Abstract
Lineage progression in osteoblasts and chondrocytes is stringently controlled by the cell-fate–determining transcription factor Runx2. In this study, we directly addressed whether microRNAs (miRNAs) can control the osteogenic activity of Runx2 and affect osteoblast maturation. A panel of 11 Runx2-targeting miRNAs (miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) is expressed in a lineage-related pattern in mesenchymal cell types. During both osteogenic and chondrogenic differentiation, these miRNAs, in general, are inversely expressed relative to Runx2. Based on 3′UTR luciferase reporter, immunoblot, and mRNA stability assays, each miRNA directly attenuates Runx2 protein accumulation. Runx2-targeting miRNAs differentially inhibit Runx2 protein expression in osteoblasts and chondrocytes and display different efficacies. Thus, cellular context contributes to miRNA-mediated regulation of Runx2. All Runx2-targeting miRNAs (except miR-218) significantly impede osteoblast differentiation, and their effects can be reversed by the corresponding anti-miRNAs. These findings demonstrate that osteoblastogenesis is limited by an elaborate network of functionally tested miRNAs that directly target the osteogenic master regulator Runx2.
Keywords: osteogenesis, chondrogenesis, post-transcriptional regulation
Cell-fate determination and subsequent lineage progression of phenotype-committed cells are mediated by master regulatory transcription factors that integrate multiple cell-signaling inputs and generate epigenetic changes in chromatin to modulate gene expression. Transcription factors are components of positive and negative feedback loops that initiate or maintain the acquisition of distinct biological states. Epigenomic mechanisms, including attenuation of mRNA and protein expression by small noncoding microRNAs (miRNAs) (1), permit effective control of gene expression beyond genomic interactions between transcription factors and their cognate elements in gene promoters. The biological potency of miRNAs, which are generated by the RNA processing enzyme Dicer, is based on their ability to control mRNA accumulation and/or protein synthesis through specific interactions with the 3′UTRs of target genes (1). Gene regulatory networks involving transcription factors and miRNAs may mutually reinforce cell fates and support phenotypic maturation of lineage-committed cells.
Osteogenic differentiation provides an effective cell model in which to define both epigenetic and epigenomic mechanisms required for cell-fate determination and phenotypic differentiation. Differentiation of multipotent mesenchymal stem cells into the osteoblast lineage and maturation of osteoprogenitors are controlled by multiple extracellular ligands [e.g., BMPs, WNTs, and FGFs] (2–4) that direct the activities of key transcription factors, including Runx2, Osterix, and different classes of homeodomain proteins (5–8). Runx2 is a critical regulator of the osteogenic lineage, and its epigenetic functions modulate expression of bone-related genes (9, 10). Bone-specific transcription of Runx2 (5, 11) is autoregulated (12) and controlled by networks of homeodomain proteins (13, 14), helix–loop–helix factors (15, 16), and Ets factors and Zn finger proteins (17, 18) during osteoblastogenesis. Runx2 is also physiologically regulated by several cell-signaling pathways, including vitamin D3 (19), TGFβ/BMP2 (20, 21), and Wnt (22). These studies have resulted in a detailed molecular blueprint that clarifies how Runx2 gene expression is stimulated during mesenchymal stem cell differentiation. However, posttranscriptional mechanisms, including miRNA-dependent control (23, 24), also play critical roles during osteogenesis.
We recently have shown that conditional ablation of the Dicer gene in the osteoblast lineage, which prevents formation of mature miRNAs, causes overt skeletal phenotypes (25). Hence, miRNAs expressed in osteoblasts (referred to as “osteo-miRNAs”) are critical for osteoblast differentiation and regulated bone formation. Initial studies on the functional activity of specific miRNAs in bone have been reported (23, 24, 26–31), and now the fundamental roles of miRNAs and target protein networks during bone development must be defined in a broader context. In this study, we directly addressed the mechanisms that support posttranscriptional regulation of Runx2 by miRNAs. We examined the full complement of miRNAs predicted to target Runx2 mRNAs and assessed their roles in osteoblast maturation. Our findings define a program of miRNAs that control Runx2 in the mesenchymal lineage as a component of an intricate regulatory network of miRNAs that controls osteogenesis.
Results
Mesenchymal Lineage-Related Expression of miRNAs Predicted to Target the Osteogenic Transcription Factor Runx2.
To assess whether miRNAs control the osteogenic activity of Runx2, we applied three miRNA target prediction tools (i.e., TargetScan, PicTar, and RNA22) (32–34) to search for Runx2-targeting miRNAs. We identified 11 miRNAs that are predicted to target the mouse Runx2 3′UTR. All but one (miR-137) of these miRNAs also potentially target the human RUNX2 3′UTR (Fig. S1A), which is 80% conserved in mammals, although some of the seed sequences are present in different locations within the 3′UTR (Fig. 1A and Fig. S1 A and B).
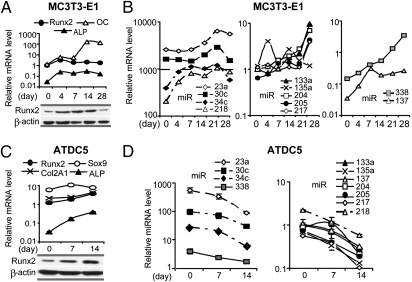
Fig. 1.
Expression profiles of predicted Runx2-targeting miRNAs in mouse osseous and nonosseous cells. (A) Seed regions of predicted Runx2-targeting miRNAs. (B) miRNA expression levels were analyzed in MC3T3-E1 osteoblasts, ATDC5 chondrocytes, and NIH 3T3 fibroblasts. miR-23a, miR-30c, and miR-34c are highly expressed in all cells, whereas miR-137, miR-218, and miR-338 display cell-type–specific expression. The remaining miRNAs are moderately expressed in all three cell types. Expression was normalized to U6 small RNA, and values reported are relative to miR-133a in MC3T3-E1 cells (set as 1). All values represent means ± SE (n = 3).
To test whether these predicted miRNAs are expressed in various mesenchymal lineages, we performed quantitative RT-PCR (qRT-PCR) analysis of total RNA in premature osteoblasts (MC3T3-E1), chondrocytes (ATDC5), and fibroblasts (NIH 3T3). Each of the three cell lines expresses characteristic cell-type–specific markers (Fig. S1C). Eight miRNAs are consistently expressed at either high (miR-23a, miR-30c, and miR-34c) or medium (miR-133a, miR-135a, miR-204, miR-205, and miR-217) levels (Fig. 1B, Left and Center), with the highly expressed miRNAs present on average at >1,000-fold higher levels in all three cell types. Three miRNAs are expressed in a clear cell-type–specific manner (miR-137, miR-218 and miR-338) (Fig. 1B, Right). Specifically, miR-137 and miR-338 are expressed at barely detectable levels in MC3T3-E1 osteoblasts but show ∼1,000-fold higher levels in ATDC5 chondrocytes (Fig. 1B). In contrast, miR-218 is highly expressed in MC3T3-E1 osteoblasts and NIH 3T3 fibroblasts but not in ATDC5 chondrocytes. Together, these data show that there appear to be several distinct classes of putative Runx2-targeting miRNAs that differ in their expression at basal or lineage-restricted levels.
Stage- and Cell-Type–Specific Modulation of Runx2-Related miRNAs During Osteogenic and Chondrogenic Lineage Progression.
We investigated whether there is a temporal correlation between expression of Runx2 and predicted miRNAs during differentiation of osteoblasts and chondrocytes. During MC3T3 differentiation, bone marker genes such as Runx2, alkaline phosphatase (ALP), and osteocalcin are significantly up-regulated (Fig. 2A, Upper). Runx2 protein is strongly increased with the onset of osteoblast differentiation (day 4) and maintained at high levels until day 28 (mineralization stage), at which point levels dramatically decrease (Fig. 2A, Lower). This modulation in expression and the discrepancy between Runx2 protein and mRNA accumulation at late stages of osteoblast maturation may be linked to miRNA expression targeting Runx2 mRNA.
Fig. 2.
Expression profiles of predicted Runx2-targeting miRNAs during differentiation of osteoblasts and chondrocytes. (A Upper) Differentiation markers of MC3T3-E1 cells including Runx2, ALP, and osteocalcin (OC) were detected by qRT-PCR. Values normalized to GAPDH are expressed relative to the Runx2 level on day 0 (set as 1). (Lower) Runx2 protein expression. (B) MiRNA expression during MC3T3-E1 osteoblast differentiation (day 0 to day 28). (C Upper) Differentiation of chondrocyte ATDC5 cells as reflected by activation of Sox9, collagen type II (Col2A1), Runx2, and ALP. Values normalized to GAPDH are expressed relative to the Runx2 level on day 0 (set as 1). (Lower) Runx2 protein expression. (D) Expression of Runx2-targeting miRNAs during ATDC5 chondrocyte differentiation (days 0, 7, and 14). Expression was normalized to U6 small RNA, and values reported are relative to miR-133a on day 0 (set as 1). All values represent means ± SE (n = 3).
During osteoblast differentiation, all 11 predicted Runx2-targeting miRNAs are up-regulated by day 28, compared with day 0 (Fig. 2B). However, the up-regulation of these miRNAs does not proceed concurrently. Highly expressed miRNAs (miR-34c and miR-218) are up-regulated at early stages of differentiation (days 0–7) and are increased by ∼10-fold (Fig. 2B, Left). As the osteoblasts mature (days 7–21), two other abundant miRNAs (miR-23a and miR-30c) are up-regulated (Fig. 2B, Left). In contrast, miRNAs expressed at lower levels are increased more robustly during differentiation. Four of these miRNAs (miR-133a, miR-204, miR-205, and miR-217) increase modestly up to day 21, then are up-regulated ∼10-fold on day 28 (Fig. 2B, Center). Notably, miR-137 and miR-338, which are barely expressed in proliferating MC3T3 cells, show an ∼100-fold increase in expression during differentiation (Fig. 2B, Right). These temporal and selective differences in miRNA expression suggest specific functions at diverse stages of differentiation. Up-regulation of a distinct set of miRNAs concomitant with decreased Runx2 protein levels at the late differentiation stage is consistent with the prediction that these miRNAs may directly regulate Runx2 during osteogenesis.
We also assessed expression of our miRNA panel during ATDC5 chondrocyte differentiation, reflected by increased mRNA expression of late markers Runx2 and ALP as differentiation proceeds (Fig. 2C, Upper). Runx2 protein levels are clearly up-regulated at day 14 (Fig. 2C, Lower), consistent with its role in hypertrophic chondrocytes. Remarkably, all miRNAs analyzed are dramatically down-regulated during ATDC5 differentiation (Fig. 2D). Expression of miRNAs decreases (Fig. 2D) while Runx2 protein levels increase (Fig. 2C, Lower) in differentiating ATDC5 cells. Thus, analogous to results with MC3T3-E1 osteoblasts, ATDC5 chondrocytes exhibit the same inverse correlation with respect to Runx2 levels. The consistent biological relationship between miRNA expression and Runx2 protein accumulation in both osteoblasts and chondrocytes corroborates the prediction that these miRNAs target Runx2 mRNA to attenuate protein translation.
3′UTR of Runx2 mRNA Is Directly Controlled by Multiple miRNAs.
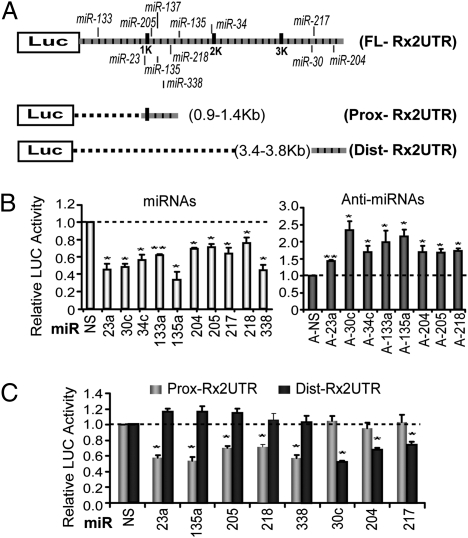
To investigate whether the 3′UTR of Runx2 mRNA is directly targeted by multiple miRNAs, we fused the full-length 3′UTR to the luciferase (LUC) reporter (FL-Rx2UTR). Because miRNA seed sequences cluster in the proximal and distal regions of the Runx2 3′UTR, we also separately tested the functional contributions of these regions in two other LUC/Runx2 3′UTR constructs, Prox-Rx2UTR and Dist-Rx2UTR (Fig. 3A). Endogenous miRNAs repress expression of all three UTR reporter constructs by ∼30–50% (Fig. S2A). All of the miRNAs showed repression of the full-length Runx2 3′UTR reporter activity by ∼20–60%, compared with a nonspecific (NS) miRNA (Fig. 3B, Left). Similar inhibitory effects of miRNAs on Runx2 3′UTR reporter activity were observed in mouse primary calvarial osteoblasts (Fig. S2B). Inhibition of endogenous miRNAs by addition of anti-miRNAs to the cells resulted in increased Runx2 3′UTR reporter activity (Fig. 3B, Right). Anti-miRNAs of miR-217 and miR-338 were not applied because their endogenous expression levels are very low in proliferating MC3T3-E1 cells (Fig. 1B). To verify the specificity of miRNA activity, we tested the proximal and distal Runx2 3′UTR reporters. The Prox-Rx2UTR reporter covers only five miRNA seed sites (for miR-23, 135, miR-205, miR-218, and miR-338). Transient expression of these miRNAs resulted in significant repression of Prox-Rx2UTR activity but did not affect Dist-Rx2UTR activity (Fig. 3C). Similarly, the Dist-Rx2UTR reporter contains seed sites for three other miRNAs (miR-30c, miR-204, and miR-217); these miRNAs inhibited the activity of the Dist-Rx2UTR reporter but not that of the Prox-Rx2UTR reporter. These results together show that miRNAs binding the distal and proximal Runx2 3′UTR selectively modulate expression by directly targeting distinct UTR domains.
Fig. 3.
Repression of Runx2 3′ UTR reporter activity by miRNAs in MC3T3-E1 cells. (A) LUC reporter vectors fused to full-length, proximal, and distal segments of the 3′ UTR of mouse Runx2 mRNA (FL-Rx2UTR, Prox-Rx2UTR, and Dist-Rx2UTR, respectively). (B) Specific miRNAs repress FL-Rx2UTR activity, whereas the corresponding anti-miRNAs increase LUC activity, compared with NS controls. (C) Each miRNA specifically inhibits the LUC activity of constructs containing the corresponding segment of the Runx2 3′ UTR containing the putative seed site. All values represent means ± SE (n = 3). *P < 0.05 and **P < 0.01, statistical significance compared with control groups treated with empty vector or NS miRNAs.
Endogenous Runx2 Protein Levels Are Diminished in a Cell-Type–Specific Manner by miRNAs That Target the 3′UTR of Runx2 mRNA.
To determine whether these miRNAs affect endogenous Runx2 expression in distinct cell types, we transfected these miRNAs into MC3T3-E1 osteoblasts and ATDC5 chondrocytes and examined Runx2 mRNA and protein levels. Except for miR-30c in MC3T3 and miR-133a in ATDC5 cells, these miRNAs repress Runx2 expression at the protein level but not the mRNA level (Fig. S3). It has been reported that miRNAs can exert translation repression or affect mRNA stability (35). We detected whether miR-30c and miR-133a alter Runx2 mRNA half life by inducing mRNA degradation. In MC3T3 cells that were treated with actinomycin D to inhibit transcription, neither miRNA affected Runx2 mRNA half life, suggesting that these two miRNAs cannot affect Runx2 mRNA stability (Fig. S4A).
Several miRNAs (miR-30c, miR-34c, miR-133a, miR-135a, and miR-338) strongly inhibit Runx2 protein expression in both MC3T3-E1 and ATDC5 cells, whereas miR-204 and miR-205 only modestly repress Runx2 protein expression in both cell lines (Fig. 4A). Interestingly, miR-23a and miR-218 diminish Runx2 expression only in ATDC5 cells, whereas miR-217 is a modest inhibitor of Runx2 in MC3T3 cells but more robust in ATDC5 cells (Fig. 4A). This more pronounced efficacy of miRNAs that target Runx2 in ATDC5 cells is generally consistent with the lower Runx2 protein levels in ATDC5 chondrocytes compared with MC3T3-E1 osteoblasts (Fig. 4B). We noted that miR-218 modestly increased Runx2 protein expression in MC3T3-E1 cells, which is inconsistent with its direct repression in the UTR assay (Fig. 3B). This finding suggests that Runx2 is also indirectly regulated by miR-218 in osteoblasts. Thus, although the miRNAs that are the focus of our study are directly rate-limiting for Runx2 protein expression, cell-type–specific cooperating factors and cellular context may also contribute to their inhibitory functions.
Fig. 4.
Cell-type–selective regulation of Runx2 protein expression by miRNAs. (A) Transfected miRNAs differentially regulate Runx2 protein expression in MC3T3-E1 and ATDC5 cells. After 72 h, samples were collected for Western blot analysis. Compared with NS miRNA, most of the miRNAs repress Runx2 expression in ATDC5 cells, but their relative efficacy is different in MC3T3-E1 cells. (B) Western blot analysis of Runx2 expression in MC3T3-E1 and ATDC5 cells. β-Actin is shown as a loading control. The data presented are representative of three experiments.
Runx2-Targeting miRNAs Inhibit Osteogenic Differentiation.
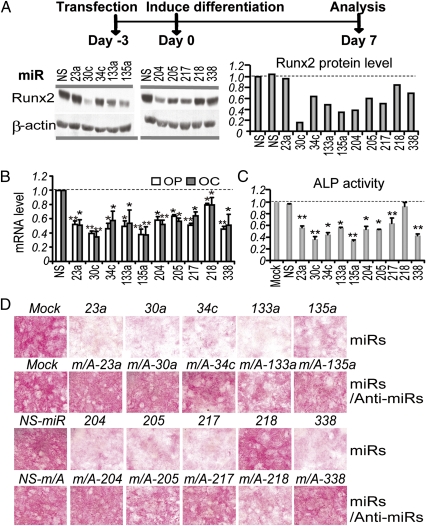
Because Runx2 is a master regulator of osteoblast differentiation, we investigated whether the Runx2-targeting miRNAs regulate osteogenic differentiation. We induced differentiation in MC3T3-E1 osteoblasts that were pretreated for 3 d with our miRNAs (Fig. 5A). After 7 d of differentiation, there was an ∼20–80% reduction of Runx2 protein by these miRNAs except for miR-23a (Fig. 5A). Compared with the NS miRNA, these miRNAs clearly repressed both the osteopontin and osteocalcin genes (Fig. 5B), which are classical markers of osteoblast differentiation as well as direct targets of Runx2. Furthermore, quantification of ALP activity revealed up to ∼50% reduction of this marker of osteoblasts, compared with the NS miRNA (Fig. 5C). In an independent study, histological analysis of miRNA-treated cells provided further corroborating evidence that all miRNAs except for miR-218 strongly repress ALP activity (Fig. 5D), consistent with the effects of these miRNAs on Runx2 expression (Fig. 5A). In addition, cotreatment of osteoblasts with each miRNA and its corresponding anti-miRNA (fourfold excess) reversed the miRNA inhibitory effects on osteogenic differentiation (Fig. 5D). Remarkably, although miR-23a does not reduce Runx2 protein expression in either proliferating (Fig. 4A) or differentiating MC3T3-E1 cells (Fig. 5A), it strongly inhibits ALP activity during osteogenic differentiation (Fig. 5 C and D) as well as expression of the osteoblast markers osteopontin and osteocalcin (Fig. 5B). We note that inhibition of osteogenesis by miR-23a is linked to other targets (30). More importantly, our results establish that at least 10 miRNAs that directly target Runx2 significantly inhibit osteogenic differentiation.
Fig. 5.
Runx2-targeting miRNAs inhibit osteoblast differentiation. MC3T3-E1 cells were transfected with miRNAs or combinations of miRNAs and their anti-miRNAs (day −3), and differentiation was induced when cells reached confluence (day 0). Differentiated cells (day 7) were collected for analyses of protein and RNA levels or histochemical assay and staining. (A) miRNA-dependent inhibition of Runx2 protein expression. (Right) Western blot quantification by AlphaImager 2200, normalized to the level of β-actin. The NS miRNA group was set as 1. (B) Repression of differentiation markers osteopontin (OP) and osteocalcin (OC) mRNA by miRNA. (C) Repression of ALP activity by miRNAs on day 7 of differentiation. Values represent means ± SE (n = 3). *P < 0.05 and **P < 0.01, statistical significance compared with the NS group. (D) Histochemical staining of ALP activity on day 7.
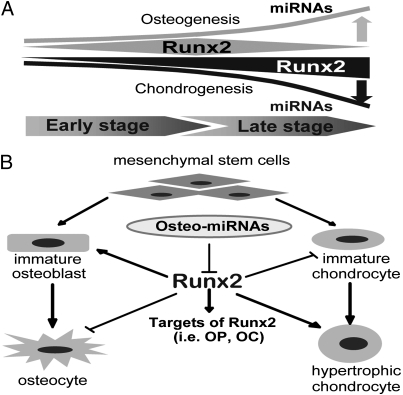
Our findings suggest a biological model of miRNA-dependent control of the protein levels and skeletal functions of Runx2 during osteoblast differentiation (Fig. 6). Expression of the majority of Runx2-targeting miRNAs is up-regulated during osteoblast differentiation, whereas the same panel of miRNAs is negatively regulated during chondrocyte differentiation. These profiles are consistent with the necessity to down-regulate Runx2 in osteocytes (final differentiation stage, day 28) and to up-regulate Runx2 for hypertrophic chondrocyte differentiation (day 14) (Fig. 6A). Runx2-targeting miRNAs are expressed at different levels during osteogenesis. Elevated levels of Runx2-targeting miRNAs during late stages of osteoblast maturation may account for the discordance between high Runx2 mRNA and low Runx2 protein levels. We conclude that Runx2 is a central node within a complex regulatory network involving 10 miRNAs that together provide a key mechanism for posttranscriptional regulation of physiological levels of Runx2 protein for normal osteogenesis and chondrocyte maturation (Fig. 6B).
Fig. 6.
Runx2-targeting miRNAs modulate osteogenesis and chondrogenesis. (A) miRNA regulation of Runx2 supports osteogenesis (gray) and chondrogenesis (black). Up-regulation of miRNA expression (upward line) during late stages of osteoblast differentiation may be responsible for the inhibition of Runx2 (narrowing polygon) that permits final stages of maturation. Conversely, decreased miRNA expression (downward line) may contribute to the up-regulation of Runx2 during chondrogenesis (widening triangle). (B) miRNAs that are selectively expressed in osteoblasts (osteo-miRNAs) regulate osteogenesis and chondrogenesis by directly targeting Runx2, a master gene during key stages in these two distinct biological processes. Runx2 is stimulatory for osteogenic lineage commitment (arrow) but inhibitory (blunted arrow) at late stages of osteoblast maturation. Attenuation of Runx2 by miRNAs inhibits bone marker gene expression (i.e., osteopontin and osteocalcin) during osteoblast differentiation.
Discussion
Here we investigated miRNA-mediated posttranscriptional control of Runx2 expression in mesenchymal cells. We characterized the full complement of miRNAs predicted to target the 3′UTR of Runx2 mRNA and showed that each miRNA selectively and directly controls Runx2 expression. Remarkably, both the expression and activity of this Runx2–miRNA network are exquisitely specific for cell type and developmental stage. Each Runx2-targeting miRNA (except miR-218) inhibits osteoblast differentiation. These findings corroborate our recent studies indicating that osteoblast-specific loss of the miRNA processing enzyme Dicer causes a dramatic high bone mass phenotype and that miRNAs are normally necessary for attenuating osteoblast maturation in vivo (25).
Because multiple Runx2-targeting miRNAs are potent inhibitors of Runx2 protein accumulation in either osteoblasts or chondrocytes, and are also effective inhibitors of osteoblast differentiation, one key question that emerges is why there is such a high level of functional redundancy in Runx2 inhibition by miRNAs. One advantage of using multiple miRNAs is that it provides broad options for biological control. The multiplicity of miRNAs that can target Runx2 ensures that there is always a subset of miRNAs available to support translational control at each stage of the osteoblast lineage. Several genetic mouse models have shown that nonphysiological levels of Runx2 protein or dosage insufficiency can have negative effects on bone. For example, transgenic mice expressing Runx2 develop osteopenia (36), and mice with a hypomorphic Runx2 allele exhibit cranial abnormalities (37). Thus, tight regulation of Runx2 protein by miRNAs is critical for normal bone formation.
Effective inhibition of mRNAs by miRNAs is mediated by multiple target sites instead of a single miRNA site (38). Furthermore, the putative seed sequences for miRNAs in the 3′UTRs of mRNAs are often clustered (39). Each of the Runx2-targeting miRNAs is predicted to have hundreds of other targets. Thus, regulation of Runx2 protein accumulation may be embedded within large networks linked to other biological processes that control skeletal development. The multiplicity of Runx2-targeting miRNAs suggests that there may be collaboration among distinct miRNAs that target the 3′UTR of the Runx2 mRNA. The potential for cooperation is exemplified by the coordinate expression of Runx2-targeting miRNAs during diverse stages of osteoblast differentiation.
We find clear differences in the efficacy with which the Runx2-targeing miRNAs block Runx2 protein expression in osteoblasts and chondrocytes. In general, our panel of different miRNAs exhibits more significant inhibition of Runx2 in chondrocytes, in which Runx2 is expressed at low levels, than in osteoblasts, in which Runx2 is highly expressed. The potency of miRNAs to reduce target protein levels could be altered in different cell types by generating differences in the 3′UTR of target mRNA (e.g., by alternative polyadenylation) (40). The seed sequences for some miRNAs (e.g., miR-30, miR-204, and miR-217) are all located downstream from a putative conserved polyadenylation site that could reduce the length of the Runx2 3′UTR and eliminate suppression of Runx2 by these three miRNAs. However, this postulated mechanism does not account for the observed cell-type dependence of miRNA activity in our studies because each of these miRNAs is effective in reducing Runx2 protein accumulation (albeit to different degrees) in osteoblasts and chondrocytes. Another possibility is that tissue-specific factors linked to miRNA-regulatory networks may provide a unique cellular context that alters the availability of seed regions or the inhibitory efficiency of miRNAs. Considerations of context in cells are necessary to fully understand the unique mechanisms by which miRNAs regulate gene expression. Thus, the finding that Runx2-targeting miRNAs down-regulate Runx2 more effectively in chondrocytes than in osteoblasts is consistent with the biological requirement for reducing Runx2 protein expression in the mesenchymal stem cell or the bipotential osteochondroprogenitor cell for commitment to chondrogenesis (9).
Our panel of 10 miRNAs defines a set of inhibitors that have as a common function suppression of the biological activity of a key lineage commitment factor in supporting cell maturation. Previous studies identified two of the miRNAs (miR-133 and miR-204) as direct regulators of Runx2 (23, 24). Only a few of the Runx2-targeting miRNAs have been characterized in other biological contexts. Our earlier studies revealed that miR-133a and miR-135a each inhibit BMP2-induced osteogenesis of C2C12 mesenchymal cells by targeting Runx2 and Smad5, respectively (41). Here we extend these results by showing that miR-135a mediates significant repression of osteogenesis by targeting not only Smad5 but also Runx2. Other studies revealed that a subset of Runx2-targeting miRNAs (i.e., miR-30, miR-34, miR-203, and miR-205) are associated with multiple cancers, including metastasis of breast and prostate tumors (41–44). Because Runx2 also contributes to breast and prostate cancer metastasis (45, 46), the possibility arises that Runx2 and its targeting miRNAs form a cancer-related biological network beyond the role of this network in osteochondrogenesis.
In conclusion, Runx2 expression must be stringently controlled at multiple gene regulatory levels to avoid skeletal phenotypes linked to either dosage insufficiency or overexpression. The network of Runx2-targeting miRNAs may be critical to ensure that Runx2 levels are attenuated at key stages of osteoblast lineage progression to accommodate its biological functions in mesenchymal cell-fate determination and maturation of osteoblasts.
Materials and Methods
Primers used for cloning are listed in Table S1. Detailed methods for cell culture and LUC sensor assays, RNA isolation, miRNA expression detection, and Western blot analysis are provided in SI Materials and Methods. Additional references are also provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our laboratory colleagues, including Yang Lou, Jonathan Gordon, Kimberly LeBlanc, Mohammad Hassan, and Dana Frederick, for advice on experimental procedures. We also thank Judy Rask for manuscript preparation. We appreciate the gift from Anne Terry and Jim Neil of a cDNA construct containing mouse Runx2 3′UTR sequences. This work was supported by National Institutes of Health Grants R01AR039588 (to G.S.S. and J.B.L.), R01AR049069 (to A.J.v.W.), and R37DE012528 (to J.B.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018493108/-/DCSupplemental.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canalis E, Deregowski V, Pereira RC, Gazzerro E. Signals that determine the fate of osteoblastic cells. J Endocrinol Invest. 2005;28(Suppl):3–7. [PubMed] [Google Scholar]
- 3.Bergenstock MK, Partridge NC. Parathyroid hormone stimulation of noncanonical Wnt signaling in bone. Ann N Y Acad Sci. 2007;1116:354–359. doi: 10.1196/annals.1402.047. [DOI] [PubMed] [Google Scholar]
- 4.Marie PJ. Fibroblast growth factor signaling controlling osteoblast differentiation. Gene. 2003;316:23–32. doi: 10.1016/s0378-1119(03)00748-0. [DOI] [PubMed] [Google Scholar]
- 5.Lian JB, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima K, et al. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 7.Hassan MQ, et al. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 9.Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010;339:189–195. doi: 10.1007/s00441-009-0832-8. [DOI] [PubMed] [Google Scholar]
- 10.Stein GS, et al. Transcription-factor-mediated epigenetic control of cell fate and lineage commitment. Biochem Cell Biol. 2009;87:1–6. doi: 10.1139/o08-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruzat F, et al. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009;48:7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drissi H, et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Sierra OL, Towler DA. Runx2 trans-activation mediated by the MSX2-interacting nuclear target requires homeodomain interacting protein kinase-3. Mol Endocrinol. 2010;24:1478–1497. doi: 10.1210/me.2010-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan MQ, et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J Cell Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The Notch-responsive transcription factor Hes-1 attenuates osteocalcin promoter activity in osteoblastic cells. J Cell Biochem. 2009;108:651–659. doi: 10.1002/jcb.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) J Biol Chem. 2009;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hesse E, et al. Zinc finger protein 521, a new player in bone formation. Ann N Y Acad Sci. 2010;1192:32–37. doi: 10.1111/j.1749-6632.2009.05347.x. [DOI] [PubMed] [Google Scholar]
- 19.Drissi H, et al. 1,25-(OH)2-vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Exp Cell Res. 2002;274:323–333. doi: 10.1006/excr.2002.5474. [DOI] [PubMed] [Google Scholar]
- 20.Selvamurugan N, Kwok S, Alliston T, Reiss M, Partridge NC. Transforming growth factor-β1 regulation of collagenase-3 expression in osteoblastic cells by cross-talk between the Smad and MAPK signaling pathways and their components, Smad2 and Runx2. J Biol Chem. 2004;279:19327–19334. doi: 10.1074/jbc.M314048200. [DOI] [PubMed] [Google Scholar]
- 21.Jun JH, et al. BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J Biol Chem. 2010;285:36410–36419. doi: 10.1074/jbc.M110.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaur T, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaur T, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340:10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inose H, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem. 2009;284:19272–19279. doi: 10.1074/jbc.M109.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan MQ, et al. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oskowitz AZ, et al. Human multipotent stromal cells from bone marrow and microRNA: Regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 33.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 35.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geoffroy V, Kneissel M, Fournier B, Boyde A, Matthias P. High bone resorption in adult aging transgenic mice overexpressing Cbfa1/Runx2 in cells of the osteoblastic lineage. Mol Cell Biol. 2002;22:6222–6233. doi: 10.1128/MCB.22.17.6222-6233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lou Y, et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum Mol Genet. 2009;18:556–568. doi: 10.1093/hmg/ddn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John B, et al. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, et al. MiRNA–miRNA synergistic network: Construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. 2011;39:825–836. doi: 10.1093/nar/gkq832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu F, et al. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 43.Saini S, et al. Regulatory role of miR-203 in prostate cancer progression and metastasis. Clin Cancer Res. December 15, 2010 doi: 10.1158/1078-0432.CCR-10-2619. 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 44.Merkel O, Asslaber D, Piñón JD, Egle A, Greil R. Interdependent regulation of p53 and miR-34a in chronic lymphocytic leukemia. Cell Cycle. 2010;9:2764–2768. [PubMed] [Google Scholar]
- 45.Pratap J, Lian JB, Stein GS. Metastatic bone disease: Role of transcription factors and future targets. Bone. 2011;48:30–36. doi: 10.1016/j.bone.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akech J, et al. Runx2 association with progression of prostate cancer in patients: Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–821. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.