncertainty of 24 ± 4 nm (Fig. 3 C and D and SI Appendix, Table S1). Their apparent diffusion coefficient calculated from ensemble mean square displacements (MSD) was 0.21 ± 0.03 μm2/s, which is in good agreement with coefficients expected for transmembrane proteins in cells (SI Appendix, Table S1 and SI Appendix, Fig. S6). A more detailed analysis by probability distribution of the square displacements (Pr2) (19) revealed that the coefficient derived from MSD actually comprises two GFP-CD4 subpopulations differing by about 10-fold in their diffusion coefficients: 0.29 ± 0.02 μm2/s (64%) and 0.021 ± 0.009 μm2/s (36%), respectively (SI Appendix, Table S1). These two subpopulations are consistent with the putative dynamic partitioning of CD4 into membrane raft microdomains (20, 21).
Similar tracking experiments were successfully carried out for individual GFP 1–10(h)-GPI fusion proteins, which diffused two times faster than GFP-CD4 as expected for lipid-anchored plasma membrane proteins (0.37 ± 0.02 μm2/s) (SI Appendix, Table S1 and Figs. S6 and S7 and Movie S7). As for GFP-CD4, Pr2 analysis revealed two diffusing subpopulations of GFP(h)-GPI proteins, with diffusion coefficients of 0.53 ± 0.03 μm2/s (69%) and 0.036 ± 0.008 μm2/s (31%), respectively. These fast and slow subpopulations are fully consistent with the dynamic partitioning of this GPI-anchor in and out of membrane raft microdomains previously observed in HeLa cells (22).
Single cav1-GFP 1–10(h) could also be imaged by CALM inside living cells. Expressing cells identified by coexpression of a nucleus-localized CFP-LacI-NLS marker were microinjected with biotin-M3 (∼5 μM final intracellular concentration) and biotin-Alexa 647, used here as a microinjection marker (Fig. 4A). As an alternative to microinjection, M3 complementary peptides were efficiently translocated to the cell cytoplasm using cell penetrating Pep-1 peptides (23) and simple addition to the cell media (SI Appendix, Fig. S8). Within minutes, single diffraction-limited GFP spots of complemented cav1-GFP(h) were detected by TIRF at the ventral plasma membrane, and an increasing number of individual cav1-GFP(h) lit up with increasing incubation times (Fig. 4B and Movie S8). No GFP fluorescence was observed for noninjected cells expressing cav1-GFP 1–10(h) or microinjected cells that did not express the fusion protein (Fig. 4B and SI Appendix, Fig. S9). Single membrane-anchored cav1-GFP(h) could be tracked with a mean localization uncertainty of 14 ± 4 nm (Fig. 4 C and D and SI Appendix, Table S1) and diffused with an apparent ensemble MSD diffusion coefficient of 0.02 ± 0.003 μm2/s. Pr2 analysis further indicated that a very slow and dominant subpopulation of caveolae-associated cav1-GFP(h) (0.002 ± 0.001 μm2/s, 74%) was present together with a faster-diffusing second subpopulation (0.059 ± 0.009 μm2/s, 26%) at the plasma membrane. These values are in excellent agreement with ensemble and caveolae-specific fluorescence recovery after photobleaching (FRAP) measurements of cav1 diffusion in the plasma membrane of a variety of other cells lines (24).
Fig. 4.
SM imaging and tracking of intracellular cav1-GFP 1–10(h) proteins by CALM. (A) A U2OS cell coexpressing cav1-GFP 1–10(h) and the nucleus-localized CFP-LacI-NLS coexpression marker (+) are microinjected with biotin-M3 peptides (∼5 μM final intracellular concentration) together with a biotin-Alexa 647 injection marker (star). A region of interest (white square) is then imaged for GFP fluorescence by TIRF microscopy. (Scale bar: 10 μm.) (B) Pixel-based maximum intensity projection (ΣImax) TIRF images of all complemented cav1-GFP(h) detected at the ventral intracellular plasma membrane for the region of interest in A 3, 5, and 10 min after injection of M3 peptides (Movie S8). When overlaid with the wide-field fluorescence image, the cumulative 3- to 10-min maximum intensity projection image shows the high specificity of complementation. (Scale bar: 5 μm.) (C) 3D rendering of raw diffraction-limited spots corresponding to membrane-associated single cav1-GFP(h) proteins (white square in B). (D) Representative trajectories from single cav1-GFP(h) diffusing in the cytoplasmic side of the plasma membrane of U2OS cells during CALM imaging.
Thus, by fine-tuning the concentration of complementary peptides and their incubation time with cells, CALM allowed rapid, controllable, and highly specific detection and tracking of single biomolecules in living cells.
Split-GFPs as Posttranslational Protein-Targeting Platforms in Living Cells.
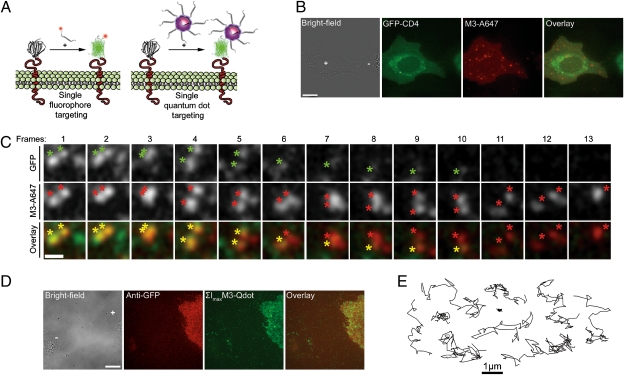
Gel electrophoresis assays, (Fig. 1 and SI Appendix, Fig. S11), live cell imaging (Fig. 2B), and additional fluorescence cross-correlation spectroscopy (FCCS) measurements all indicated that functionalized M3 peptides were irreversibly bound to complemented GFPs. This irreversible binding, consistent with prior reports on this split-GFP (11) and most BiFc systems (9), provided a means to use GFP 1–10 as a targeting platform. Thus, we used functionalized synthetic M3 peptides as vehicles for site-directed posttranslational modification of split-GFP fusion proteins in living cells, using GFP fluorescence as an effective signal of the targeting specificity (Fig. 5A). Cells expressing GFP 1–10-CD4 proteins were efficiently complemented by M3-A647 peptide conjugates, and GFP fluorescence was specifically activated at the plasma membrane, which was colabeled with M3-A647 (Fig. 5B). Relying on the absence of colocalizing GFP signal, we could also easily identify a few intracellular vesicles containing endocytosed and noncomplemented M3-A647 peptides observed in the Alexa 647 channel for both expressing and nonexpressing cells. The 1:1 stochiometry complementation of GFP 1–10-CD4 by M3-A647 was clearly observed by dual-color TIRF imaging. At the plasma membrane, single complemented A647-GFP-CD4 proteins emitted in both detection channels and diffused as individual colocalizing spots in a correlated manner before photobleaching in a single step (Fig. 5C and Movies S9 and S10). The presence of a few noncolocalizing GFP and M3-A647 molecules was due to incidental photobleaching of one of two probes in A647-GFP-CD4 proteins during imaging or to rare nonspecific membrane binding of M3-A647 peptides. Using the complementation-induced coincident SM detection of both probes as a selection criterion, we could focus our analysis only on specifically targeted M3-A647 peptides, rejecting fluorophores nonspecifically bound to the plasma membrane. A second advantage of this dual detection scheme was that single proteins could be tracked with brighter fluorophores and for longer times than afforded by the complemented GFP alone. For instance, tracking M3-A647 sometimes allowed for a doubling of the tracking time, even after photobleaching of GFP in A647-GFP-CD4 proteins (Fig. 5C).
Fig. 5.
Addressable live cell targeting and tracking of single fluorophores and qdots by CALM. (A) Schematic of CALM with fluorescent M3 peptide conjugates and M3 peptide-coated qdots. (B) Labeling of a GFP 1–10-CD4–expressing U2OS cells (+) with M3-A647 peptide conjugates. (Scale bar: 10 μm.) (C) Coincident dual-color detection of single diffusing A647-GFP-CD4 proteins by TIRF in U2OS cells. Colocalizing diffraction-limited spots are simultaneously detected in the GFP and M3-A647 channels (colored asterisks). During diffusion, GFP photobleaches in a single step (frames 6 and 11), but A647-GFP-CD4 proteins can still be tracked in the M3-A647 channel before disappearing on M3-A647 photobleaching (frame 13). The diffraction-limited spots are intentionally expanded to facilitate visualization (Movie S10). (Scale bar: 1 μm.) (D) Wide-field fluorescence imaging of peptide-coated CdSe/ZnS M3-qdots specifically targeted to U2OS cells expressing GFP 1–10-CD4 fusion proteins (+). The M3-qdot image is a pixel-based maximum intensity projection of diffusing M3-qdots (ΣImax) for all frames of Movie S11. (Scale bar: 15 μm.) (E) Representative trajectories from qdot tracking of single complemented qdot-GFP-CD4 proteins diffusing in the plasma membrane of U2OS cells.
Minutes-long tracking of single split-GFP fusion proteins was also achieved by complementation-induced targeting of highly photostable qdot probes. M3-qdots were produced by coating hydrophobic CdSe/ZnS qdots (545 nm emission, ∼4 nm core/shell diameter) with synthetic peptides containing the C-terminal M3 sequence and an N-terminal semiconductor binding domain (25, 26) (FCC-M3). These compact peptide-coated M3-qdots (∼10 nm diameter) specifically labeled cells expressing the transmembrane GFP 1–10-CD4 fusion proteins (Fig. 5D), and complemented qdot-GFP-CD4 could be tracked for extended periods (Fig. 5E and Movie S11). Despite their size and the reduced conformational freedom of surface-attached M3 peptides, small M3-qdots could efficiently target split-GFP fusion proteins in living cells, and diffusing qdots were easily identified on labeled cells (Movie S11). The presence of a few qdots bound to the fibronectin-coated coverslips or nonexpressing cells was due to the 1-h-long incubation time. The complementation efficacy was, however, significantly reduced for CdSe/ZnS qdots with core/shell sizes above ∼4–5 nm, suggesting that M3 peptide accessibility and flexibility need to be further optimized for larger qdots. Although the insufficient spectral separation between qdots and GFP in complemented qdot-GFP-CD4 prevented SM coincident imaging in cells, coincident detection of one or multiple copies of complemented GFP-biotin bound to individual red-emitting streptavidin-coated qdots was achieved by in vitro TIRF imaging (SI Appendix, Fig. S10).
Single Biomolecule Tracking by Complementation-Induced Intramolecular spFRET in Living Cells.
An additional imaging modality afforded by CALM involved intramolecular spFRET between a complemented split-GFP fusion protein and an acceptor fluorophore attached to an M3 peptide. Although we intentionally chose the red-shifted Alexa 647 dye to limit spectral overlap between complemented GFPs and M3-A647 conjugates (SI Appendix, Fig. S11), significant FRET from A647-GFP complexes was observed in native gels (Fig. 6A and SI Appendix, Fig. S11). The mean FRET efficiency in solution, determined from changes in GFP fluorescence lifetime was E ∼ 0.3, indicating that the M3-attached Alexa 647 fluorophore is less than 4 nm away from the GFP chromophore after complementation. In living cells, intramolecular spFRET was confirmed by detecting Alexa 647 fluorescence at the plasma membrane of U2OS using 488-nm TIRF excitation of GFP 1–10-CD4 fusion proteins complemented with M3-A647 peptides (Fig. 6B and Movie S12). Under the same 488-nm excitation, cells complemented with nonfluorescent M3-biotin showed no FRET but an expected brighter GFP signal. Single complemented A647-GFP-CD4 proteins were identified as diffusing colocalized spots that emitted in both detection channels (Movie S13), and direct evidence for spFRET was obtained by plotting GFP-CD4 and M3-A647 fluorescence intensity time traces along the diffusion path of individual A647-GFP-CD4 proteins (Fig. 6C). Consistent with intramolecular spFRET, FRET signal was altogether lost on single-step GFP photobleaching (Fig. 6C). When M3-A647 photobleached before GFP-CD4, a large anticorrelated increase in GFP intensity was immediately observed (Fig. 6C).
Fig. 6.
Cell imaging and single biomolecule tracking by complementation-induced intramolecular spFRET. (A) In vitro native gel shift assay of M3-A647 peptide binding to soluble GFP 1–10 (+) or in TGN buffer (−). The gel is sequentially imaged for M3-A647, GFP, and A647-GFP intramolecular FRET emission. (B) Live cell imaging of GFP 1–10-CD4 proteins complemented with fluorescent M3-A647 (Left) or nonfluorescent M3-biotin (Right) peptides and imaged by dual-color TIRF microscopy using only 488-nm excitation. Images are pixel-based maximum intensity projections of diffusing A647-GFP-CD4 proteins (ΣImax) for all frames of Movie S12. (Scale bar: 5 μm.) (C) GFP (green) and M3-A647 fluorescence time traces (red) along the diffusion path (Right) of individual A647-GFP-CD4 proteins showing intramolecular spFRET. Fluorescence background traces (gray) are taken in the immediate vicinity of the trajectories. The single-step photobleaching of GFP (Upper, green arrow) or M3-A647 (Lower, red arrow) induces an arrest of intramolecular spFRET (Movie S13). (D) Live cell TIRF imaging and tracking of individual A647-GFP-CD4 proteins by spFRET at high M3-A647 concentrations (0.7 μM) without washing. A cell is sequentially imaged using direct M3-A647 excitation at 638 nm (Left) and then, indirect spFRET excitation at 488-nm laser (Right). Individual A647-GFP-CD4 proteins diffusing in the plasma membrane can be tracked in the M3-A647 channel using indirect spFRET excitation (red trajectories and white squares) but are lost in the saturating surrounding fluorescent signal when directly excited at 638 nm (red squares). Three representative examples of A647-GFP-CD4 trajectories are presented (from white squares). (Scale bar: 10 μm.)
By combining CALM and indirect spFRET excitation of M3-A647, we could specifically label and track CD4 proteins in living cells using very high fluorophore concentrations and no washing. Indeed, the large-excitation Stoke shift (>150 nm) afforded by 488-nm spFRET excitation prevented the excitation of nonbound M3-A647, which remained dark when excited by TIRF despite concentrations of ∼1 μM in the imaging buffer of GFP 1–10-CD4–expressing cells. When complemented A647-GFP-CD4 proteins were directly excited at 638 nm, the elevated M3-A647 concentration in the buffer prevented the detection of individual proteins (Fig. 6D). However, on 488-nm excitation, individual A647-GFP-CD4 proteins were specifically detected in both the GFP and Alexa 647 channels, with no interference from excess nonbound M3-A647 peptides (Fig. 6D and Movie S14). In the Alexa 647 channel, diffusing A647-GFP-CD4 proteins undergoing spFRET could easily be tracked, despite M3-A647 concentration orders of magnitude higher than normally required for SM imaging.
Discussion
CALM is a versatile approach for very high confidence targeting and imaging of single biomolecules in living cells. Its use of an asymmetric split-GFP is advantageous, because one of the fragments can be produced synthetically with great flexibility for advanced designs. The ability to reconstruct and activate dark split-GFPs fusion proteins into bright GFPs by simple external addition of self-complementary synthetic peptides provides a flexible means for low-background and ultra-specific imaging of a controlled subset of biomolecules, even when they are highly expressed. Indeed, in CALM, both the stochastic binding of M3 peptides and the chromophore maturation time, which drive GFP appearance, are key to spreading protein activation in time and space and to facilitating low-density tracking. Thus, by simply adjusting the concentration of complementary peptides and incubation times, hundreds to thousands of individual biomolecules can be continuously recorded, or the entire population can be imaged.
As shown for three different proteins in a variety of cell lines, CALM imaging requires a simple GFP 1–10 fusion and relatively inexpensive synthetic peptides. The proper organelle association and diffusive behaviors of the N- and C-terminal split-GFP fusion proteins that we have tested indicate that CALM should easily apply to other extra- and intracellular proteins. CALM is, in principle, simpler than cell-labeling techniques such as Bir-A ligase (27), PRIME (28), ACP-tag (29), or Sortase (30), which necessitate enzyme-assisted ligation. An additional advantage of CALM over these techniques and other labeling methods (31–34) is that no washing of excess probes is necessary, because fluorescence is only generated on GFP complementation and specific detection of only the targeted proteins can be achieved by CALM-spFRET. In this respect, CALM resembles the recently introduced fluorogen-activating single-chain antibodies (35), with the important advantages that our approach allows irreversible linkage, does not require extensive protein or probe engineering for multiplexing, enables SM imaging, and works efficiently inside living cells, where the reducing cytoplasmic environment might interfere with the activity of some single-chain antibodies (35).
Compared with tracking with photoswitchable or photoactivable FPs (6, 36) (sptPALM), where a large number of SMs can be imaged in live cells immediately after laser activation, the complementation kinetics of our current split-GFP can generate a short delay (∼2–3 min) between the addition of complementary peptides and the detection of statistically significant SM events during CALM imaging. However, at longer incubation times and appropriate peptide concentrations, the average SM activation and tracking density are similar to those of sptPALM. In addition, CALM has a few advantages over sptPALM. For instance, it does not require additional lasers other than the imaging 488-nm laser for SM detection, which simplifies optical setups. Importantly, split-GFP fusion proteins are dark and do not undergo self-activation, even at elevated protein expression, which is contrary to some photoswitchable FPs (37). Finally, the stochastic binding of peptides and maturation time in CALM make it unlikely that multiple split-GFP fusion proteins packed in a subcellular structure be simultaneous activated. Thus, CALM might offer a favorable alternative to sptPALM for live imaging of single biomolecules associated with highly crowded cellular nanostructures, which is shown by our ability to track individual cav1-GFP(h) within 50- to 100-nm caveolae containing ∼150 copies of cav1 (38). Current limitations of CALM include the need for peptide carriers (SI Appendix, Fig. S8) or microinjection for intracellular delivery and imaging and limited spatial control of the activation compared with sptPALM. Some of these issues might be solved by advanced engineering of complementary synthetic peptides.
An important specificity of CALM is that individual proteins are not only detected and imaged with nanometer precision but are also irreversibly modified, posttranslationally, within living cells. Engineered complementary peptides can, thus, be used as vectors for site-directed and stochiometric targeting of exogenous chemical moieties and probes to proteins of interest in living cells, as shown here with M3-biotin, M3-A647, and M3-qdots. With CALM, the built-in feedback on targeting efficiency provided by the appearance of GFP allows for a minimization of analytical artifacts related to nonspecific binding of probes, which is often unpredictable at the SM level in cells. Using coincident SM detection, mislocalized probes can be filtered out of the analysis or altogether ignored if imaging is performed by CALM-spFRET. Although we showed CALM-spFRET for external proteins only, this approach should greatly simplify SM imaging inside living cells and in living animals. Indeed, by allowing specific tracking of single biomolecules independently of their expression level and at very high probe concentrations, CALM-spFRET alleviates issues associated with inappropriate protein expression levels or inadequate intracellular or intravital probe delivery.
As for all imaging methodologies that use FPs, SM tracking by CALM or CALM-spFRET is limited by the photostability of GFP. However, as we have shown, longer tracking can be achieved using CALM-targeted fluorophores or qdots. CALM should be easily extended to other photostable SM probes like quantum rods, nanodiamonds, or fluorescent beads functionalized with M3 peptides, and it will be useful for light-assisted targeting of other nanomaterials. For these nanomaterials, including qdots, CALM readily solves the complicated issues of surface monofunctionalization (39, 40). Indeed, as we have shown, counting complemented GFPs on a single probe provides a direct readout of its valency and can be used as a criterion to filter out cross-linked probes from the analysis or correlate the diffusive behavior of a protein with its clustering state.
Here we have used CALM with split-GFP only, but the technique is, in principal, extendable to other variants such as split-CFP or split-YFP, which are also derived from Aequorea Victoria GFP. These variants should facilitate multicolor SM imaging in living cells using a unique M3 complementary synthetic peptide common to all these split-FPs. Additional developments of red-shifted split-FPs will also broaden the panel of genetically encoded probes for CALM. Sets of split-FPs, sets of complementary M3 peptide fluorophore conjugates, and sets of M3-qdots will constitute a modular toolkit for easy tailoring of SM probes to specific biological applications in vitro, in cells, and in vivo.
In conclusion, CALM uniquely combines targeting, imaging, and addressable posttranslational synthetic modification of biomolecules in living cells with SM sensitivity and nanometer precision. In addition to allowing ever more controlled SM imaging in complex biological environments, CALM also provides new bioimaging and manipulation modalities for cellular biology.
Methods
Design of Synthetic Split-GFP M3 Complementation Fragments.
Complementary peptides to GFP 1–10 and GFP 1–10 fusion proteins were designed based on the GFP 11 M3 sequence published by Cabantous et al. (10). All synthetic peptides were obtained at >70% or >95% purity (Biomatik or New England Peptide), and their identity and purity were confirmed by MS. A biotinylated M3 peptide (biotin-M3) was designed with an N-terminal biotin, an amidocaproyl (acp) linker, a short 8-aa linker, and the C-terminal M3 amino acid sequence (underlined) biotin-acp-GSGGGSTSRDHMVLHEYVNAAGIT (MW = 2,756 Da). A cysteine-terminated M3 peptide (Cys-M3) was used for conjugation to fluorophores such as Alexa Fluor 647 C2 maleimide (Invitrogen). Cys-M3 contains an N-terminal cysteine residue, an acp linker, a short 8-aa peptide linker, and the C-terminal M3 sequence (underlined) C-acp-GSGGGSTSRDHMVLHEYVNAAGIT (MW = 2,633 Da). For the functionalization of qdots with M3 peptides, we designed a synthetic peptide based on engineered cysteine-rich peptides capable of specifically binding on the semiconductor surface of CdSe/ZnS qdots (25, 26). This peptide (FCC-M3) contains an acetylated N terminus, an N-terminal qdot binding domain (in bold), a flexible and pegylated amino acid linker, and the C-terminal M3 sequence (underlined) ac-FCCFCCFCCFGGSESG-peg6-GSGGGSTSRDHMVLHEYVNAAGIT (MW = 4,476 Da).
Cell Transfection, Staining, and Imaging.
All cell lines (U20S, HEK, and COS-7) were cultured in DMEM + 10% fetal calf serum (FCS) at 37 °C in 5% CO2. Transient and stable transfections with GFP 1–10-CD4, GFP 1–10(h)-GPI, and cav1-GFP 1–10(h) expression vectors were performed with lipofectamine (Invitrogen) or fugene (Roche) reagents. To visualize cells expressing cav1-GFP 1–10(h) before microinjections of the complementary M3 peptides, the cav1-GFP 1–10(h)-N1 expression plasmid was transiently cotransfected with either a plasmid encoding the nuclear CFP-LacI-NLS or a plasmid encoding the actin-binding peptide mCherry-LifeAct. No cotransfection was performed for immunolabeling of cav1-GFP 1–10(h)–expressing cells. Detailed experimental protocols for ensemble and SM extracellular and intracellular staining with biotin-M3, Cys-M3, FCC-M3, and M3-A647 peptides, M3-qdots, Alexa-647–labeled anti-GFP, and Alexa-647–labeled streptavidin are available in SI Appendix, SI Methods. Cells grown on fibronectin-coated glass coverslips to 70–80% confluency were imaged in Tyrode's (136 mM NaCl, 10 mM KCl, 0.4 mM MgCl2, 1.0 mM CaCl2, 5.6 mM Glucose, 10.0 mM Hepes, pH 7.8) or Hepes-buffered HBSS (145 mM NaCl, 5 mM KCl, 1.2 mM MgSO4, 2.0 mM CaCl2, 1.2 mM NaH2PO4, 10 mM glucose, 20.0 mM Hepes, pH 7.6) at 37 °C.
Optical Setups.
Wide-field epifluorescence imaging was performed on an IX70 Olympus inverted microscope equipped with a ×100, 1.45 NA objective, a UV lamp, and appropriate optical filters for imaging CFP, GFP, mCherry, or Alexa Fluor 647. Fluorescence was detected on a QuantEM:512SC EMCCD camera (Photometrics). TIRF imaging was performed on the same microscope using a custom-built optical setup allowing simultaneous dual-color laser excitation at 488 and 638 nm and simultaneous dual-color detection through a DV2 Dual-View system (Photometrics) equipped with appropriate filters and mirrors for the detection of GFP and Alexa 647 on the EMCCD camera. Coincidence SM imaging and FRET imaging of A647-GFP-CD4 complexes in cells were performed by TIRF using simultaneous dual excitation at 488 and 638 nm or single excitation at 488 nm, respectively, and dual-color detection, respectively.
Tracking and Diffusion Analysis of Single Split-GFPs, Single Fluorophores, and Single Qdots in Living Cells.
All SM tracking and subsequent analyses were done using a series of homemade software called AsteriX and written in Labview (22). Tracking was done by 2D Gaussian fitting of individual diffraction-limited spots corresponding to single complemented split-GFPs, single fluorophores, or single qdots in each frame of the acquired videos. Single trajectories are represented by the fitted positions connected by a straight line. The mean trajectory lengths are reported in seconds ± SD of the mean (SI Appendix, Table S1). The localization uncertainty for single proteins was estimated as previously described (41–43) and is reported as a mean value in nanometer ± SD of the mean (SI Appendix, SI Methods and Table S1). The software also makes possible the exporting of a diffusion trajectory tracked in one channel (e.g., GFP channel) to a second channel acquired simultaneously (e.g., Alexa 647 channel). After image correction and alignment, it is possible to obtain M3-A647 intensity–time traces along the diffusion path of a complemented GFP 1–10 fusion protein. Using this approach, single-pair FRET signals from diffusing A647-GFP-CD4 proteins were obtained by correlating fluorescence intensity time traces from both Alexa 647 and GFP channels along the diffusion trajectory of single proteins.
Diffusion analyses were performed as previously described (22) on ensemble MSD curves (SI Appendix, Fig. S6) and ensemble histograms of probability distribution of the square displacements (Pr2) (19). Diffusion coefficients were obtained by fitting the MSD and Pr2 curves on the first four nonzero points of the curves (D1–D4) using a simple Brownian diffusion model with measurement error: 4σ2 + 4Dt. Diffusion coefficients are reported in micrometers squared per second ± SD of the fit. Analyses by Pr2 also provide an additional set of parameters (αi), which indicates the fraction of each subpopulation detected. These fractions are reported in percentages (SI Appendix, Table S1).
Additional information on biochemical, analytical, and synthetic methods, in vitro complementation assays, cloning, cell labeling and imaging, optical setups, and other protocols is in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Geoff Waldo for providing an aliquot of the soluble GFP 1–10 plasmid, Mathieu Piel and Xavier Darzacq for providing ABP-Cherry and CFP-LacI-NLS plasmids, respectively, Alain Joliot for the gift of the CD4-2::spGFP1-10 plasmid, Ignacio Izzedin for help with the TIRF setup, and Xavier Michalet for insightful discussions. We also thank Joerg Enderlein and members of his group for help with FCCS experiments. F.P. acknowledges financial support from Marie Curie Intra-European Fellowship MEIF-CT-2006-040210, a European Molecular Biology Organization long-term fellowship, and a fellowship from Centre de Nanoscience Ile de France. M.D. acknowledges support from Fondation pour la Recherche Médicale.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 9735.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101929108/-/DCSupplemental.
References
- 1.Lord SJ, Lee HLD, Moerner WE. Single-molecule spectroscopy and imaging of biomolecules in living cells. Anal Chem. 2010;82:2192–2203. doi: 10.1021/ac9024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 3.Yano Y, Matsuzaki K. Tag-probe labeling methods for live-cell imaging of membrane proteins. Biochim Biophys Acta. 2009;1788:2124–2131. doi: 10.1016/j.bbamem.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 5.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc Natl Acad Sci USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heilemann M, van de Linde S, Mukherjee A, Sauer M. Super-resolution imaging with small organic fluorophores. Angew Chem Int Ed Engl. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- 8.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 11.Cabantous S, Waldo GS. In vivo and in vitro protein solubility assays using split GFP. Nat Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto S, Kudo K. Supramolecular control of split-GFP reassembly by conjugation of beta-cyclodextrin and coumarin units. J Am Chem Soc. 2008;130:9574–9582. doi: 10.1021/ja802313a. [DOI] [PubMed] [Google Scholar]
- 13.Kent KP, Childs W, Boxer SG. Deconstructing green fluorescent protein. J Am Chem Soc. 2008;130:9664–9665. doi: 10.1021/ja803782x. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 15.Van Engelenburg SB, Palmer AE. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat Methods. 2010;7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothberg KG, et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 17.Hailstones D, Sleer LS, Parton RG, Stanley KK. Regulation of caveolin and caveolae by cholesterol in MDCK cells. J Lipid Res. 1998;39:369–379. [PubMed] [Google Scholar]
- 18.Bush WS, Ihrke G, Robinson JM, Kenworthy AK. Antibody-specific detection of caveolin-1 in subapical compartments of MDCK cells. Histochem Cell Biol. 2006;126:27–34. doi: 10.1007/s00418-006-0144-y. [DOI] [PubMed] [Google Scholar]
- 19.Schütz GJ, Schindler H, Schmidt T. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys J. 1997;73:1073–1080. doi: 10.1016/S0006-3495(97)78139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fragoso R, et al. Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling. J Immunol. 2003;170:913–921. doi: 10.4049/jimmunol.170.2.913. [DOI] [PubMed] [Google Scholar]
- 21.Popik W, Alce TM. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry. Identification of a novel raft localization marker in CD4. J Biol Chem. 2004;279:704–712. doi: 10.1074/jbc.M306380200. [DOI] [PubMed] [Google Scholar]
- 22.Pinaud F, et al. Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic. 2009;10:691–712. doi: 10.1111/j.1600-0854.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 24.Thomsen P, Roepstorff K, Stahlhut M, van Deurs B. Caveolae are highly immobile plasma membrane microdomains, which are not involved in constitutive endocytic trafficking. Mol Biol Cell. 2002;13:238–250. doi: 10.1091/mbc.01-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinaud F, King D, Moore HP, Weiss S. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J Am Chem Soc. 2004;126:6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer G, Pinaud F, Tsay J, Weiss S. Solubilization of quantum dots with a recombinant peptide from Escherichia coli. Small. 2007;3:793–798. doi: 10.1002/smll.200600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen I, Howarth M, Lin WY, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 28.Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci USA. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.George N, Pick H, Vogel H, Johnsson N, Johnsson K. Specific labeling of cell surface proteins with chemically diverse compounds. J Am Chem Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. Site-specific protein modification on living cells catalyzed by Sortase. ChemBioChem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 31.Martin BR, Giepmans BNG, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 32.Lata S, Gavutis M, Tampé R, Piehler J. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J Am Chem Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 33.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 34.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 35.Szent-Gyorgyi C, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 36.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 37.Nowotschin S, Hadjantonakis AK. Use of KikGR a photoconvertible green-to-red fluorescent protein for cell labeling and lineage analysis in ES cells and mouse embryos. BMC Dev Biol. 2009;9:49–61. doi: 10.1186/1471-213X-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: Domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke S, et al. Covalent monofunctionalization of peptide-coated quantum dots for single-molecule assays. Nano Lett. 2010;10:2147–2154. doi: 10.1021/nl100825n. [DOI] [PubMed] [Google Scholar]
- 40.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82:041914-1–041914-13. doi: 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalet X, Lacoste TD, Weiss S. Ultrahigh-resolution colocalization of spectrally separable point-like fluorescent probes. Methods. 2001;25:87–102. doi: 10.1006/meth.2001.1218. [DOI] [PubMed] [Google Scholar]
- 43.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]