Abstract
Significant efforts have been devoted to the development of nanoparticular delivering systems targeting tumors. However, clinical application of nanoparticles is hampered by insufficient size homogeneity, difficulties in reproducible synthesis and manufacturing, frequent high uptake in the liver, systemic toxicity of the carriers (particularly for inorganic nanoparticles), and insufficient selectivity for tumor cells. We have found that properly modified synthetic analogs of transmembrane domains of membrane proteins can self-assemble into remarkably uniform spherical nanoparticles with innate biological activity. Self-assembly is driven by a structural transition of the peptide that adopts predominantly a beta-hairpin conformation in aqueous solutions, but folds into an alpha-helix upon spontaneous fusion of the nanoparticles with cell membrane. A 24-amino acid peptide corresponding to the second transmembrane helix of the CXCR4 forms self-assembled particles that inhibit CXCR4 function in vitro and hamper CXCR4-dependent tumor metastasis in vivo. Furthermore, such nanoparticles can encapsulate hydrophobic drugs, thus providing a delivery system with the potential for dual biological activity.
Keywords: virus-like particles, CXCL12, cancer, undruggable target
Lack of means for selective delivery of therapeutic molecules to the right organ and cells remains a key problem in therapy of many diseases, including cancer. Useful lessons in specific cell and organ delivery can be learned from nature. Viral particles are able to deliver whole proteins and large nucleic acid molecules to certain types of cells. Two critical properties that enable viruses to deliver their cargo are their ability to self-assemble and disassemble when needed and their capacity to selectively enter only certain cell types. Genetically engineered virus-like particles have proved to be effective in eliciting immune responses and have been used as vaccines (1). Viral capsids have been explored as drug delivery systems (2). However, assembly of molecules generated by chemical synthesis into virus-like particles has yet to be achieved. Here, we describe fully synthetic nanoparticles that are formed by precise self-assembly of a biologically active synthetic peptide. Like viruses, nanoparticles disassemble upon fusion with the cell membrane, thus delivering their cargo to the interior of the cells. Self-assembly protects the peptide drug from degradation in the circulation and makes it effective in inhibiting tumor growth in an animal model of metastatic breast tumor disease. Self-assembling peptides provide fully synthetic fusogenic protoviral particles that can be used both as a drug and as a delivery vehicle with dual biological activity.
Synthetic analogs of transmembrane helixes have been found to inhibit the function of the corresponding integral membrane protein, allowing in several instances development of potent inhibitors for important drug targets (3–9). Synthetic helixes interfere with the correct assembly of the target membrane protein, thus rendering it inactive (6). Structure-activity studies have revealed that addition of carboxylates to the extracellular terminus significantly improves the potency of transmembrane inhibitors. Dehydration of negatively charged carboxylates requires lots of energy because they are among strongest known kosmotropes with the Jones–Dole viscosity B coefficient around 0.250 (10). That is why carboxylates pass cell membranes much less efficiently than positively charged or neutral amino acid residues. Consequently, carboxylates function as stop signals during membrane insertion and provide for correct orientation inside the membrane if placed on the extracellular end of the peptide (7, 8). We have now discovered that transmembrane peptides with additional negative charges on one of the termini self-assemble into spherical nanoparticles with surprising precision.
Results
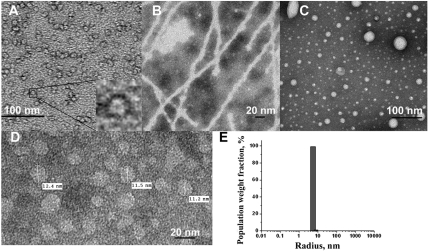
We have found that self-assembly of the transmembrane (TM) helixes of integral membrane proteins provides remarkably stable and homogeneous nanoparticles of nearly ideal size (10–20 nm) with innate biological activity. Electron microscopy has revealed that peptides derived from many membrane-spanning helices are capable of self-assembly in aqueous solution (Fig. S1). We chose a transmembrane antagonist of the CXCR4 chemokine receptor for detailed studies of TM peptide self-assembly, because CXCR4 is an important drug target and is implicated in tumor metastasis, particularly into bones and lung tissue, where expression of the CXCR4 agonist, the stromal cell-derived factor (SDF-1α) also known as C-X-C motif ligand 12 (CXCL12) is particularly high. Earlier studies have shown analogs of the second transmembrane helix of chemokine receptors to have potent and selective antagonistic effects on the corresponding receptor (7). Transmission electron microscopy studies revealed assembly of transmembrane peptides into uniform round nanoparticles. The particles, however, tend to aggregate, forming string-like structures (Fig. 1A). As a result, dynamic light scattering (DLS) data reflected two species of particles in solution with the larger one having high polydispersity (Table 1). To prevent this nanoparticle superaggregation, the original X4-2-1 sequence was augmented with polyethyleneglycol (PEG) extensions of different lengths at the C-terminus. Addition of 27 monomeric units of PEG (X4-2-6, Table 1) was sufficient to prevent superaggregation without interfering with self-assembly (Fig. 1 D and E) or biological activity. We found, however, that longer PEG tails obstructed fusion with cell membranes (Fig. S2) and thus reduced activity. To investigate the structural requirements for nanoparticle assembly, variants of the X4-2-1 peptide were synthesized and tested by DLS and transmission electron microscopy for their ability to form nanoparticles. Substitution of two C-terminal negatively charged aspartates of X4-2-1 with two positively charged lysine residues (X4-2-2, Table 1) resulted in aggregates that were very heterogeneous in size (Fig. 1C). X4-2-1 analog without two C-terminal aspartates (X4-2-3) had poor solubility and showed strong tendency to form super aggregates (Table 1). X4-2-3 derivative with PEG-27 on the C-terminus, X4-2-5 (Table 1) did form nanoparticles. However, they were more heterogeneous than the ones formed by the peptides with negative charges, X4-2-6. To test the importance of the N-terminal residues for nanoparticle assembly, N-terminal truncation mutants of the X4-2-6 peptide (X4-2-8, X4-2-9, and X4-2-10) were constructed (Table 1). Unexpectedly, removal of two N-terminal leucine residues (X4-2-9) resulted in the formation of fibrils instead of round particles (Fig. 1B), suggesting that these particular residues may be instrumental in nanoparticle assembly. We were able to observe the critical aggregation concentration (CAC) for this peptide by DLS, which was between 0.1 and 0.2 mg/mL, whereas X4-2-6 CAC was below the detection limit of DLS for this system, 0.02 mg/mL. Further shortening the peptide sequence by three amino acids (X4-2-10) resulted in nanostructures with an apparent radius of 1.6 nm. Truncation of the peptide by twelve amino acids (X4-2-8) produced a peptide that did not self-assemble at concentrations as high as 10 mg/mL.
Fig. 1.
Self-assembly of analogs of the CXCR4 second transmembrane helix. (A) Transmission electron microscopy of self-assembled X4-2-1 (LLFVITLPFWAVDAVANWYFGNDD). (B) X4-2-9 (FVITLPFWAVDAVANWYFGNDD-(CH2CH2O)27-NH2), just two residues shorter than X4-2-6, self-assembles into fibrils. (C) X4-2-2 (LLFVITLPFWAVDAVANWYFGNKK) assembled into nonuniform nanostructures. (D) Transmission electron microscopy of X4-2-6 (LLFVITLPFWAVDAVANWYFGNDD-PEG27). (E) DLS of X4-2-6, 0.4 mg/mL in PBS reveals remarkable size homogeneity.
Table 1.
Assessment of nanoparticle formation by CXCR4 TM2 derivatives
| Peptide | Structure | Particle hydrodynamic radii, nm* (population weight fraction) | Percentage polydispersity† | Apparent number of peptide molecules per particle |
| X4-2-1 | LLFVITLPFWAVDAVANWYFGNDD-OH | 6.0 (87.3%) 22.4 (12.1%) | 6.7 16.1 | 80 1746 |
| X4-2-2 | LLFVITLPFWAVDAVANWYFGNKK-NH2 | 66.5 (100%) | 68.7 | 20235 |
| X4-2-3 | LLFVITLPFWAVDAVANWYFGN-OH | 92.2 (100%) | 19.5 | 49369 |
| X4-2-5 | LLFVITLPFWAVDAVANWYFGN-(CH2CH2O)27-NH2 | 6.8 (99.9%) | 26.2 | 80 |
| X4-2-4 | LLFVITLPFWAVDAVANWYFGNDD-(CH2CH2O)11-NH2 | 5.5 (100%) | 11.8 | 42 |
| X4-2-6 | LLFVITLPFWAVDAVANWYFGNDD-(CH2CH2O)27-NH2 | 5.5 (99.9%) | 8.5 | 45 |
| X4-2-11 | LLFVITLPFWAVDAVANWYFGNDD-(CH2CH2O)27-CO-NH-(CH2CH2O)11-NH2 | 5.6 (99.9%) | 16.5 | 41 |
| X4-2-15 | LLFVITLPFWAVDAVANWYFGNDDD-(CH2CH2O)27-NH2 | 5.3 (99.9%) | 12.5 | 40 |
| X4-2-9 | FVITLPFWAVDAVANWYFGNDD-(CH2CH2O)27-NH2 | 11.1 (97.4%) 62.8 (2.5%) | 16.9 19.5 | 302 21817 |
| X4-2-10 | TLPFWAVDAVANWYFGNDD-(CH2CH2O)27-NH2 | 1.6 (100%) | 15.5 | 3 |
| X4-2-8 | AAVANWYFGNDD-(CH2CH2O)27-NH2 | N/A | N/A |
*The particle hydrodynamic radii were determined from DLS. The data presented is for 0.4 mg/mL solutions of peptides in PBS containing 1.25% (v/v) DMSO. Dynamics 6.7.7.9 software (Wyatt Technologies) was used to calculate molecular weight from the size data. The calculation was based on a globular protein model. Comparison of the calculated apparent molecular weight with the weight of the monomer provided the apparent number of peptide molecules per particle.
†Percentage polydispersity reflects the homogeneity of the nanoparticles. It is defined as a standard deviation of the size divided by the mean radius multiplied by 100%.
CXCR4 TM2 analogs with additions of 27 or more monomeric units of PEG showed remarkable size homogeneity (Fig. 1 D and E). The size of the particles showed no dependence on peptide concentration in 0.05–0.4 mg/mL concentration range (Fig. S3). The critical aggregation concentration appeared to be below 5 μM (0.02 mg/mL) and could not be determined by light scattering, titration calorimetry or analytical ultracentrifugation, because it was below the detection limits of the corresponding instrumentation. The zeta potential of X4-2-6 nanoparticles measured in 10 mM phosphate buffer at three applied voltages (100–150 V, ZetaSizer detector, Malvern Instruments, Inc.) was equal to -20.8 ± 1.0 mV, which is indicative of a stable structure with a strong negative surface charge. A peptide with an additional terminal aspartate (X4-2-15) had slightly increased zeta potential of -23.8 ± 2.0 mV and showed slightly improved size homogeneity upon prolonged storage (Fig. S4A). Dynamic light scattering revealed that nanoparticles formed by both peptides had unchanged apparent radii and dispersity for 21 d when stored at room temperature. Interestingly, peptide elongated with an additional PEG block consisting of 11 monomeric units (38 units in total), X4-2-11(Table 1) started aggregating much sooner, just in 8 d (Fig. S4A). When kept at 37 °C, self-assembled X4-2-6 showed no changes for at least 48 h (Fig. S4B), suggesting that nanoparticles remain intact at temperature conditions of biological assays. The half-life of self-assembled X4-2-6 in bovine serum was 9 h (Fig. S5). Truncated version of X4-2-6 that forms fibrils, X4-2-9 is less stable in serum and has a half-life of less than 2.5 h (Fig. S5). Increased degradation rates correlate with reduced stability of X4-2-9 self-assembling structure.
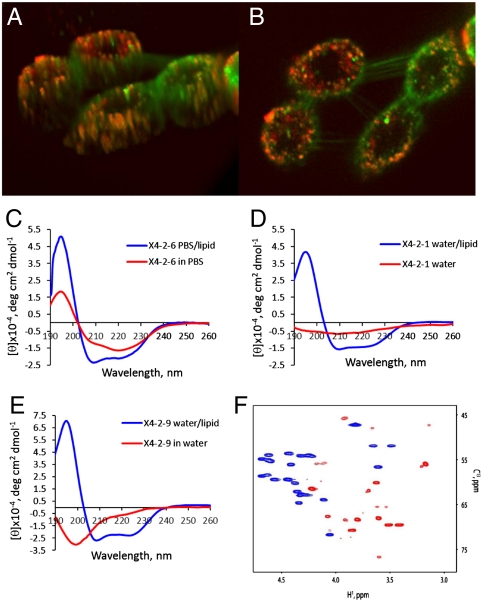
Laser scanning confocal microscopy studies with fluorescently labeled peptides showed that nanoparticular inhibitors fuse spontaneously with cell membranes and saturate all intracellular membranes within 10–15 min after application (Fig. 2 A and B). Addition of the C-terminal PEG slows down cell fusion, but does not change intracellular localization of the self-assembling peptides (Fig. S2). Spontaneous and efficient fusion of nanoparticles with a lipid bilayer was also demonstrated using dynamic light scattering and liposomes (Fig. S6). Nanoparticles (0.2 mg/mL solution) have been titrated with an increasing amount of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1, 2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG)/cholesterol liposomes. Both liposomes (with radii of 55 nm) and nanoparticles (with radii of 5.5 nm) could be identified in mixtures containing up to 20-fold excess of the peptide by weight compared to the lipids. DLS detected complete disappearance of nanoparticles when the amount of the lipids in the sample was 1/4 of the amount of the peptide, or higher suggesting an effective fusion of the nanoparticles with the liposomes.
Fig. 2.
Self-assembled particles fuse spontaneously with the cell membrane. Fusion with lipids is accompanied by transition in the peptide conformation from beta-type to helical. (A, B) Confocal laser scanning microscopy images of A549 cells labeled with membrane marker, DiOC18(3) (green) after 20 min treatment with 0.05 mg/mL rhodamine red derivative of X4-2-1 (red). Colocalization of the two appears as yellow or orange. (A) The 3D image of cells exposed to nanoparticles generated from Z-section scanning of the cells. (B) Optical planar section of the cells presented in (A). Peptide appears to saturate intracellular membranes. (C) CD spectra of X4-2-6 micelles demonstrate transition from predominantly beta-type conformation in aqueous solution into an α-helix upon fusion with the lipid. (D) CD spectra of nanoparticles-forming peptide with a tendency to super aggregation, X4-2-1 (D) CD spectra of a peptide that form fibrils, X4-2-9. (F) An overlay of Cα regions of 13C HSQC spectra of X4-2-1 in 100% DMSO - d6 (blue) and in dodecylphocphocholine-d28 micelles (red). A profound downfield shift of 13Cα resonances and an upfield shift of 1Hα resonances indicate that the X4-2-1 peptide adopts a helical conformation in dodecylphosphocholine micelles.
The presence of a distinctive negative band at approximately 220 nm and a positive band between 190 and 200 nm in circular dichroism spectra (Fig. 2C) strongly suggests that during self-assembly into homogeneous nanoparticles in aqueous solutions, X4-2-6 adopts a predominantly beta-type conformation. The peptide undergoes transformation into a roughly helical structure in membrane-mimicking dodecylphosphocholine micelles that is accompanied by the appearance of a characteristic negative band at 208 nm. CXCR4 TM2 derivative that forms nanoparticles with a tendency to form super aggregates, X4-2-1 is also predominantly helical in the micelles (Fig. 2D). However, aqueous solutions of X4-2-1 have CD spectra that suggest significant degree of disordering, which probably results from random aggregation of the nanoparticles. Truncated version of X4-2-6 that forms fibrils, X4-2-9 appears predominantly disordered in water, but is also helical in the micelles (Fig. 2E). 13C Heteronuclear single quantum coherence NMR spectra (Fig. 2F) further confirmed transformation of the predominantly beta-type fold of the nanoparticles-forming peptide into an α-helix upon fusion with lipid micelles.
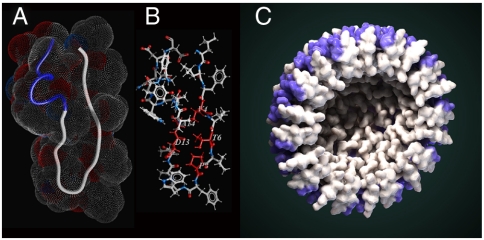
NMR studies of nanoparticular CXCR4 inhibitors revealed that X4-2-1 monomer in DMSO adopts a structure consisting of a β-like hairpin with two tight helix-like turns in the C-terminus (Fig. 3 A and B and Table S1). Amino acids responsible for stabilization of the hairpin structure include Val4, Thr6, Pro8, Asp13, and Ala14. The side-chain of Thr forms a hydrogen bond with the amide backbone hydrogen of Asp13, whereas Val4 and Ala14 are engaged in a hydrophobic interaction. The NMR spectra of nanoparticles in aqueous solution had much broader signals consistent with increased molecular weight. However, it was possible to identify nuclear Overhauser effects (NOEs) reflecting intramolecular proximity of Pro8 and Val12 as well as Pro8 and Ala11 (Fig. S7A). Both are possible only if the peptide retains the hair-pin conformation inside the nanoparticle. Light scattering studies suggest that homogeneous nanoparticles are formed by approximately forty peptide molecules (X4-2-6, Table 1). Electron microscopy images reveal almost perfectly round structures with a cavity in the center (Fig. 1A, Insert).
Fig. 3.
Structure of monomeric X4-2-1. (A) Overall fold of X4-2-1 represented as a tube with space-filling topology presented as a mesh. Blue part of the tube corresponds to the C-terminal alpha-helix. (B) The average structure of X4-2-1 showing side-chain positions. The residues stabilizing the hairpin conformation of X4-2-1 are colored red and their types and sequence positions are marked. (C) Hypothetical model of X4-2-1 self-assembly. Cross-section of the nanoparticle is presented. Alpha-helical portion of the peptide is highlighted in blue. Autodesk 3ds Max software was used for the generation of the model.
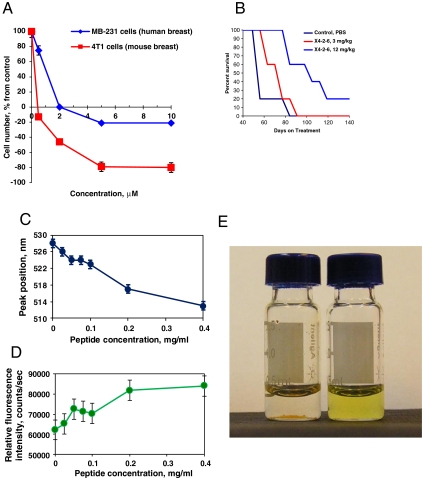
To explore the therapeutic potential of the CXCR4 TM peptide nanoparticles modified with PEG (X4-2-6 peptide-based nanoparticles), they were tested in a mouse model of metastatic breast cancer. CXCR4 is known to play a critical role in tumor metastasis in the lung and bone, in which expression of the CXCR4 agonist SDF-1α is particularly high (11). Nude mice were injected intravenously with highly metastatic MDA-MB-231 breast cancer cells. The mice were divided into three groups. The control group received phosphate buffered saline solution intraperitoneally once every 3 d. The remaining two groups were treated once every 3 d with X4-2-6 peptide-based nanoparticles at doses of 3 mg/kg and 12 mg/kg, respectively. The control animals began to succumb to tumors or had to be sacrificed about 2 mo after the start of the experiment (Fig. 4B). Necropsy revealed numerous tumor metastases, mostly in the lung. Nanoparticles slowed the growth of the tumors and considerably prolonged the life of the animals (Fig. 4B).
Fig. 4.
Self-assembled particles formed by the CXCR4 TM2 derivative have intrinsic anti-CXCR4 activity in vitro and in vivo and can encapsulate hydrophobic drugs. (A) Nanoparticles formed by X4-2-6 inhibited growth and killed human and mouse breast cancer cells grown in the presence of SDF-1α in serum-free medium. (B) X4-2-6 prolonged survival in a mouse model of metastatic breast tumor. Nude mice were injected with MDA-MB-231 breast cancer cells intravenously. Intraperitoneal treatment with X4-2-6 was started the day following the injection of cancer cells and was given every 3 d thereafter. (C, D) Change in environment-sensitive fluorescence spectra of an imidazoacridone drug upon mixing with nanoparticles demonstrates drug incorporation into nanoparticles. (C) Emission peak position; (D), emission peak intensity. (E) Self-assembling peptide nanoparticles solubilize hydrophobic anticancer agents. Comparison of HKH-40A forming a precipitate in PBS (Left) and a clear X4-2-6/PBS solution (Right).
To test the ability of the nanoparticles to encapsulate hydrophobic drugs, we used a drug developed in our lab, HKH-40A (12). HKH-40A is practically insoluble in PBS and cell media. However, it partitions into the hydrophobic nanoparticles and goes into solution easily in the presence of X4-2-6 (Fig. 4E). The HKH-40A analog, imidazoacridone WMC77 was used to prove that the compound is truly encapsulated by the particles. The fluorescent properties of this compound are environment-dependent (13, 14). Changes in fluorescence spectra of WMC77 upon addition of nanoparticles were consistent with a transfer into a hydrophobic environment (Fig. 4 C and D). Furthermore, additional evidence of WMC-77 incorporation into nanoparticles arises from a twofold decline in fluorescence intensity of particles labeled with Alexa-546 upon addition of WMC77. This decline occurs due to the proximity of the WMC77 emission maximum at 528 nm and Alexa Fluor excitation (absorption) maximum at 556 nm. The loss of WMC77 emission due to energy transfer was as substantial as 60%. Fluorescent energy transfer takes place only if fluorophores are positioned in close proximity and thus proves close physical contact of the drug with peptide forming the nanoparticles. The fact that the environment of the compound changes upon contact with the particles indicates that the drug is encapsulated by the nanoparticles. The resulting particles have the same shape and size as unloaded nanoparticles, as revealed by electron microscopy and DLS. Upon the fusion of nanoparticles with cells, the drugs get released. Confocal laser scanning microscopy allowed observation of rapid release and accumulation of bis-imidazoacridone WMC26 in the nucleus of treated cells within minutes after addition of nanoparticles to cells (Fig. S8).
Discussion
Nanoparticle-based drug delivery systems have drawn considerable attention because of their ability to lower toxicity, increase effectiveness, improve bioavailability, and reduce the cost of drug treatments (15). In the case of anticancer agents, they can be tailored to accumulate in tumor cells and tissues due to enhanced permeability and retention or by targeting with ligands that recognize cell surface receptors overexpressed in tumors (16, 17). To date, the major challenges encountered in advancing the therapeutic use of nanoparticles include difficulties in reproducible manufacturing, obtaining uniform size and composition, and minimizing toxicity, particularly for inorganic nanoparticles. Self-assembly allows for the straightforward and reproducible generation of the nanostructures, but only if the geometry of resulting structures can be controlled. Using peptides as the building blocks for self-assembling nanostructures is particularly attractive because synthetic procedures at practically any scale are well-developed and metabolic decomposition of peptides leads to physiologically neutral amino acids. Peptides have been used for the generation of many infinite self-assembling systems like fibers, films, gels, and tubes (18–23). However, development of uniform and relatively small structures (below 20 nm in diameter) that are preferred for drug delivery (24, 25) turned out to be challenging. Smaller nanoparticles size is essential for efficient tumor penetration because lack of treatment efficacy is frequently caused by limited drug distribution within tumors (25).
Self-assembly described above resulted in small nanoparticles that were more than 99.9% homogeneous in size and homogeneity was achieved without any fractionation. Intrinsic biological activity dictated by the nature of the building block of the nanoparticles is a unique feature of this system (Fig. 5). Peptides are known to have very short half-life in circulation. Self-assembling peptide was stable in serum and showed remarkable in vivo activity even when injected just twice a week suggesting that self-assembly may protect the peptide from degradation.
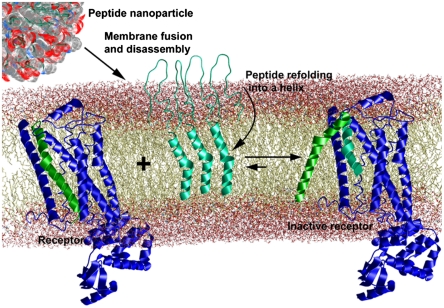
Fig. 5.
Proposed mechanism of action of fusogenic protoviral nanoparticles. The hair-pin shaped peptides (teal ribbons) self-assemble into spherical nanoparticles in the aqueous solution. Nanoparticles fuse spontaneously with the plasma membrane and disassemble upon fusion. Peptides transition into an active helical conformation in the apolar lipid environment. The helical peptides interact with the target receptor substituting the helix they are derived from (green) and thus inhibit receptor activity (7).
Polytopic membrane proteins are important drug targets (26–29). Although many of them can be inhibited by small molecules and are considered “druggable,” rational drug design is hampered by difficulties in structural studies (26). Analogs of transmembrane domains have been shown to inhibit the function of many polytopic membrane proteins in vitro (3–8). The advantage of the transmembrane inhibitors is in the fact that they can be designed in a rational way even in the absence of the tertiary structure information. We found that many transmembrane peptides are capable of self-assembly in aqueous solutions into round-shaped structures if equipped with terminal negative charges. The resulting particles can be used for effective in vivo delivery of the corresponding inhibitors, thus opening opportunities for generation of effective chemical biology tools and drug candidates. We speculate that their ability to encapsulate hydrophobic compounds could be used for generation of efficient delivery systems with dual biological activity. The fusion of particles could potentially be facilitated by anchoring them to cell surface receptors through conjugation of self-assembling peptide with a receptor ligand. The resulting particles could allow for cell-specific delivery of both the coat peptide and the encapsulated compound.
Newly developed fully synthetic fusogenic protoviral peptide nanoparticles open up opportunities for generating highly homogeneous virus-like particles with controllable size, surface properties, biological activity, and targeting ability for a wide range of applications.
Methods
Peptide Synthesis.
Peptides were synthesized by solid phase peptide synthesis on a 433A Applied Biosystems Peptide Synthesizer utilizing Fmoc amino acid derivatives. Because all peptides contain Asp residues that are prone to aspartamide formation upon piperidine treatment during deprotection, the synthesizer was reprogrammed to use 50% piperidine in NMP containing 0.25 M hydroxybenzotriazole. Peptides were cleaved from the resin with 87.5% trifluoroacetic acid containing 5% water, 5% thioanisol, and 2.5% triisopropylsilane, precipitated with cold diethyl ether, washed with ether, and dried in a vacuum overnight. Peptides dissolved in dimethylformamide were purified by HPLC on a preparative (19 × 250 mm) Atlantis C3 reverse phase column (Agilent Technologies) in a gradient of 0.05% trifluoroacetic acid in water and acetonitrile containing 0.05% trifluoroacetic acid. The fractions were analyzed by electrospray LC/MS on Agilent 1100 Series LC/MSD mod. G1946D (Agilent) with Poroshell 300SB-C18 column. The purity and structure were further confirmed by electrospray LC/MS with separation on a Zorbax 300SB-C3 analytical column.
Dynamic Light Scattering.
The peptide solutions in PBS, pH 7.2, were prepared from 32 mg/mL stock solutions of peptides in DMSO. The final concentration of DMSO in all samples was 1.25% (v/v). The solutions were sonicated for 10 min and left at ambient temperature overnight. The samples were spun at 9740 × g for 10 min and analyzed by dynamic light scattering (DLS) on a DynaPro Titan instrument equipped with Temperature-Controlled MicroSampler (Wyatt Technology Corp.) at a laser wavelength of 830 nm. To obtain the hydrodynamic radii (Rh), the intensity autocorrelation functions were analyzed by “Dynamics 6.7.7.9.” software (Wyatt ). For data analysis, viscosity, and refractive index values for a solution of 1.25% DMSO in water (v/v) were calculated using data from Nieto-Draghi et al. (30) and Viggiano et al. (31). Percentage polydispersity (a standard deviation of the size divided by the mean radius multiplied by 100%) of 15% or less is considered to indicate a homogeneous size distribution (32).
Supplementary Material
Acknowledgments.
We thank Dr. Anil Patri and Jeffrey D. Clogston (Nanotechnology Characterization Laboratory, SAIC, NCI-Frederick) for the help with measurement of Zeta potential; Stephen Lockett and Kimberly Peifley (Image Analysis Laboratory, Advanced Technology Program, SAIC, NCI-Frederick) for the help with laser scanning confocal microscopy; and Kunio Nagashima (SAIC, NCI-Frederick) for transmission electron microscopy. This work was funded by Congressionally Directed Medical Research Program, Prostate Cancer Research Award PC08116 (N.I.T.); by the Intramural Research Program of the NIH, NCI, Center for Cancer Research, with federal funds from the NCI, NIH, under Contract HHSN26120080001E; and by NIH Grant R01CA135341 (V.G.).
Footnotes
Conflict of interest statement: S.G.T. and N.I.T. are the coinventors on a patent application filed by the National Institutes of Health (NIH)/National Cancer Institute (NCI) for the self-assembling nanoparticles. Other authors declare no potential conflicts of interest.
This article is a PNAS Direct Submission. S.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014598108/-/DCSupplemental.
References
- 1.Roy P, Noad R. Virus-like particles as a vaccine delivery system: Myths and facts. Hum Vaccin. 2008;4:5–12. doi: 10.4161/hv.4.1.5559. [DOI] [PubMed] [Google Scholar]
- 2.Wu W, Hsiao SC, Carrico ZM, Francis MB. Genome-free viral capsids as multivalent carriers for taxol delivery. Angew Chem Int Ed Engl. 2009;48:9493–9497. doi: 10.1002/anie.200902426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George SR, et al. A transmembrane domain-derived peptide inhibits D1 dopamine receptor function without affecting receptor oligomerization. J Biol Chem. 1998;273:30244–30248. doi: 10.1074/jbc.273.46.30244. [DOI] [PubMed] [Google Scholar]
- 4.Hebert TE, et al. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 5.Partridge AW, Melnyk RA, Yang D, Bowie JU, Deber CM. A transmembrane segment mimic derived from Escherichia coli diacylglycerol kinase inhibits protein activity. J Biol Chem. 2003;278:22056–22060. doi: 10.1074/jbc.M210685200. [DOI] [PubMed] [Google Scholar]
- 6.Percherancier Y, et al. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J Biol Chem. 2005;280:9895–9903. doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- 7.Tarasova NI, Rice WG, Michejda CJ. Inhibition of G-protein-coupled receptor function by disruption of transmembrane domain interactions. J Biol Chem. 1999;274:34911–34915. doi: 10.1074/jbc.274.49.34911. [DOI] [PubMed] [Google Scholar]
- 8.Tarasova NI, et al. Transmembrane inhibitors of P-glycoprotein, an ABC transporter. J Med Chem. 2005;48:3768–3775. doi: 10.1021/jm049065t. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 10.Collins KD. Charge density-dependent strength of hydration and biological structure. Biophys J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Lee VC, Chevalier E, Hwang ST. Chemokine receptors as targets for cancer therapy. Curr Pharm Des. 2009;15:742–757. doi: 10.2174/138161209787582165. [DOI] [PubMed] [Google Scholar]
- 12.Hariprakasha HK, et al. Optimization of naphthalimide-imidazoacridone with potent antitumor activity leading to clinical candidate (HKH40A, RTA 502) J Med Chem. 2007;50:5557–5560. doi: 10.1021/jm7009777. [DOI] [PubMed] [Google Scholar]
- 13.Tarasov SG, et al. Bisimidazoacridones: 2. Steady-state and time-resolved fluorescence studies of their diverse interactions with DNA. Photochem Photobiol. 2003;78:313–322. doi: 10.1562/0031-8655(2003)078<0313:bsatfs>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Tarasov SG, Casas-Finet JR, Cholody WM, Michejda CJ. Bisimidazoacridones: Effect of molecular environment on conformation and photophysical properties. Photochem Photobiol. 1999;70:568–578. [PubMed] [Google Scholar]
- 15.Zhang L, et al. Nanoparticles in medicine: Therapeutic applications and developments. Clin Pharmacol Ther. 2008;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 16.Alexis F, et al. New frontiers in nanotechnology for cancer treatment. Urol Oncol. 2008;26:74–85. doi: 10.1016/j.urolonc.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Heath JR, Davis ME. Nanotechnology and cancer. Annu Rev Med. 2008;59:251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazit E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem Soc Rev. 2007;36:1263–1269. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- 19.Haines LA, et al. Light-activated hydrogel formation via the triggered folding and self-assembly of a designed peptide. J Am Chem Soc. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung AM, Stupp SI. Simultaneous self-assembly, orientation, and patterning of peptide-amphiphile nanofibers by soft lithography. Nano Lett. 2007;7:1165–1171. doi: 10.1021/nl062835z. [DOI] [PubMed] [Google Scholar]
- 21.Lamm MS, Rajagopal K, Schneider JP, Pochan DJ. Laminated morphology of nontwisting beta-sheet fibrils constructed via peptide self-assembly. J Am Chem Soc. 2005;127:16692–16700. doi: 10.1021/ja054721f. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Zhang S. Molecular designer self-assembling peptides. Chem Soc Rev. 2006;35:1105–1110. doi: 10.1039/b511336a. [DOI] [PubMed] [Google Scholar]
- 23.Semino CE. Self-assembling peptides: From bio-inspired materials to bone regeneration. J Dent Res. 2008;87:606–616. doi: 10.1177/154405910808700710. [DOI] [PubMed] [Google Scholar]
- 24.Wong C, et al. Multistage nanoparticle delivery system for deep penetration into tumor tissue. Proc Natl Acad Sci USA. 2011;108:2426–2431. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 26.Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: The largest class of drug targets. Drug Discov Today. 2009;14:1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gether U, Andersen PH, Larsson OM, Schousboe A. Neurotransmitter transporters: Molecular function of important drug targets. Trends Pharmacol Sci. 2006;27:375–383. doi: 10.1016/j.tips.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 29.Rajendran L, Knolker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010;9:29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 30.Nietro-Draghi C, Bonet Ávalos J, Rousseau B. Transport properties of dimethyl sulfoxide aqueous solutions. J Chem Phys. 2003;119:4782–4789. [Google Scholar]
- 31.Viggiano G, Ragozzino E, D’Ambrosio L, Santamaria R. Water-DMSO and water-DMSO-urea systems. Boll Chim Farm. 1973;112:746–752. [PubMed] [Google Scholar]
- 32.Collins B. A trick of light:novel technologies for sizing liposomal drug-delivery particles. The Biochemist. 2006;28(2):25–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.