Abstract
Speciation in animals commonly involves an extrinsic barrier to genetic exchange followed by the accumulation of sufficient genetic variation to impede subsequent productive interbreeding. All-female species of whiptail lizards, which originated by interspecific hybridization between sexual progenitors, are an exception to this rule. Here, the arising species instantaneously acquires a novel genotype combining distinctive alleles from two different species, and reproduction by parthenogenesis constitutes an effective intrinsic barrier to genetic exchange. Fertilization of diploid parthenogenetic females by males of sexual species has produced several triploid species, but these instantaneous speciation events have neither been observed in nature nor have they been reconstituted in the laboratory. Here we report the generation of four self-sustaining clonal lineages of a tetraploid species resulting from fertilization of triploid oocytes from a parthenogenetic Aspidoscelis exsanguis with haploid sperm from Aspidoscelis inornata. Molecular and cytological analysis confirmed the genetic identity of the hybrids and revealed that the females retain the capability of parthenogenetic reproduction characteristic of their triploid mothers. The tetraploid females have established self-perpetuating clonal lineages which are now in the third generation. Our results confirm the hypothesis that secondary hybridization events can lead to asexual lineages of increased ploidy when favorable combinations of parental genomes are assembled. We anticipate that these animals will be a critical tool in understanding the mechanisms underlying the origin and subsequent evolution of asexual amniotes.
Keywords: hybrid speciation, polyploidy, Cnemidophorus, unisexual
New species ordinarily arise over many generations through the gradual accumulation of incremental differences that eventually result in self-sustaining populations phenotypically distinctive and reproductively isolated from other species including contemporary representatives of their progenitor species (1, 2). With few exceptions (e.g., ref. 3), interspecific hybridization has been viewed as detrimental to the process of speciation in animals rather than a driving force for it. However, the recent application of molecular tools in Heliconius butterflies (4), tephritid fruit flies (5), and several other taxa has led to the realization that hybrid speciation may be more common in animals than previously thought (6). At the extreme of instant speciation, hybridization combined with parthenogenesis has given rise to almost all unisexual lizards (7). The incidence of such speciation events varies widely among families and is unusually high in Caucasian rock lizards (genus Darevskia) and North American whiptail lizards (Aspidoscelis; ref. 8). For example, of the 12 Aspidoscelis species found in New Mexico, 7 are parthenogenetic, and 5 of these are triploid (9, 10).
Karyotypic and molecular evidence revealed that diploid parthenogenetic Aspidoscelis species arose from hybridization events between sexual progenitors (11–14). Subsequent secondary hybridization events between diploid parthenogenetic females and males of sympatric sexual species produced triploid unisexuals. Hybrid origin of parthenogenetic species has also been documented in several other lizard families including geckos (15) and appears to be the most common, if not universal, path by which unisexual vertebrates arise (8). How the unisexual mode of reproduction is induced in diploid hybrids and maintained in triploids remains unknown. Several lines of evidence suggest that hybridization events resulting in new species are exceedingly rare. Firstly, histocompatibility studies support that single hybridization events have given rise to each of several parthenogenetic Aspidoscelis species (16–20). Secondly, de novo hybridization events between closely related species or subspecies result in offspring that reproduce sexually and are not reproductively isolated from the progenitor species (21). Hybridization between more divergent sexual species appears to occur much less frequently and results in sterile progeny (e.g., ref. 22). In contrast, quite a few first-generation hybrids between parthenogenetic Aspidoscelis species and males of sexual species have been observed in field studies over the past 40 y. When hybridization occurs between a diploid parthenogenetic female and a sexual male, the hybrid offspring are triploid (e.g., ref. 23); whereas hybridization events involving triploid parthenogenetic females produce tetraploid hybrids (24–27). Notably, in no case has successful reproduction of a hybrid been documented; and with one exception (24) the animals were clearly infertile where examined (e.g., ref. 28). In addition, a 29-y study aimed at creating a hybrid species in the laboratory involving 74 males and 156 females of nine species produced five confirmed hybrids, which were all sterile (22). In summary, these findings indicate that in most cases ploidy elevation coincides with a loss of the ability to reproduce parthenogenetically in the offspring.
Results
Generation of Tetraploid Hybrids.
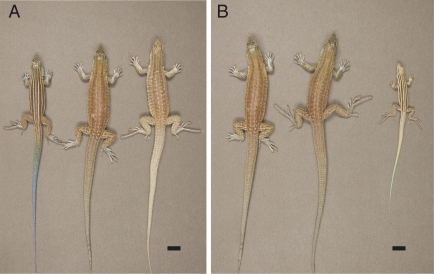
To gain more insight into the relationship between hybridization and infertility, we paired males of the diploid sexual species Aspidoscelis inornata with females of the triploid parthenogenetic species Aspidoscelis exsanguis. This choice was inspired by the description of an apparent hybrid between A. inornata and A. exsanguis that was captured in August 1967. While in captivity, this animal laid two fully yolked eggs, but desiccation made it impossible to determine whether the eggs could have produced viable offspring (24). In our present study, the A. inornata male was observed mating with A. exsanguis females on several occasions. Three clutches totaling six eggs were recovered from the enclosure and incubated at 28 °C. Subsequent genotyping showed that all three clutches had been produced by the same A. exsanguis female following fertilization by a single A. inornata male. Hatching occurred after 63–67 d, and the six offspring appeared morphologically similar to A. exsanguis with the exception of subtle blue pigmentation visible especially on the tail and indicative of a hybrid origin (Fig. 1).
Fig. 1.
Morphology of parental species and tetraploid hybrid animals. (A) Dorsal view of A. inornata (Left), A. exsanguis (Right), and the A. exsanguis/A. inornata hybrid (Center). (Scale bar, 10 mm.) (B) Individuals representing the first (H1, Left), second (H2, Center), and third (H3, Right) hybrid generation of the tetraploid species. The H1 and H2 individuals are adults photographed on day 1,168 and 645 after hatching, respectively. The H3 individual is shown at an age of 44 d and displays the color and pattern typical for juveniles.
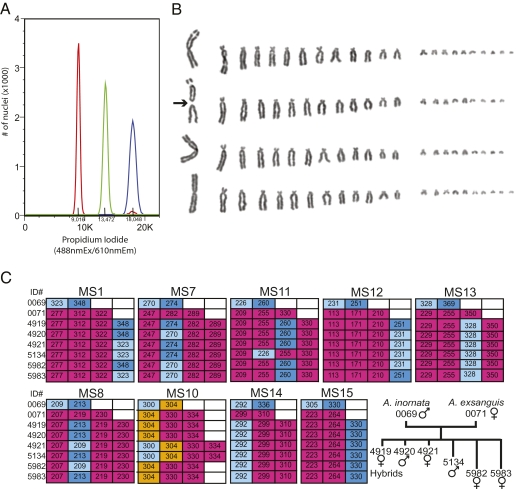
The ploidy of the animals was determined by quantifying the DNA content in nucleated erythrocytes by flow cytometry. Blood samples from A. inornata and A. exsanguis served as diploid and triploid controls, respectively. The analysis revealed a 4C DNA content in somatic cells of the hybrid lizards (Fig. 2A). Tetraploidy was further confirmed by karyotyping cultured fibroblasts isolated from the heart of a hybrid female that died at 20 mo of age. The somatic cell karyotype comprised 90–92 chromosomes (Fig. 2B), consistent with a combination of the haploid (n = 23) chromosome complement of an A. inornata sperm with the unreduced triploid (3n ∼ 69) chromosome complement of parthenogenetic A. exsanguis (Fig. S1).
Fig. 2.
DNA and chromosome analysis. (A) Determination of DNA content of whole blood nuclei by propidium-iodide staining followed by FACS analysis. A comparison between diploid A. inornata (red), triploid A. exsanguis (green) and a putative hybrid (blue) indicates a tetraploid DNA content for the hybrid. (B) The karyotype of the A. exsanguis/A. inornata hybrid was determined from metaphase chromosomes of cultured cells. Each row shows a haploid chromosome set, with chromosomes arranged by decreasing size. The centric fission of one of the three large chromosomes is indicated with an arrow. (C) Microsatellite analysis at nine loci in the A. exsanguis and A. inornata parents and the six hybrid progeny. The tree depicts the relationships between the eight animals. Unique alleles of A. exsanguis and A. inornata are highlighted in red and blue, respectively. In some experiments, an additional peak at 261 was observed for MS15, but was not reproducible in repeat runs and is considered a technical artifact.
Microsatellite Analysis.
We next developed a panel of microsatellite markers to examine the parentage and genetic fingerprint of each hybrid lizard. Microsatellite analysis for nine highly polymorphic loci consistently reflected the parentage of the six animals and identified the parent individuals (Fig. 2C). The A. exsanguis mother had three different alleles at each of the MS1, -7, -8, and -10 to -13 loci, but only two at loci MS14 and -15, the latter presumably reflecting the presence of the same allele on two of the three homeologous chromosomes. The male A. inornata was heterozygous at each of the nine loci, but at locus MS10 one of the two alleles was the same as one of the three alleles in A. exsanguis. All alleles present at the nine loci in A. exsanguis were detected in the six hybrid offspring, a finding consistent with the mother ovulating eggs carrying the unreduced somatic chromosome complement as previously observed in other Aspidoscelis species (29, 30). Importantly, all six hybrids had received A. inornata alleles from the fertilizing sperm. Two of the animals had enlarged femoral pores and more intense blue pigmentation characteristic of A. inornata males. Based on these criteria as well as the presence of hemipenal bulges at the base of the tail we surmise that these two individuals are males, whereas the other four hybrids are females. The reproductive status of the presumed males is still under investigation and will be reported in due course.
Preservation of Meiotic Mechanism.
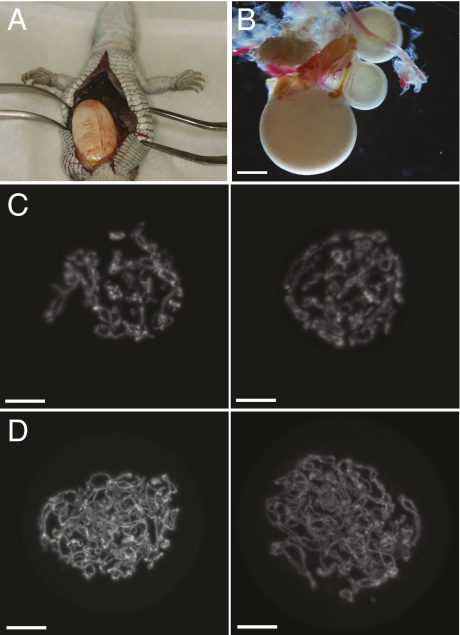
At 20 mo of age, one of the hybrid females died of unknown cause. A large egg was removed from the oviduct during necropsy (Fig. 3A) and incubated unsuccessfully. However, unlike previous hybrids examined by others, the ovaries of this lizard appeared normal and contained numerous developing follicles (Fig. 3B). Germinal vesicles (GVs) were isolated, stained with 4’,6-diamidino-2-phenylindole (DAPI), and examined by confocal microscopy to compare the chromosome content with GVs from diploid sexual and parthenogenetic species.
Fig. 3.
Ovaries and mechanism of oogenesis. (A) A large yolked and shelled egg was found in the body cavity of a deceased tetraploid hybrid. (B) One of two fully developed ovaries from the same lizard shown in A. (Scale bar, ∼0.5 mm.) (C) Three-dimensional projections of DAPI-stained germinal vesicles (GV) in prophase I of meiosis from a diploid parthenogenetic A. tesselata. (Scale bar, 20 μm.) (D) GVs from the tetraploid hybrid.
In diploid parthenogenetic Aspidoscelis species, meiosis commences with twice the number of chromosomes found in sexual species so that diploid oocytes are produced following the two meiotic divisions (30). Whether transient ploidy elevation is accomplished via two rounds of replication without intervening mitosis, by failed cytokinesis or by oogonial fusion is still unclear. If sexual reproduction occurred in the tetraploid hybrids, the premeiotic nuclei would contain twice the amount of DNA found in prophase I of meiosis in a sexually reproducing diploid species or the same amount as in a GV from a diploid parthenogenetic species (Fig. 3C). In contrast, if the tetraploid hybrids were capable of parthenogenetic reproduction, their GVs should contain twice the amount of DNA found in GVs of a diploid parthenogenetic species such as A. tesselata, an outcome that is consistent with our experimental findings (Fig. 3D). These observations indicate that the deceased tetraploid hybrid had been capable of producing eggs with a tetraploid chromosome content by using the same mechanism previously characterized for diploid parthenogenetic species in this genus (30). Indeed, genotyping later revealed that the deceased lizard had previously laid an egg from which a viable tetraploid offspring had hatched and grown to maturity.
Establishment of Four Tetraploid Lineages.
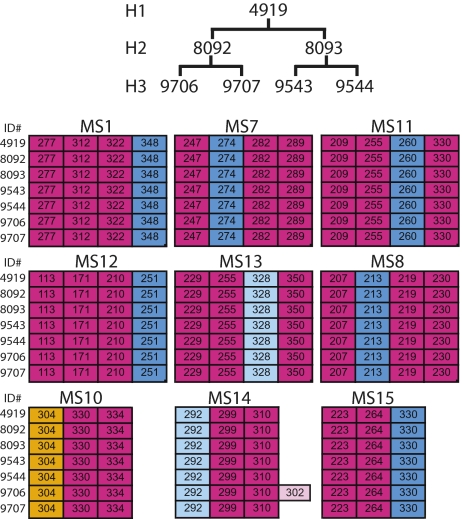
Between April 2009 and October 2010, 25 offspring were produced in aggregate by the four tetraploid females. The microsatellite analysis was first extended to these second-generation (H2) animals and later to 22 third-generation (H3) animals that hatched between April and December 2010. With one exception (see below), all alleles present at the nine loci were identical between the first-generation hybrids (H1) and their respective daughters and granddaughters, providing evidence for four independent parthenogenetic lineages. The example shown in Fig. 4 includes one H1 hybrid, two of its daughters and four granddaughters. The single exception to clonal inheritance occurred at the MS14 locus, where the H3 animal 9706 deviates from its siblings and progenitors by the appearance of a new allele 302 not otherwise found in the lineage. This allele appears to be the result of a repeat expansion confined to a single individual.
Fig. 4.
Maintenance of heterozygosity over three generations. Microsatellite analysis at nine loci for a first-generation hybrid (H2), two of its daughters (H2), and four granddaughters (H3). Alleles originally inherited from A. inornata are highlighted in blue, those from A. exsanguis in red. A single novel allele of MS14 is highlighted in pink.
It should be noted that the two H2 animals represented in Fig. 4 were produced by the H1 female while she was housed with two H1 males (4920 and 5134), which are distinguishable at two and four loci from the H1 female, respectively (Fig. 2C). Nevertheless, only maternal alleles were detected in the daughters providing further evidence for unisexual reproduction. The other members of the H2 generation thus far were daughters of H1 females housed separately from males, and microsatellite analysis revealed each to be genetically identical to its mother. Together with the increased DNA content in meiotic prophase and the fact that all H2 and H3 animals are female, the genotyping results therefore strongly support a parthenogenetic mode of reproduction.
Discussion
As of March 2011, the breeding experiment described here has produced 68 confirmed tetraploid animals representing three generations with more forthcoming as eggs hatch and additional eggs are laid. The maintenance of reproductive competence following ploidy elevation was highly unexpected, because other Aspidoscelis hybrids (both field-collected and laboratory-generated) have failed to reproduce. It has even been suggested that the decline of parthenogenetic A. dixoni in Antelope Pass, NM, is a result of fertilization of already diploid dixoni eggs by sexual A. tigris males resulting in sterile triploid hybrids (31). We have now described a case where ploidy elevation in a reptile has not resulted in embryonic lethality or infertility, providing the proof of principle for how triploid parthenogenetic species are likely to have arisen in nature.
Although evidence from field and laboratory studies (22, 32) indicates that speciation by ploidy elevation is exceedingly rare in reptiles, unisexual lineages of some fish and amphibians are polyclonal and in a few cases are readily reconstituted. For example, laboratory hybridization of Poeciliopsis monacha females and P. lucida males reconstituted a hybridogenetic species of fish, Poeciliopsis monacha-lucida, found in northwestern Mexico (33, 34). Similarly, the hemiclonal frog Rana esculenta was recreated in the laboratory by crossing the two parental species Rana ridibunda and Rana lessonae (35, 36). Inherent to the hemiclonal mechanism of hybridogenetic reproduction, eggs only contain one of the parental genomes and diploidy must be restored via fertilization by sperm from sympatric males each generation. Even in clonally reproducing (i.e., gynogenetic) unisexual salamanders and fish where the sperm genome is not incorporated into the offspring, sperm from a related species is required to trigger embryogenesis in eggs carrying the full somatic chromosome complement (37, 38). This absolute requirement for males from related sexual species is shared by all unisexual anamniotes and prevents establishment of reproductively independent unisexual species in these taxa.
The four lineages described here constitute a laboratory-generated vertebrate species that can reproduce independently of its progenitors. The absence of gene flow both within and between unisexual taxa has fueled debate about the taxonomic treatment of parthenogenetic animals (39–41). Although the lack of interbreeding within populations prevents strict application of the biological species concept, parthenogenetic lizards exist as phenotypically and genetically discrete, self-reproducing entities, that are defined by the unique combination of two or more haploid genomes derived from ancestral species. For the most part, they have been recognized as valid taxonomic units with the same status as sexually reproducing species (e.g., ref. 10). The proposal that each clonal lineage should be treated as a separate species (40) based on reproductive isolation and unique combination of alleles seems impractical as distinct origin can be exceedingly difficult to verify (42). The tetraploid species reported here is phenotypically distinct from the triploid ancestor by virtue of its possession of an additional haploid genome from A. inornata. In contrast, representatives of the four different lineages cannot be distinguished morphologically or based on ploidy. Although microsatellite and sequencing analysis permit distinction between lineages, these differences are no more pronounced than molecular differences observed among individuals of a sexual species. We therefore propose to treat the tetraploid lineages described here as one taxonomic unit.
The origin of reproducing lines of tetraploid whiptail lizards in the laboratory raises the question whether this species could survive in nature in competition with other Aspidoscelis species. Or is it comparable to domestic species that depend on husbandry under captive conditions to persist? It is premature to speculate on answers, but tetraploids of all three generations in the laboratory pursue and capture live crickets as effectively as their progenitors and exhibit no obvious competitive disadvantage when housed with individuals of sexual or parthenogenetic Aspidoscelis species. In experiments where a single food item (larvae of the darkling beetle Tenebrio molitor) was offered to a group comprised of four A. exsanguis and four tetraploids, the item was consumed as frequently by a tetraploid as by an A. exsanguis. On several occasions, we observed a tetraploid removing the food item from the mouth of an A. exsanguis (Fig. S2).
The laboratory synthesis of a tetraploid Aspidoscelis species, coupled with the collection of a tetraploid hybrid between A. inornata and A. exsanguis in Alamogordo more than 40 y ago (24), raises the question of why a tetraploid species derived from hybridization between these two species has not yet been found in nature. The apparent health and vigor of the tetraploids and their parthenogenetically produced offspring in captivity does not ensure their ability to succeed in nature, but it does suggest that sporadic mating of A. inornata with A. exsanguis could result in self-sustaining tetraploid lineages in locations where both species are sympatric. The tetraploid species synthesized in captivity may be the prototype of a species that might eventually emerge in the deserts of the southwestern US or northern Mexico. Perhaps its existence in the laboratory, together with recognition of the subtle phenotypic differences that distinguish it from its triploid progenitor, will stimulate a productive search for its counterpart in nature.
Materials and Methods
Animals.
Laboratory colonies of A. exsanguis, A. inornata, A. tesselata, and A. exsanguis x A. inornata hybrids were established from animals collected in Socorro, Sierra, and Otero Counties, NM, under a permit from the New Mexico Department of Game and Fish (permit numbers 3199 and 3395). Animals were propagated and maintained in the Reptile and Aquatics Facility under conditions similar to a previously published description of captive lizard husbandry (43) and in compliance with protocols approved by the Institutional Animal Care and Use Committee of the Stowers Institute for Medical Research.
Flow Cytometry.
Blood was isolated from tail-clips in acid citrate dextrose anticoagulant (45 mM sodium citrate, 22.8 mM citric acid, 81.5 mM dextrose). Cells were centrifuged at 500 × g and resuspended in citrate buffer, pH 7.6 (0.25 M sucrose, S-0389 from Sigma, 38.6 mM trisodium citrate, C-8532 from Sigma, and 5% DMSO) then pelleted again at 500 g. After decanting the supernatant, the cells were resuspended in citrate buffer at a density of 2.5 × 106 per mL. One hundred microliters of cell suspension was transferred into a 15 mL conical tube and incubated with 900 μL 30 mg/L trypsin, pH 7.6 (T-0134, Sigma), diluted in buffer S (3.4 mM trisodium citrate, 0.1% Triton X-100, T-6878, 1.5 mM spermine, S-1141 Sigma and 0.38 mM Tris·HCl, T-7149 from Sigma) for 10 min with gentle rotation, followed by the addition of 750 μL of trypsin inhibitor solution (0.5 mg/mL trypsin inhibitor, T9003 from Sigma, 0.1 mg/mL RNase A, R-5500 from Sigma prepared in buffer S) for an additional 10 min with gentle rotation. An additional 750 μL of propidium iodide solution (propidium iodide 0.42 mg/mL P-4170 from Sigma, 3.33 mM spermine in buffer S) was added and incubated with gentle agitation for 10 min while protected from light. Events were collected with an Influx instrument (BD Biosciences) by excitation at 488 nm and collection at 610 nm with a threshold set to exclude small debris. No gates were used.
Cell Culture.
Primary cell lines were established from embryos (A. exsanguis and A. inornata) or heart tissue (tetraploid hybrid) using a procedure modified from ref. 44. Briefly, A. exsanguis and A. inornata eggs were incubated at 29 °C for 30–40 d, sterilized in an ethanol-iodine mixture (90% ethanol, 120 mM potassium iodide and 39 mM iodine), and embryos were removed under sterile conditions and immediately decapitated. Minced embryos or, in the case of the hybrid animal, heart tissue was rinsed with cold PBS and agitated for 15 min at room temperature in the presence of trypsin-EDTA solution (T4049, Sigma). The suspension was passed through sterile cheesecloth for A. inornata and A. exsanguis samples into a 50 mL Falcon tube containing 2 mL of ice-cold M199 cell culture medium (Sigma) supplemented with 20% FBS, 50 μg/mL gentamycin (Sigma), glutamax (Invitrogen), MEM nonessential amino acids (Invitrogen), MEM vitamin solution (Invitrogen), 56 U/mL nystatin (Sigma), 100 U/mL penicillin and 100 μg/mL streptomycin (Sigma). For the A. exsanguis × A. inornata hybrid sample larger tissue fragments were manually removed using forceps before the addition of 2 mL of ice-cold M199 cell culture medium plus supplements as above. The filtered cell suspensions were kept on ice and the remaining tissue was trypsinized for another 15 min. Larger tissue fragments were again removed and the cell suspensions were combined and centrifuged to pellet cells, then washed in M199 media plus supplements and finally resuspended in 2–6 mL of M199 media plus supplements. Cells were seeded in six-well dishes (Falcon, 353046) and cultured at 30 °C, 5% O2 and 2% CO2. When cells exceeded 85% confluency, the cultures were passaged at a 1 in 3 dilution.
Karyotyping.
At 50–70% confluency, cultured cells were treated with 0.5 μg/mL Karyomax colcemid (Invitrogen) and incubated for 3 h at 30 °C, 5% O2 and 2% CO2. The cells were harvested by trypsinization and subjected to hypotonic swelling in 0.075 M KCl at 37 °C for 10 min. The cells were then pelleted, washed twice in PBS, and resuspended in methanol:acetic acid fixative (3:1). Coverslips were cleaned with a 1:1 ethanol:ethyl ether solution, air-dried and stored in water at 4 °C. Cells were dropped onto the coverslips and immediately washed with 1–3 mL of fixative. Coverslips were then incubated on a heat block at 75 °C for 1 min. Coverslips were further processed by RNase treatment (0.5 mg/mL in PBS) for 30 min at 37 °C, washed twice briefly in PBS, then fixed for 2 min in 4% formaldehyde (Sigma F8775) in PBS. After rinsing briefly in PBS three times, the coverslips were incubated with 1 mg/mL Pepsin (Sigma P6887, 3,200–4,500 units/mg) for 10 min at 37 °C, rinsed twice in PBS, and fixed again in 4% formaldehyde. Following three washes in PBS, the samples were dehydrated in an ethanol series (70%, 90%, 100%) and air-dried. Giemsa staining was performed by mixing giemsa (VWR, 15204–144) and phosphate buffer, pH 6.8 (VWR, 34171–002), in a 1:12 ratio, then filtering the mix through a 0.2-μm filter with an attached 18-gauge needle onto coverslips. The samples were incubated for 10–15 min, rinsed in water and air-dried in a dust-free environment before mounting. Samples were imaged on an Axiovert microscope equipped with an Axiocam HRm camera using a 100×, 1.3 NA Fluar objective. Images were analyzed with AxioVision software and karyotypes were assembled in Adobe Photoshop.
Microsatellite Analysis.
Microsatellites MS1, MS7, and MS8 were isolated from a genomic library prepared from A. tesselata liver tissue. Genomic DNA was isolated using QIAGEN Genomic-tips and the library was constructed using the Lambda FIX II library construction kit (Stratagene). Briefly, BAMH1-digested gDNA was ligated into the Lambda FIX II vector and packaged according to the Gigapack III manual (Stratagene). Plaque lifts were performed and membranes were hybridized with a 32P-labeled CA(10) probe. Positive plaques were isolated and phage DNA was purified using the Wizard Lambda Prep DNA Purification System (Promega) and sequenced by primer walking to identify microsatellites and flanking sequences. Microsatellites MS12 to -15 were isolated from genomic DNA libraries enriched for tetranucleotide repeats that were generated by Genetic Identification Services using genomic DNA isolated from liver of A. exsanguis. MS10 primers were modified from ref. 45 to amplify the Cvanμ7 microsatellite in Aspidoscelis species and MS11 (Ai5062) was as described (46). For each primer set one primer was 6-carboxyfluorescein-labeled at the 5′ end. Primer sequences are listed in Table S1.
One-millimeter lizard tail-clips were placed in 300 μL of a solution consisting of 50 mM Tris (pH 8.8), 1 mM EDTA, 0.5% Tween-20, and 100 μg/mL proteinase K. Samples were incubated for 12–18 h at 55 °C then placed in a 95 °C heat block for 10 min followed by direct storage at −80 °C until use. One microliter of each sample was used as template and PCR was performed with Biolase DNA polymerase (Bioline) or Taq DNA polymerase (New England Biolabs). PCR products were detected by capillary electrophoresis on a 3730 DNA Analyzer and analyzed with GeneMapper Version 4.0. Size ranges used to bin each allele are listed in Table S2.
Confocal Microscopy.
Germinal vesicles were isolated using jeweler's forceps, incubated with 40 ng/mL 4',6-diamidino-2-phenylindole (DAPI) and imaged using a LSM 510 META (Carl Zeiss, Jena) system equipped with a C-Apochromat 40×, NA 1.2 water immersion lens. A 405-nm laser was used to excite the fluorescent dye and signal was collected using a long-pass 420-nm filter. Images were cropped in Photoshop to digitally remove the nuclear envelope. Noise was removed by smoothing in Imaris with a 3 × 3 × 3 median filter.
Supplementary Material
Acknowledgments
We thank David Jewell, Rick Kupronis, Christina Piraquive, Jill Schieszer, and Kristy Winter for outstanding animal care; Nehemiah Alvarez for assisting in the development of microsatellite markers; the Stowers Institute Microscopy Center and the Cytometry and Molecular Biology Facilities for excellent support; and Jay Cole, Carol Townsend, and the members of the Baumann laboratory for helpful discussions. We are also very grateful to Scott Hawley and Jerry Workman for comments on the manuscript. This work was funded by the Stowers Institute for Medical Research. P.B. is an Early Career Scientist with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9733.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102811108/-/DCSupplemental.
References
- 1.Mayr E. Animal Species and Evolution. Cambridge, MA: Belknap Press; 1963. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 3.White MJ, Contreras N, Chency J, Webb GC. Cytogenetics of the parthenogenetic grasshopper Warramaba (formerly Moraba) virgo and its bisexual relatives. II. Hybridization studies. Chromosoma. 1977;61:127–148. doi: 10.1007/BF00327397. [DOI] [PubMed] [Google Scholar]
- 4.Mavárez J, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- 6.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 7.Neaves WB, Baumann P. Unisexual reproduction among vertebrates. Trends Genet. 2011;27:81–88. doi: 10.1016/j.tig.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Kearney M, Fujita MK, Ridenour J. Lost sex in the reptiles: Constraints and correlations. In: Schön I, Martens K, van Dijk P, editors. Lost Sex. Berlin: Springer; 2009. pp. 447–474. [Google Scholar]
- 9.Degenhardt WG, Painter CW, Price AH. Amphibians and Reptiles of New Mexico. Albuquerque: U. New Mexico Press; 1996. [Google Scholar]
- 10.Reeder TW, Cole CJ, Dessauer HC. Phylogenetic relationships of Whiptail Lizards of the Genus Cnemidophorus (Squamata: Teiidae): A test of monophyly, reevaluation of karyotypic evolution, and review of hybrid origins. Am Mus Novit. 2002;3365:1–61. [Google Scholar]
- 11.Lowe CH, Wright JW. Evolution of parthenogenetic species of Cnemidophorus (whiptail lizards) in Western North America. J Ariz Acad Sci. 1966;4:81–87. [Google Scholar]
- 12.Dessauer HC, Cole CJ. Clonal inheritance in parthenogenetic whiptail lizards: biochemical evidence. J Hered. 1986;77:8–12. [Google Scholar]
- 13.Neaves WB, Gerald PS. Gene dosage at the lactate dehydrogenase b locus in triploid and diploid teiid lizards. Science. 1969;164:557–559. doi: 10.1126/science.164.3879.557. [DOI] [PubMed] [Google Scholar]
- 14.Neaves WB, Gerald PS. Lactate dehydrogenase isozymes in parthenogenetic teiid lizards (Cnemidophorus) Science. 1968;160:1004–1005. doi: 10.1126/science.160.3831.1004. [DOI] [PubMed] [Google Scholar]
- 15.Moritz C. Parthenogenesis in the endemic Australian lizard Heteronotia binoei (Gekkonidae) Science. 1983;220:735–737. doi: 10.1126/science.220.4598.735. [DOI] [PubMed] [Google Scholar]
- 16.Cuellar O. Intraclonal histocompatibility in a parthenogenetic lizard: evidence of genetic homogeneity. Science. 1976;193:150–153. doi: 10.1126/science.779030. [DOI] [PubMed] [Google Scholar]
- 17.Maslin TP. Skin grafting in the bisexual teiid lizard Cnemidophorus sexlineatus and in the unisexual C. tesselatus. J Exp Zool. 1967;166:137–149. doi: 10.1002/jez.1401660114. [DOI] [PubMed] [Google Scholar]
- 18.Taylor HL, Cole CJ, Dessauer HC, Parker ED., Jr Congruent patterns of genetic and morphological variation in the parthenogenetic lizard Aspidoscelis tesselata (Squamata: Teiidae) and the origins of color pattern classes and genotypic clones in eastern New Mexico. Am Mus Novit. 2003;3424:1–40. [Google Scholar]
- 19.Cordes JE, Walker JM. Skin histocompatibility between syntopic pattern classes C and D of parthenogenetic Cnemidophorus tesselatus in New Mexico. J Herpetol. 2003;37:185–188. [Google Scholar]
- 20.Cuellar O. Genetic homogeneity and speciation in the parthenogenetic lizards Cnemidophorus velox and C. neomexicanus: Evidence from intraspecific histocompatibility. Evolution. 1977;31:24–31. doi: 10.1111/j.1558-5646.1977.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 21.Dessauer HC, Cole CJ, Townsend CR. Hybridization among western whiptail lizards (Cnemidophorus tigris) in southwestern New Mexico: Population genetics, morphology, and ecology in three contact zones. Bull. Am. Mus. Nat. Hist. 2000;246:1–146. [Google Scholar]
- 22.Cole CJ, Hardy LM, Dessauer HC, Taylor HL, Townsend CR. Laboratory hybridization among North American whiptail lizards, including Aspidoscelis inornata arizonae × A. tigris marmorata (Squamata: Teiidae), ancestors of unisexual clones in nature. Am Mus Novit. 2010;3698:1–43. [Google Scholar]
- 23.Cuellar O, McKinney CO. Natural hybridization between parthenogenetic and bisexual lizards: detection of uniparental source of skin grafting. J Exp Zool. 1976;196:341–350. doi: 10.1002/jez.1401960308. [DOI] [PubMed] [Google Scholar]
- 24.Neaves WB. Tetraploidy in a Hybrid Lizard of the Genus Cnemidophorus (Teiidae) Brev. Mus. of Comp. Zool. 1971;381:1–25. [Google Scholar]
- 25.Lowe CH, Wright JW, Cole CJ, Bezy RL. Natural hybridization between the teiid lizards Cnemidophorus sonorae (parthenogenetic) and Cnemidophorus tigris (bisexual) Syst Zool. 1970;19:114–127. [Google Scholar]
- 26.Cole CJ. Chromosome inheritance in parthenogenetic lizards and evolution of allopolyploidy in reptiles. J Hered. 1979;70:95–102. [Google Scholar]
- 27.Walker JM, Parker ED, Jr, Taylor HL, Cordes JE, Abuhteba RM. Hybridization between all-female Cnemidophorus tesselatus and gonochoristic Cnemidophorus sexlineatus. J Herpetol. 1990;24:388–396. [Google Scholar]
- 28.Hardy LM, Cole CJ. Morphology of a sterile, tetraploid, hybrid whiptail lizard (Squamata: Teiidae: Cnemidophorus) Am Mus Novit. 1998;3228:1–16. [Google Scholar]
- 29.Cuellar O. Reproduction and the mechanism of meiotic restitution in the parthenogenetic lizard Cnemidophorus uniparens. J Morphol. 1971;133:139–165. doi: 10.1002/jmor.1051330203. [DOI] [PubMed] [Google Scholar]
- 30.Lutes AA, Neaves WB, Baumann DP, Wiegraebe W, Baumann P. Sister chromosome pairing maintains heterozygosity in parthenogenetic lizards. Nature. 2010;464:283–286. doi: 10.1038/nature08818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole CJ, Painter CW, Dessauer HC, Taylor HL. Hybridization between the endangered unisexual gray-checkered whiptail lizard (Aspidoscelis dixoni) and the bisexual western whiptail lizard (Aspidoscelis tigris) in southwestern New Mexico. Am Mus Novit. 2007;3555:1–31. [Google Scholar]
- 32.Taylor HL, Walker JM, Cordes JE, Manning GJ. Application of the evolutionary species concept to parthenogenetic entities: Comparison of postformational divergence in two clones of Aspidoscelis tesselata and between Aspidoscelis cozumela and Aspidoscelis maslini (Squamata: Teiidae) J Herpetol. 2005;39:266–277. [Google Scholar]
- 33.Schultz RJ. Unisexual fish: laboratory synthesis of a “species”. Science. 1973;179:180–181. doi: 10.1126/science.179.4069.180. [DOI] [PubMed] [Google Scholar]
- 34.Wetherington JD, Kotora KE, Vrijenhoek RC. A test of the spontaneous heterosis hypothesis for unisexual vertebrates. Evolution. 1987;41:721–731. doi: 10.1111/j.1558-5646.1987.tb05848.x. [DOI] [PubMed] [Google Scholar]
- 35.Hotz H, et al. Rana ridibunda varies geographically in inducing clonal gametogenesis in interspecies hybrids. J Exp Zool. 1985;236:199–210. [Google Scholar]
- 36.Hotz H, Semlitsch RD, Gutmann E, Guex GD, Beerli P. Spontaneous heterosis in larval life-history traits of hemiclonal frog hybrids. Proc Natl Acad Sci USA. 1999;96:2171–2176. doi: 10.1073/pnas.96.5.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogart JP, Bi K, Fu J, Noble DW, Niedzwiecki J. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome. 2007;50:119–136. doi: 10.1139/g06-152. [DOI] [PubMed] [Google Scholar]
- 38.Lamatsch DK, Stöck M. Sperm-dependent parthenogenesis and hybridogenesis in teleost fish. In: Schön I, Martens K, van Dijk P, editors. Lost Sex. Berlin: Springer; 2009. pp. 399–432. [Google Scholar]
- 39.Cole CJ. Taxonomy of Parthenogenetic Species of Hybrid Origin. Syst Zool. 1985;34:359–363. [Google Scholar]
- 40.Frost DR, Wright JW. The taxonomy of uniparental species, with special reference to parthenogenetic Cnemidophorus (Squamata: Teiidae) Syst Zool. 1988;37:200–209. [Google Scholar]
- 41.Walker JM. The taxonomy of parthenogenetic species of hybrid origin: cloned hybrid populations of Cnemidophorus (Sauria: Teiidae) Syst Zool. 1986;35:427–440. [Google Scholar]
- 42.Cole CJ. When is an individual not a species? Herpetologica. 1990;46:104–108. [Google Scholar]
- 43.Townsend CR. Establishment and maintenance of colonies of parthenogenetic whiptail lizards: Cnemidophorus spp. Int. Zoo. Yrbk. 1979;19:80–86. [Google Scholar]
- 44.Moore MK, Work TM, Balazs GH, Docherty DE. Preparation, cryopreservation, and growth of cells prepared from the green turtle (Chelonia mydas) Methods Cell Sci. 1997;19:161–168. [Google Scholar]
- 45.Rowe G. St. Lucia whiptail lizard Cnemidophorus vanzoi (Sauria: Teiidae) microsatellite primers. Mol Ecol Notes. 2002;2:124–126. [Google Scholar]
- 46.Crawford NG, et al. Thirteen polymorphic microsatellite DNA loci from whiptails of the genus Aspidoscelis (Teiidae: Squamata) and related cnemidophorine lizards. Molecular Ecology Resources. 2008;8:219–223. doi: 10.1111/j.1471-8286.2007.01930.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.