Abstract
The assembly of progenitor cells is a crucial step for organ formation during vertebrate development. Kupffer's vesicle (KV), a key organ required for the left–right asymmetric body plan in zebrafish, is generated from a cluster of ∼20 dorsal forerunner cells (DFCs). Although several genes are known to be involved in KV formation, how DFC clustering is regulated and how cluster formation then contributes to KV formation remain unclear. Here we show that positive feedback regulation of FGF signaling by Canopy1 (Cnpy1) controls DFC clustering. Cnpy1 positively regulates FGF signals within DFCs, which in turn promote Cadherin1-mediated cell adhesion between adjacent DFCs to sustain cell cluster formation. When this FGF positive feedback loop is disrupted, the DFC cluster fails to form, eventually leading to KV malformation and defects in the establishment of laterality. Our results therefore uncover both a previously unidentified role of FGF signaling during vertebrate organogenesis and a regulatory mechanism underlying cell cluster formation, which is an indispensable step for formation of a functional KV and establishment of the left–right asymmetric body plan.
Keywords: left–right patterning, ciliogenesis
Fibroblast growth factor (FGF) signaling plays crucial roles in multiple morphogenetic processes of vertebrate development, including gastrulation movement, mesoderm formation, and left–right (LR) patterning (1–3). Because gain or loss of function of FGF signaling results in morphological changes in the embryo, some mechanism must ensure appropriate FGF signal levels in space and time for proper morphogenesis throughout development. FGF effectors acting as positive or negative regulators show a wide range of expression patterns and activities, contributing to the precise regulation of FGF signal activity (1, 4). Although most effectors identified to date act as negative regulators of FGF signaling, a few that positively regulate FGF activity have been reported (1, 4).
We recently identified in zebrafish a positive regulator of FGF signaling named canopy1 (cnpy1), which is required for maintenance of the midbrain–hindbrain boundary (MHB) (5). Expression of cnpy1 was restricted to the MHB at late-somitogenesis stages, whereas cnpy1 was broadly distributed in earlier embryos (5) (SI Appendix, Fig. S1A), suggesting an additional role(s) for Cnpy1-mediated FGF signaling beyond the regulation of MHB formation. In this study, we characterize cnpy1 in detail during early zebrafish development and show that a Cnpy1-mediated positive feedback loop of FGF signaling promotes cell cluster formation between dorsal forerunner cells (DFCs) during gastrulation. We also demonstrate that the failure of DFCs to cluster when this FGF positive loop is disrupted eventually leads to Kupffer's vesicle (KV) malformation and randomization of LR asymmetric patterning.
Results
Positive Feedback Loop of FGF Signaling Mediated by Cnpy1 Is Activated Specifically in DFCs During Zebrafish Gastrulation.
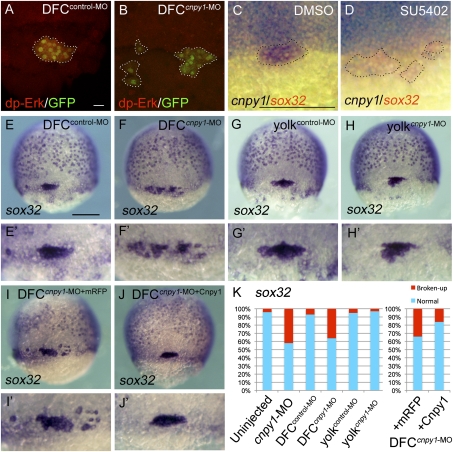
To reveal the role of Cnpy1-mediated FGF signaling in early zebrafish embryos, we first looked for the specific regions and cells in which Cnpy1 positively regulates FGF signaling, by monitoring FGF signal activity using an anti–di-phosphorylated Erk (dp-Erk) antibody. FGF signal activity was observed in the blastoderm margin and DFCs at midgastrulation (SI Appendix, Fig. S2A), whereas knockdown of cnpy1 with an antisense morpholino (cnpy1-MO) reduced the FGF activity in DFCs (SI Appendix, Fig. S2B). To test whether Cnpy1 is required autonomously for the FGF activation in DFCs, we next knocked down cnpy1 in DFCs but not in the rest of the embryo by using a DFC-specific MO delivery method (6–8). Similar to cnpy1 morphants, DFC-specific knockdown of cnpy1 (DFCcnpy1-MO) reduced the FGF activity in DFCs (Fig. 1B and SI Appendix, Fig. S2C). Because cnpy1 expression is induced by Fgf8 in the MHB (5), we checked whether FGF signaling is also required for cnpy1 expression in DFCs. We found that cnpy1 expression in DFCs could indeed be blocked by knockdown of fgf8 (SI Appendix, Fig. S2G) or by treatment with the FGF receptor inhibitor SU5402 (Fig. 1D). These results imply that a positive feedback loop between FGF and Cnpy1 is activated specifically in DFCs at midgastrulation.
Fig. 1.
Cnpy1 within DFCs regulates DFC clustering. (A and B) dp-Erk staining in DFCcontrol-MO–injected (A) or DFCcnpy1-MO–injected (B) Tg[sox17:GFP] embryos at 60% epiboly stage. (Scale bar: 20 μm.) dp-Erk signals (red) were down-regulated in GFP-positive DFCs (green). (C and D) cnpy1 (purple) and sox32 (red) expression in DMSO-treated (C) or SU5402-treated (D) embryos at 60% epiboly stage. (Scale bar: 200 μm.) Dotted lines in A–D mark the outlines of DFC populations. (E–J) sox32 expression in DFCcontrol-MO (E), DFCcnpy1-MO (F), yolkcontrol-MO (G), yolkcnpy1-MO (H), DFCcnpy1-MO+mRFP (I), or DFCcnpy1-MO+Cnpy1 (J) embryos at 70% epiboly stage. Dorsal view, anterior to the top. (Scale bar: 200 μm.) (E′–J′) Higher-magnification images highlight DFCs. (K) Percentages of normal (clustered) or broken-up DFCs were scored by using the sox32 expression pattern in uninjected (n = 68), cnpy1-MO (n = 77), DFCcontrol-MO (n = 61), DFCcnpy1-MO (n = 78), yolkcontrol-MO (n = 56), yolkcnpy1-MO (n = 62), DFCcnpy1-MO+mRFP (n = 119), or DFCcnpy1-MO+Cnpy1 (n = 123) embryos. Statistically significant (P < 0.05) differences could be seen in uninjected versus cnpy1-MO, DFCcontrol-MO versus DFCcnpy1-MO, and DFCcnpy1-MO+mRFP versus DFCcnpy1-MO+Cnpy1 embryos.
Cnpy1 Function Within DFCs Is Required for DFC Clustering.
DFCs are progenitor cells of KV, which is a key organ required for LR patterning (9–11). At midgastrulation, a cluster of ∼20 DFCs appears adjacent to the embryonic shield (12, 13). The DFC cluster then moves to the vegetal pole and forms a more compact and oval-shaped cluster by late gastrulation (7, 11, 14). At the end of gastrulation, DFCs differentiate into ciliated epithelial cells of the KV, which generates the nodal flow required for LR patterning (7, 12, 13). Recent studies have shown that FGF signaling is required for morphogenesis and ciliogenesis of the KV as well as for LR patterning (2, 8, 15). Although knockdown of the FGF target genes ier2 and fibp1 is known to interfere with DFC formation (15), the contribution of FGF signaling before KV formation is poorly understood.
To investigate the role of Cnpy1 in DFC/KV formation, we analyzed the expression of markers specific for DFC fate specification (sox32) or differentiation (no tail) in cnpy1-MO–injected embryos. We found that the DFC cluster was broken up into multiple groups of cells (Fig. 1K and SI Appendix, Fig. S3 B, C, and E), and the broad distribution of endoderm cells marked by sox32 was disrupted (SI Appendix, Fig. S3B) in cnpy1 morphants. Even though cnpy1 morphants showed a failure of DFC clustering, neither cell fate specification nor total cell number in DFCs was affected by cnpy1 knockdown (SI Appendix, Fig. S3B and Table S1). Similar to cnpy1 morphants, DFC-specific knockdown of cnpy1 resulted in a broken-up DFC phenotype, whereas DFC specification and cell number were unaffected (Fig. 1 F and K and SI Appendix, Fig. S3 C and G and Table S1). When embryos were coinjected with cnpy1-MO and MO-resistant cnpy1 mRNA (DFCcnpy1-MO+Cnpy1), the broken-up DFC phenotype was significantly rescued (53%; P = 0.00174; Fig. 1 I–K). Because, in the DFC-specific MO delivery method, the MO is also delivered to the yolk and the yolk syncytial layer (YSL), it was possible that effects of cnpy1 in yolk/YSL might be essential for DFC clustering. To address this, we knocked down cnpy1 in yolk/YSL but not in DFCs (yolkcnpy1-MO) and found no DFC defects in terms of specification, cell number, or cluster formation (Fig. 1 H and K and SI Appendix, Table S1). Live confocal imaging revealed that the sparse DFC populations in DFCcnpy1-MO embryos never assembled into a compact cluster, although normal downward migration was observed (SI Appendix, Fig. S4 and Movies S1 and S2), indicating that Cnpy1 regulates formation of the cell cluster itself, rather than controlling directed cell migration.
Cnpy1 Function Within DFCs Is Essential for KV Ciliogenesis and LR Patterning.
Observation of DFCs using zebrafish embryos from transgenic line Tg[sox17:GFP] revealed that broken-up DFC phenotypes did not generate multiple clusters at the end of gastrulation. In DFCcnpy1-MO–injected embryos, a rosette-like structure containing a small number of DFCs was formed, around which fragmented GFP signals that might signify dead cells could be observed, whereas a proper rosette structure containing a larger number of DFCs was evident in DFCcontrol-MO embryos (SI Appendix, Fig. S5 C–E). These results suggest that the broken-up DFC clusters seen in DFCcnpy1-MO embryos reflect a failure in the recruitment of DFCs to the KV.
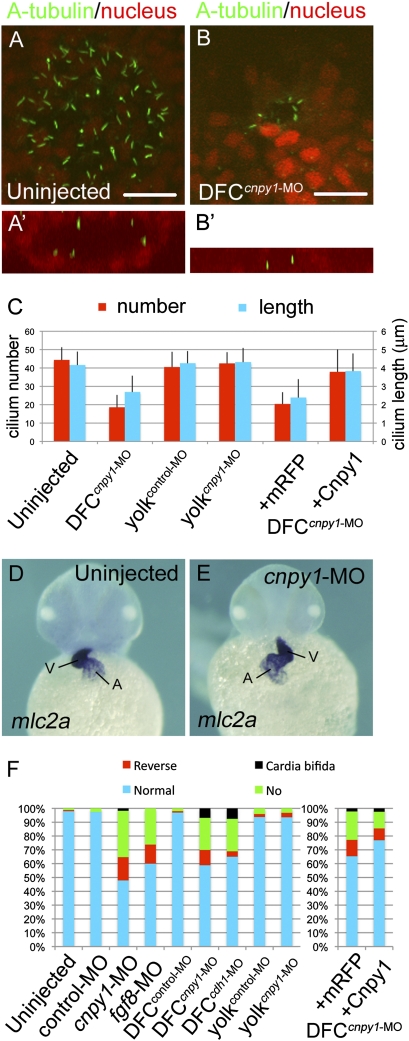
To examine how the failure of DFC cluster formation influences KV organogenesis and function, we investigated the presence and characteristics of primary cilia in the KV in DFCcnpy1-MO morphants by using an anti-acetylated tubulin (A-tubulin) antibody. DFC-specific knockdown of cnpy1 resulted in 60% and 35% reductions in the number and length, respectively, of primary cilia in the KV at early somitogenesis (Fig. 2 B and C). In addition to this disruption of ciliogenesis, lumen formation in the KV was incomplete (Fig. 2B′), suggesting that Cnpy1-mediated DFC clustering is required for proper formation of the KV. This idea is supported by the observation that the horseshoe-shaped pattern of charon expression in the caudal region of the KV was lost in DFCcnpy1-MO morphants (SI Appendix, Fig. S6 B and C). Consistent with these defects, knockdown of cnpy1 altered the left-sided expression of southpaw (spaw) in the lateral plate mesoderm at late somitogenesis (SI Appendix, Fig. S6 E and F) and led to defects in cardiac laterality at later stages (Fig. 2 E and F). Defective ciliogenesis and cardiac laterality in DFCcnpy1-MO embryos could be rescued by coinjection of MO-resistant cnpy1 mRNA (Fig. 2C and SI Appendix, Fig. S7D). Collectively, these results suggest essential roles for cnpy1 in KV ciliogenesis and LR patterning.
Fig. 2.
Cnpy1 function within DFCs is essential for ciliogenesis and LR patterning. (A and B) A-tubulin (green) and nucleus (red) staining in uninjected (A) or DFCcnpy1-MO–injected (B) embryos at the six-somite stage. Vegetal pole view. (Scale bars: 20 μm.) (A′ and B′) X–Z view around the KV. Lumen formation was not completed in DFCcnpy1-MO–injected embryos (B′). (C) Number (red) or length (blue) of KV primary cilia in uninjected (n = 10 or 49), DFCcnpy1-MO (n = 10 or 48), yolkcontrol-MO (n = 11 or 77), yolkcnpy1-MO (n = 11 or 58), DFCcnpy1-MO+mRFP (n = 10 or 61), or DFCcnpy1-MO+Cnpy1 (n = 11 or 85) embryos. (Error bars show SEM.) Statistically significant (P < 0.05) differences could be seen in uninjected versus DFCcnpy1-MO and DFCcnpy1-MO+mRFP versus DFCcnpy1-MO+Cnpy1 embryos. (D and E) Representative images of mlc2a expression demonstrating normal looping (uninjected; D) or reversed looping (cnpy1-MO; E) of the heart in embryos at the high pec stage. Ventral view, anterior to the top. A, atrium; V, ventricle. (F) Percentages of normal looping, reversed looping, no looping, or cardia bifida of the heart in uninjected (n = 164), control-MO (n = 118), cnpy1-MO (n = 119), fgf8-MO (n = 65), DFCcontrol-MO (n = 95), DFCcnpy1-MO (n = 146), DFCcdh1-MO (n = 106), yolkcontrol-MO (n = 96), yolkcnpy1-MO (n = 94), DFCcnpy1-MO+mRFP (n = 136), and DFCcnpy1-MO+Cnpy1 (n = 165) embryos. Statistically significant (P < 0.05) differences could be seen in uninjected versus cnpy1-MO, DFCcontrol-MO versus DFCcnpy1-MO, and DFCcnpy1-MO+mRFP versus DFCcnpy1-MO+Cnpy1 embryos.
Amplification of FGF Signaling by Cnpy1 Is Required for DFC Clustering.
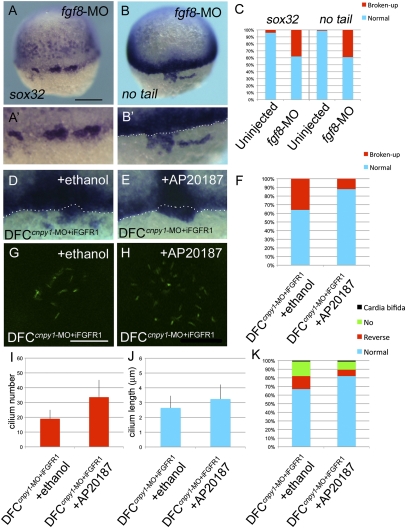
The above phenotypes in DFCcnpy1-MO embryos are reminiscent of the defects seen in embryos in which FGF signaling has been disrupted, such as fgf8, fgfr1, ier2, and fibp1 morphants (Fig. 2F and SI Appendix, Fig. S6F) (8, 15). To test for a functional relationship between FGF signaling and cnpy1 in DFC clustering, we analyzed whether the loss of FGF signaling function could phenocopy cnpy1 morphants. Intriguingly, ace/fgf8 mutations lead to failures of KV formation and LR patterning (2). Although fgf8 is expressed in and around DFCs (2), overlapping with cnpy1 expression, the role of fgf8 in DFC clustering is uncertain. We therefore examined the contribution of fgf8 to the formation of the DFC cluster. As for cnpy1-MO–injected embryos, fgf8 morphants exhibited the broken-up DFC phenotype (Fig. 3 A–C and SI Appendix, Table S1). fgf8 knockdown also resulted in defects in KV formation (SI Appendix, Fig. S6C) and LR patterning (Fig. 2F and SI Appendix, Fig. S6F). We also found that 57% of the ace/fgf8 mutants displayed the broken-up DFC phenotype (SI Appendix, Fig. S8 C and D). These results suggest that fgf8 plays an essential role in DFC clustering and that Cnpy1 contributes to this role.
Fig. 3.
FGF signaling plays crucial roles in DFC clustering and KV ciliogenesis. (A and B) sox32 (A) or no tail (B) expression in fgf8-MO–injected embryos. Dorsal view, anterior to the top. (Scale bar: 200 μm.) (A′ and B′) Higher-magnification images highlight DFCs. (B′, D, and E) The white dotted lines mark the boundary between DFCs and the blastoderm margin. (C) Percentages of normal or broken-up DFCs were scored by using the sox32 or no tail expression patterns in uninjected (n = 68 or 89) or fgf8-MO (n = 61 or 69) embryos. Statistically significant (P < 0.05) differences could be seen in uninjected versus fgf8-MO embryos. (D–K) Transient activation of FGF signaling restored the broken-up DFC phenotype (D–F), ciliogenesis (G–J), and cardiac laterality (K) in DFCcnpy1-MO embryos. (D and E) Expression of no tail in DFCcnpy1-MO+iFGFR1 embryos treated with ethanol (D) or AP20187 (E). (F) Percentages of broken-up DFC phenotype in ethanol-treated (n = 84) or AP20187-treated (n = 93) DFCcnpy1-MO+iFGFR1 embryos. The conditional activation of Fgfr1 after treatment with AP20187 significantly decreased the broken-up DFC phenotype (67%; P < 0.05) (G–J) A-tubulin (green) staining in ethanol-treated (G) or AP20187-treated (H) DFCcnpy1-MO+iFGFR1 embryos at the six-somite stage. (Scale bar: 20 μm.) (I and J) Number (I) or length (J) of KV primary cilia in ethanol-treated DFCcnpy1-MO+iFGFR1 (n = 9 or 36) or AP20187-treated DFCcnpy1-MO+iFGFR1 (n = 8 or 34) embryos at the six-somite stage. (Error bars show SEM.) Statistically significant (P < 0.05) differences could be seen in ethanol-treated versus AP20187-treated DFCcnpy1-MO+iFGFR1 embryos. (K) Percentages of cardiac laterality defect in ethanol-treated (n = 89) or AP20187-treated (n = 102) DFCcnpy1-MO+iFGFR1 embryos. The conditional activation of Fgfr1 after treatment with AP20187 alleviated the cardiac laterality defect (48%; P < 0.05).
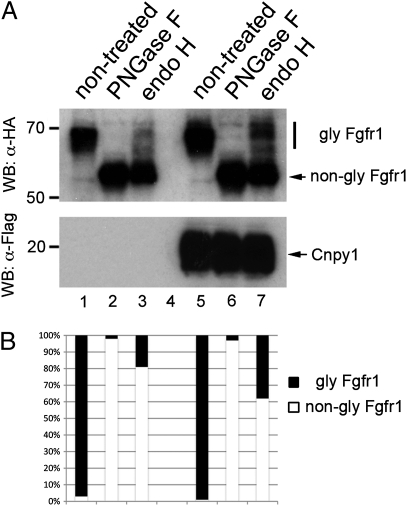
We have shown that Cnpy1 is a protein localized to the endoplasmic reticulum (ER) that can interact with Fgfr1 (5). However, it is still unclear how Cnpy1 modulates FGF signaling. Because the ER is a quality-control system that ensures maturation of secreted and membrane-bound proteins (16, 17), we reasoned that Cnpy1 might assist in the maturation of Fgfr1 in the ER, and we tested this with in vitro glycosylation assays (SI Appendix, Materials and Methods). Mature forms of Fgfr1 increased up to twofold in Cnpy1-overexpressing cells (Fig. 4 A and B), suggesting that Cnpy1 enhances FGF signaling by promoting the maturation of its receptor in the ER. This idea was further supported by proteomic data showing that a human Cnpy1 homolog binds to ER chaperones and folding-assisting enzymes (SI Appendix, Table S2).
Fig. 4.
Cnpy1 enhances FGF receptor maturation within the ER. (A and B) Fgfr1 N-glycosylation level was examined by PNGase F (lanes 2 and 6) or endo H (lanes 3 and 7) treatment. Lanes 1–3, mock control cells; lanes 5–7, Cnpy1-overexpressing cells. Fgfr1 and Cnpy1 were tagged with HA and Flag, respectively (SI Appendix, Materials and Methods). Upper indicates the glycosylation levels of Fgfr1, and Lower shows expression of Cnpy1 protein. (B) Ratio of glycosylated (black) and nonglycosylated (white) forms of Fgfr1. The amount of the endo H-resistant mature form of Fgfr1 in Cnpy1-overexpressing cells (lane 7) was twice that in mock control cells (lane 3).
If the amplification of FGF signals via Cnpy1-mediated Fgfr1 maturation is required for DFC clustering, it seemed possible that forced activation of Fgfr1 would restore the failure of DFC clustering in DFCcnpy1-MO embryos. Using iFGFR1, a conditional activation system for Fgfr1 that depends on AP20187-induced dimerization (5, 18), we activated Fgfr1 spatially and temporally in DFCs (SI Appendix, Materials and Methods). AP20187-mediated conditional activation of Fgfr1 in DFCs led to a 67% reduction in the broken-up DFC phenotype relative to vehicle (ethanol)-treated controls (P = 2.89 × 10−4; Fig. 3 D–F). Despite the conditional activation being restricted to DFCs during gastrulation, this manipulation partially restored deficiencies in cilium number (P = 9.23 × 10−3) and length (P = 7.77 × 10−3) in the KV (Fig. 3 G–J) and in cardiac laterality at later stages (P = 0.0114; Fig. 3K). These results therefore indicate that Cnpy1 function reinforces FGF signal activity within DFCs and suggest that DFC clustering mediated by this positive loop is a prerequisite for formation of a functional KV and proper LR patterning.
Induction of cadherin1 (cdh1) by Cnpy1-Mediated FGF Signaling Is Responsible for Generating Cell Adhesion Between DFCs.
To investigate the cellular function of Cnpy1 in DFC clustering, we analyzed cytoskeletal organization in DFCs of DFCcnpy1-MO embryos. Phalloidin staining showed that F-actin accumulated to a high level at the cell–cell contact sites between DFCs containing control-MO (SI Appendix, Fig. S9A), meaning that DFCs adhered tightly to each other in control embryos. In contrast, cell–cell adhesion between DFCs containing cnpy1-MO, evaluated by F-actin accumulation, was weaker than that between control-MO–containing DFCs (SI Appendix, Fig. S9B). These results suggest that Cnpy1-mediated FGF signaling modulates cell adhesions between DFCs during the control of cell clustering.
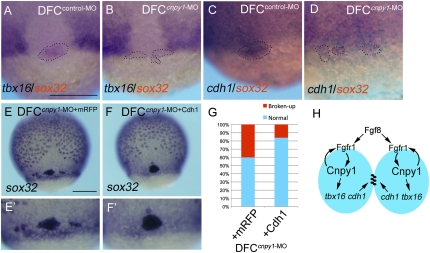
Recent studies have shown that the T-box transcription factor Tbx16 regulates DFC/KV formation in a cell-autonomous manner, although the underlying mechanism is still unclear (7). Tbx16 is also a mediator of FGF signaling, a function that is implicated in the control of cell adhesions via the transcriptional regulation of paraxial protocadherin (papc) (7, 19). Although papc expression is not detected in DFCs, cdh1 expression is (7, 20); thus, we hypothesized that tbx16 and cdh1 are downstream effectors of FGF signaling during the control of DFC clustering. To test this possibility, we analyzed whether Cnpy1-mediated FGF signaling affects expression of tbx16 or cdh1 within DFCs. DFCcnpy1-MO embryos showed reduced tbx16 or cdh1 expression in sparse DFC populations (Fig. 5 B and D and SI Appendix, Fig. S10 B and D). Importantly, DFC-specific knockdown of tbx16 (DFCtbx16-MO) also led to a reduction of cdh1 expression within DFCs (SI Appendix, Fig. S10 E and G), suggesting that tbx16 plays an important role in cdh1 expression within DFCs.
Fig. 5.
A Cnpy1-mediated FGF positive loop regulates cell adhesion through the control of cdh1 expression. (A and B) cdh1 (purple) and sox32 (red) expression in DFCcontrol-MO (A) or DFCcnpy1-MO (B) embryos at 65% epiboly stage. (C and D) tbx16 (purple) and sox32 (red) expression in DFCcontrol-MO (C) or DFCcnpy1-MO (D) embryos at 65% epiboly stage. Dotted lines in A–D mark the outlines of DFC populations. (Scale bar: 200 μm.) (E and F) Expression of sox32 in DFCcnpy1-MO+mRFP (E) or DFCcnpy1-MO + Cdh1 (F) embryos at 80% epiboly. (Scale bar: 200 μm.) (G) Percentage of broken-up DFC phenotype in mRFP-overexpressing (n = 82) or Cdh1-overexpressing (n = 103) DFCcnpy1-MO embryos. Overexpression of Cdh1 rescued the broken-up DFC phenotype in DFCcnpy1-MO embryos (60%; P < 0.05). (H) Diagram illustrating the FGF-dependent cell–cell communication control mechanisms of the forerunner cell cluster during early development. The model depicts the activation of intracellular FGF signaling via binding of Fgf8 ligands and Fgfr1 on the cell surface of two adjacent DFCs (blue ovals). The amplified FGF signal, through Cnpy1-mediated maturation of Fgfr1 within DFCs, subsequently activates the expression of tbx16 and cdh1 to organize forerunner cells as a cluster.
We next investigated whether DFC-specific knockdown of tbx16 (DFCtbx16-MO) or cdh1 (DFCcdh1-MO) could phenocopy DFCcnpy1-MO morphants. DFC-specific knockdown of either tbx16 or cdh1 led to the broken-up DFC phenotype (SI Appendix, Fig. S11 B–D and Table S1), outcomes similar to those observed in DFCcnpy1-MO morphants. These results suggested that a genetic cascade including tbx16 and cdh1 mediates FGF signal-dependent DFC clustering and prompted us to examine whether the broken-up DFC phenotype in DFCcnpy1-MO embryos could be rescued by overexpressing Cdh1. This restored DFC clustering in 60% of the manipulated embryos, relative to overexpression of monomeric red fluorescent protein (mRFP) as a control (Fig. 5 E–G). Hence, our results demonstrate that the Cnpy1-mediated FGF positive feedback loop regulates tbx16 and cdh1 to assemble cells into a tight cluster.
Taking into account all of these results, we propose the following stepwise regulatory mechanism underlying DFC cluster formation (Fig. 5H). First, FGF signaling is initiated in DFCs by Fgf8. Second, the up-regulated Cnpy1 within DFCs modulates FGF signal strength by enhancing Fgfr1 maturation in the ER. Third, the amplified FGF signals then promote cell–cell adhesion between adjacent DFCs through the action of Cdh1, eventually leading to the generation of a tight and stable cluster of DFCs.
Discussion
Accumulated evidence points to crucial roles of FGF signaling in several processes of LR asymmetric patterning (2, 8, 9, 15, 21, 22). Two recent studies, in particular, have shown that FGF signaling regulates KV ciliogenesis during LR pattering (8, 15). However, we uncover the importance of this signal pathway for the regulation of progenitor cell clustering at a stage before KV ciliogenesis: DFC-specific knockdown of cnpy1, tbx16, or cdh1 results in broken-up DFC clusters during gastrulation. The cause of such a discrepancy may originate from the differences of regulatory mechanisms underlying DFC cluster formation and ciliogenesis. DFC clustering requires activities of FGF-dependent effectors such as tbx16 and cdh1, as shown in this study. In contrast, ciliogenesis depends on the intraflagellar transport pathway regulated by the coordinated action of various signals, including FGF, Sonic hedgehog, and/or Wnt pathways (8, 15, 23, 24).
In this study, we have proposed that Cnpy1 controls DFC clustering, KV formation, and ciliogenesis by promoting Fgfr1 maturation. However, Neugebauer et al. (8) showed a different and specific role of fgfr1 in ciliogenesis and KV formation: DFC-specific knockdown of fgfr1 (DFCfgfr1-MO) leads to short cilia without affecting cilium number and KV size. This discrepancy may explain the redundant action between fgfr1 paralogs. A recent study has shown that the fgfr1 that was knocked down by Neugebauer et al. (8) and a second fgfr1 (fgfr1b) can functionally compensate for each other during early development (25). We reasoned that DFC-specific knockdown of cnpy1 might lead to defects more severe than those seen in DFCfgfr1-MO embryos because Cnpy1 can modulate the maturation of both receptors within DFCs. To test this possibility, we used a dominant-negative form of Fgfr1 (dn-Fgfr1), which lacks the cytoplasmic domain, and attempted to inhibit the functions of both receptors. Because injection of dn-fgfr1 mRNA into one-cell embryos led to severe defects in mesoderm formation and axis elongation, as shown previously (1), we used DFC-specific gene-transfer methods. As seen in DFCcnpy1-MO embryos, DFCdn-Fgfr1 embryos resulted in a broken-up DFC phenotype (SI Appendix, Fig. S12 B and C). Treatment with SU5402 (100 μg/mL) also led to broken-up DFC clusters (Fig. 1D). These results therefore suggest that strong loss-of-function effects on Fgfr1, such as cnpy1 knockdown, dn-Fgfr1 overexpression, and SU5402 treatment, prevent DFCs from organizing into a tight cell cluster, and that Cnpy1 may assist the maturation of both receptors within DFCs. On the other hand, mild loss of Fgfr1 function, including the single knockdown of fgfr1 performed by Neugebauer et al. (8), may yield the specific defect in cilium length.
Our results do not support data showing that loss of FGF signaling function—by SU5402 treatment (6–7 μg/mL), genetic disruption of fgf8 and/or fgf24, or ectopic expression of dn-Fgfr1 using hsp70:dn-fgfr1 transgenic zebrafish—leads to a specific defect in cilium length (8). This discrepancy may arise from variable loss-of-function efficiency caused by different inhibitor concentrations, genetic backgrounds, or experimental protocols. Regarding the role of fgf8 in LR asymmetry, severe KV defects—including partial or complete loss of KV formation, short cilia, and a reduced number of cilia—have been observed in ace/fgf8 mutants or knockdown embryos of fgf8 or fgf8 effectors (ier2 and fibp1) (2, 15). In addition, Hong and Dawid (15) have reported that severe KV defects in knockdown embryos of ier2 and fibp1 may be associated with disorganization of the DFC cluster. These findings also differ from those of Neugebauer et al. (8) but are consistent with our observations that either ace/fgf8 mutants or fgf8 morphants display failures of DFC clustering, KV formation, and LR patterning. Although particular issues remain to be resolved, these results clearly demonstrate that FGF signaling plays important roles in DFC clustering, KV formation, and ciliogenesis.
Contact between DFCs and the overlying surface ectoderm is known to be important for DFCs to migrate toward the vegetal pole (14). Because loss of function of FGF signal components (fgf8 and cnpy1) and downstream effectors (tbx16 and cdh1) showed the broken-up DFC clusters but normal migration of these disrupted DFCs to the vegetal pole during gastrulation, FGF signal-dependent cell adhesion may specifically contribute to the interaction among DFCs themselves. However, in these phenotypes, some DFCs remained capable of interacting with others to form small groups of cells, implying that other factor(s) may contribute to DFC clustering. It has been reported that integrin αV and integrin β1b have a role in DFC clustering (26) and that planar cell polarity signaling regulates cell adhesion between DFCs (27). Additional experiments to clarify the relationship between FGF signaling and integrins or planar cell polarity signaling during DFC cluster formation will be important to understand the entire mechanism underlying DFC cluster formation.
Conclusions
We have discovered the cells (DFCs) in which Cnpy1 functions and further added an insight into the molecular mechanism by which Cnpy1 regulates cell signaling in the ER. We identified an essential signal cascade—ligand, receptor, mediator, and downstream effector—that is required for proper cluster formation by progenitor cells. In addition, our findings reveal that progenitor clustering regulated by a positive feedback loop of cell signaling contributes to the formation of a functional organ to establish the LR asymmetric body plan during vertebrate development.
Materials and Methods
Zebrafish and Whole-Mount in Situ Hybridization.
A wild-type strain (RIKEN-Wako), Tg[sox17:GFP] (28), and aceti282a (2) were used in this study. Single- or double-color whole-mount in situ hybridization was performed as described previously (29, 30). cDNA fragments of cdh1, cnpy1, mlc2a, no tail, sox32, and spaw were used as templates for the antisense probes.
Other Methods.
Detailed methods for immunofluorescence analyses, pharmacological experiments, and rescue experiments are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Thomas N. Sato, Yasukazu Nakahata, and Kinichi Nakashima for advice, helpful discussions, and critical reading of the manuscript. We also thank Ian Smith for help in preparing the manuscript; Ryutaro Akiyama, Maiko Yokouchi, Takeshi Fujimuro, and Naoyuki Tahara for technical assistance; Tatsuro Matsuta for help with statistical analysis; Bruce W. Draper for advice; Kazushige Sakaguchi, Masahiko Hibi, and Masatoshi Takeichi for sharing reagents; Kota Yanagitani, Michiko Saito, and Kenji Kohno for sharing reagents and discussions; National BioResource Project Zebrafish, Core Institution for zebrafish lines Tg[sox17:GFP] and aceti282a; and ARIAD for AP20187. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas “WAKATE(B)” and “Systems Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; the Nakajima Foundation; and the Global Center of Excellence Program of Nara Institute of Science and Technology, MEXT, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017248108/-/DCSupplemental.
References
- 1.Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 2.Albertson RC, Yelick PC. Roles for fgf8 signaling in left–right patterning of the visceral organs and craniofacial skeleton. Dev Biol. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- 4.Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 5.Hirate Y, Okamoto H. Canopy1, a novel regulator of FGF signaling around the midbrain-hindbrain boundary in zebrafish. Curr Biol. 2006;16:421–427. doi: 10.1016/j.cub.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 6.Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer's vesicle in zebrafish. Dev Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 8.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Kramer-Zucker AG, et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 11.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 12.Cooper MS, D'Amico LA. A cluster of noninvoluting endocytic cells at the margin of the zebrafish blastoderm marks the site of embryonic shield formation. Dev Biol. 1996;180:184–198. doi: 10.1006/dbio.1996.0294. [DOI] [PubMed] [Google Scholar]
- 13.Melby AE, Warga RM, Kimmel CB. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development. 1996;122:2225–2237. doi: 10.1242/dev.122.7.2225. [DOI] [PubMed] [Google Scholar]
- 14.Oteíza P, Köppen M, Concha ML, Heisenberg CP. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–2813. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- 15.Hong SK, Dawid IB. FGF-dependent left–right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc Natl Acad Sci USA. 2009;106:2230–2235. doi: 10.1073/pnas.0812880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 17.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: Folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 18.Pownall ME, et al. An inducible system for the study of FGF signalling in early amphibian development. Dev Biol. 2003;256:89–99. doi: 10.1016/s0012-1606(02)00120-3. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto A, et al. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 1998;125:3389–3397. doi: 10.1242/dev.125.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esguerra CV, et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007;134:4381–4393. doi: 10.1242/dev.000026. [DOI] [PubMed] [Google Scholar]
- 21.Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left–right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 22.Raya A, Izpisúa Belmonte JC. Left–right asymmetry in the vertebrate embryo: From early information to higher-level integration. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 23.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oishi I, Kawakami Y, Raya A, Callol-Massot C, Izpisúa Belmonte JC. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat Genet. 2006;38:1316–1322. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- 25.Rohner N, et al. Duplication of fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr Biol. 2009;19:1642–1647. doi: 10.1016/j.cub.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 26.Ablooglu AJ, Tkachenko E, Kang J, Shattil SJ. Integrin αV is necessary for gastrulation movements that regulate vertebrate body asymmetry. Development. 2010;137:3449–3458. doi: 10.1242/dev.045310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oteiza P, et al. Planar cell polarity signalling regulates cell adhesion properties in progenitors of the zebrafish laterality organ. Development. 2010;137:3459–3468. doi: 10.1242/dev.049981. [DOI] [PubMed] [Google Scholar]
- 28.Mizoguchi T, Verkade H, Heath JK, Kuroiwa A, Kikuchi Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development. 2008;135:2521–2529. doi: 10.1242/dev.020107. [DOI] [PubMed] [Google Scholar]
- 29.Matsui T, et al. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 2005;19:164–175. doi: 10.1101/gad.1253605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.