Abstract
Malaria parasite cell motility is a process that is dependent on the dynamic turnover of parasite-derived actin filaments. Despite its central role, actin's polymerization state is controlled by a set of identifiable regulators that is markedly reduced compared with those of other eukaryotic cells. In Plasmodium falciparum, the most virulent species that affects humans, this minimal repertoire includes two members of the actin-depolymerizing factor/cofilin (AC) family of proteins, P. falciparum actin-depolymerizing factor 1 (PfADF1) and P. falciparum actin-depolymerizing factor 2. This essential class of actin regulator is involved in the control of filament dynamics at multiple levels, from monomer binding through to filament depolymerization and severing. Previous biochemical analyses have suggested that PfADF1 sequesters monomeric actin but, unlike most eukaryotic counterparts, has limited potential to bind or depolymerize filaments. The molecular basis for these unusual properties and implications for parasite cell motility have not been established. Here we present the crystal structure of an apicomplexan AC protein, PfADF1. We show that PfADF1 lacks critical residues previously implicated as essential for AC-mediated actin filament binding and disassembly, having a substantially reduced filament-binding loop and C-terminal α4 helix. Despite this divergence in structure, we demonstrate that PfADF1 is capable of efficient actin filament severing. Furthermore, this severing occurs despite PfADF1’s low binding affinity for filaments. Comparative structural analysis along with biochemical and microscopy evidence establishes that severing is reliant on the availability of an exposed basic residue in the filament-binding loop, a conserved minimal requirement that defines AC-mediated filament disassembly across eukaryotic cells.
Keywords: crystallography, circular dichroism spectroscopy, total internal reflection fluorescence microscopy, gliding motility, tight junction
The eukaryotic parasites from the genus Plasmodium that cause malaria disease require rapid cell movement to complete development, a process dependent on a parasite-derived actomyosin motor (1, 2). Short, dynamic actin filaments engage with an internal single-headed myosin, generating force that propels parasites along substrates or into host cells (reviewed in ref. 3). Drugs that stall actin filament growth or stabilize them from depolymerization prevent parasite motility when used at high concentrations, demonstrating the importance of dynamic actin (4–8). Despite this central role, however, actin filament turnover across all Apicomplexa, the phylum to which malaria parasites belong, is controlled by only a minimal set of identifiable regulators (3, 9). In Plasmodium falciparum, the most virulent species causing malaria disease, this minimal set includes two members of the actin-depolymerization factor/cofilin (AC) family of proteins, actin-depolymerization factor 1 (ADF1) and actin-depolymerization factor 2 (ADF2) (10).
AC proteins function as key regulators of actin turnover in diverse cellular processes (reviewed in ref. 11). Their function is linked to an ability to interact, to varying degrees, with both monomeric (G)- and filamentous (F)-actin. This activity is regulated by phosphorylation (12, 13), phosphoinositide binding (14, 15), and spatial concentration within the cell (16). Structurally, the G-actin sequestering activity depends on possession of a conserved binding site involving the AC protein N terminus, a long α3 helix [following yeast cofilin (Saccharomyces cerevisiae cofilin, ScCOF) nomenclature (17)] and turn connecting the β6 strand and α4 helix (17–19). Equally important is possession of a filament-binding site, thought to involve charged residues located at the extended filament-binding loop (F-loop) and C-terminal α4 helix as well as the C-terminal tail, which fold together to form a highly conserved F-actin binding motif (19–21).
Although ADF2 is thought to function in sexual stages of parasite development (10, 22), studies in the mouse malaria parasite Plasmodium berghei and malaria-related parasite Toxoplasma gondii have established that ADF1 (and its ortholog TgADF) is essential for parasite viability (10) and actin filament turnover (23). Biochemical studies of recombinant ADF1 from the most virulent human parasite, P. falciparum, have suggested several unique properties. Although it can sequester G-actin, P. falciparum ADF1 (PfADF1) is unable to disassemble F-actin, a feature attributed to predicted divergence in its structure compared with other better-characterized AC proteins (10). Furthermore, and uniquely, PfADF1 can stimulate nucleotide exchange in vitro, a property usually associated with profilin (10). Neither the structural basis for these properties nor the in vivo functions of ADF1 have been established.
Here we present biochemical and microscopy evidence that PfADF1 can mediate actin filament severing at nanomolar concentration. The crystal structure of P. falciparum ADF1 presented here, however, reveals that this severing occurs even though PfADF1 possesses a markedly reduced F-loop and C-terminal α4 helix. Importantly, conserved charged residues implicated in the F-actin binding and severing motif are absent in the PfADF1 structure with the exception of a single “exposed” basic residue. Using comparative structural analysis of PfADF1 with other AC members and analysis of mutants for this residue, we establish that the exposed basic residue presented by the F-loop defines a minimal motif, conserved across the entire family of AC proteins, required for filament severing.
Results
Spatiotemporal Localization of PfADF1 in Asexual Blood-Stage Parasites.
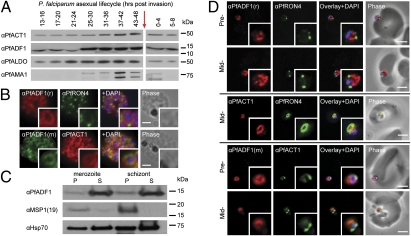
Of the two AC proteins expressed in malaria parasites, previous reports have suggested that ADF1 is a monomer-sequestering actin regulator with minimal filament disassembly or binding activity (10). To dissect further the cellular and biochemical properties of AC proteins from P. falciparum, we expressed both PfADF1 and P. falciparum ADF2 (PfADF2) in Escherichia coli and used these to generate AC-specific antisera (Fig. S1 A and B). Immunoblot analyses on whole-parasite lysates revealed that PfADF1 is expressed broadly, reaching maximal expression late in the 48-h asexual blood-stage lifecycle (Fig. 1A), a time when motile merozoite forms are maturing for cell reinfection (1). This peak of expression is similar to that of P. falciparum actin1 (PfAct1) (Fig. 1A) and other proteins that function in erythrocyte invasion (1). Expression of PfADF2 was not detected (Fig. S1 A and B), consistent with its having a role in extraerythrocytic or sexual stages (22). Immunofluorescence assays (IFA) of schizont stages (before merozoite release) demonstrated broad labeling across the parasite cytosol with the exception of the nucleus (marked by DAPI) (Fig. 1B). This distribution was consistent with that of PfAct1 (Fig. 1B). Solubility of PfADF1 in cell extracts following hypotonic lysis suggested a free association in the cytoplasm rather than association with detergent-soluble membrane fractions (Fig. 1C). In support of this notion, recombinant PfADF1 displayed low micromolar affinity for phosphatidyl inositol derivatives (Fig. S1C), lipid groups used to recruit AC proteins to the plasma membrane in other eukaryotes (14, 15).
Fig. 1.
Expression and localization of PfADF1 and PfAct1 in P. falciparum. (A) Immunoblot of whole-parasite lysates across the 48-h lifecycle (0 h indicates reinvasion, red arrow) probed with antiserum against PfADF1, PfAct1, P. falciparum aldolase (PfALDO; loading), and P. falciparum apical membrane antigen 1 (PfAMA1; timing). (B) IFA of mature P. falciparum schizont stages labeled with PfAct1, PfADF1, and PfRON4 antisera. r, rabbit; m, mouse. (Scale bars, 2 μm.) (C) Solubility of schizont and merozoite lysate following hypotonic lysis. Control immunolabeling with merozoite surface protein-1(19) (MSP1-19, a GPI-anchored protein) and 70-kDa heat shock protein (Hsp70) as a loading control. P, pellet fraction; S, supernatant fraction. (D) IFA of merozoite invasion probed with anti-PfADF1, PfAct1, and PfRON4 during merozoite invasion. (Scale bars, 2 μm.)
Erythrocyte invasion is a time when actin dynamics are required for parasite movement (5, 6) and therefore is a time when actin regulators are expected to be active (3). We explored PfADF1 localization by IFA in free merozoites captured mid-invasion and colabeled with rhoptry neck protein 4 (RON4), a marker of the tight junction (24) that is the point of close apposition between host and parasite membranes during invasion. The tight junction was shown recently to be the site at which a ring of actin forms (24), a feature consistent with engagement of the actomyosin motor at the junction. PfADF1 retained its cytosolic localization during invasion (Fig. 1D) with the exception that labeling was absent at the tight junction and its associated actin ring (Fig. 1D). This distribution would support a predominant role for the majority of PfADF1 molecules in actin monomer sequestration, although it does not directly address its function with respect to filaments.
X-Ray Crystal Structure of PfADF1 Confirms Presence of G-actin Binding Site.
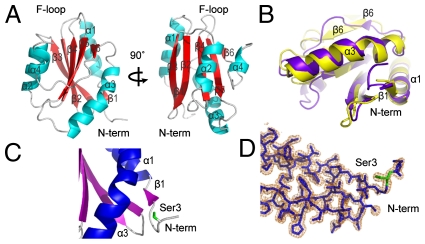
To assess the biochemical properties of PfADF1 in greater detail, we prepared recombinant PfADF1 for crystallographic structure determination. X-ray diffraction from PfADF1 crystals permitted structure determination and refinement to 1.6 Å (Table S1). Defined electron density of the whole protein chain was interpretable except for the disordered C-terminal Lys122, which was omitted. PfADF1 adopts the conserved α/β fold of the AC family, a six-stranded β-sheet in which four antiparallel β strands (β2–β5) are flanked on either edge by a shorter parallel strand along with four α helices surrounding the β-sheet (Fig. 2A and Fig. S2). Structural alignment of PfADF1 with budding yeast cofilin, ScCOF, showed that the G-actin–binding sites (Fig. S2) (18, 25, 26) superimpose closely (rmsd on 41 Cα atoms = 1.6 Å) (Fig. 2B). β6 is markedly shorter in PfADF1than in ScCOF and, significantly, loops out from the sheet. Because residues from β6 provide contact sites for subdomain 3 of the actin monomer identified in the crystal structure of the mouse-twinfilin-actin complex, it is possible that divergence in this region in PfADF1 may provide the basis for promoting nucleotide exchange (10), inducing a more open configuration in the cleft between subdomains 2 and 4 of the actin monomer. A conserved serine at the N terminus that is frequently the target for AC phosphoregulation (12, 13) is well defined in the PfADF1 electron density map (Fig. 2 C and D). Given its exposed position and overlap with the G-actin–binding site of the N-terminal tail, phosphorylation at this residue would prevent complex formation with actin, suggesting the possibility of phosphoregulation in PfADF1.
Fig. 2.
Crystal structure of PfADF1 confirms the conserved G-actin binding fold. (A) Schematic representations of the overall fold of PfADF1 related by 90° rotations. (B) Overlay of crystal structures of PfADF1 (3Q2B; purple) and ScCOF (PDB ID code 1COF; yellow) orientated to show the G-actin binding face conserved between two proteins. (C) Cartoon representation of PfADF1 structure showing Ser3 at the N-terminal tail. (D) 2|Fo|-|Fc| electron density map depicting the N-terminal tail of PfADF1. Residue Ser3 is shown as a green stick. The map is contoured at 1.2 σ. Water molecules have been removed for clarity.
F-Actin Binding Motifs and Associated Residues Are Absent in PfADF1.
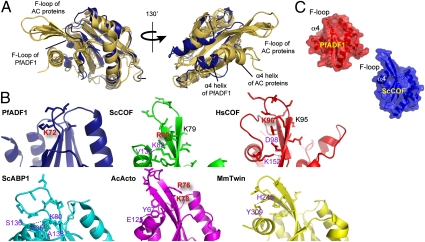
Comparison of the PfADF1 structure with other AC proteins (including PfADF2) revealed significant changes in a key structural motif implicated in actin filament binding and disassembly (17, 19–21), that is, the F-loop and C-terminal helix (α4), which together form the putative F-actin–binding fold (21, 27). Although the basis for AC protein–filament interactions are not known (27) several charged residues in this motif have been implicated in the process (17, 19–21). Chief among these are Lys79, Arg80, and Lys82 of ScCOF located on β5 (or Lys95 and Lys96 of Homo sapiens cofilin, HsCOF), and Glu134, Arg135, and Arg138 of ScCOF located on α4 (19, 21) (Fig. S2).
In PfADF1, truncation of α4 has eliminated the latter cluster of charged residues (Fig. 3A and Figs. S2 and S3). Moreover, strands β4 and β5 are substantially shorter than in ScCOF, so that the tips of the connecting F-loops in superimposed Pf and Sc structures are separated by a distance of 16 Å (Fig. 3 A and C and Fig. S2). Importantly, this combination of features alters the molecular geometry to preclude direct interaction between the F-loop and C terminus. A key difference is that, although in ScCOF Lys82 is sufficiently close to make an intramolecular hydrogen bond to the α4 main chain (via Val136; Fig. 3B), the homologous residue in PfADF1 (Lys72) instead makes an intermolecular hydrogen bond to an adjacent molecule in the crystal. Because the distance between Lys72 and the C-terminal Ile121 is ∼13.4 Å, Lys72 would be solvent exposed in solution (Fig. 3B). Notably, although the conserved lysine is positioned and oriented similarly in the two structures, Lys82 lies midway along β5 in ScCOF, but in PfADF1 it is the very first residue of a shortened β5. Overall, this variation leads to a pronounced difference in the morphology and electrostatic appearance of PfADF1. The Plasmodium protein has an exposed hydrophobic crevice on an otherwise flattened face (Fig. 3C) that in ScCOF is enclosed by α4 and the extended F-loop.
Fig. 3.
F-loop and C-terminal α4 helix implicated in filament disassembly are substantially reduced in PfADF1. (A) Overlay of the crystal structures of PfADF1 (PDB ID code 3Q2B) with AC proteins [MmTwin (PDB ID code 3DAW), SpCOF (PDB ID code 2I2Q), and ScCOF (PDB ID code1COF)] shown from two different viewpoints rotated by 130°. PfADF1 and AC proteins are shown as blue and yellow, respectively. (B) Schematic representations of the F-loops from AC proteins. Basic residues in the exposed state present in the F-loops are highlighted in red text. Residues involved in polar interactions between the F-loop and the α4 helix are highlighted in purple text. Hydrogen bonds are indicated by dashed lines. (C) Transparent surface representation of PfADF1 and ScCOF structures.
PfADF1 Can Mediate Filament Severing Despite Substantial Divergence in the F-Actin Binding Motif.
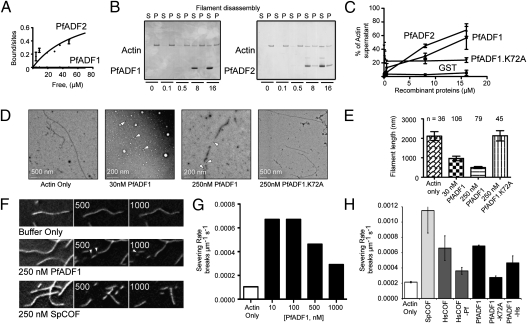
To investigate its ability to regulate actin dynamics in vitro, we first examined the binding affinity of PfADF1 for rabbit muscle F-actin. A fixed concentration of PfADF1 was incubated with varying concentrations of preformed filaments followed by ultracentrifugation to determine the dissociation constant (Kd) between PfADF1 and filaments. SDS/PAGE and densitometry analysis showed that all PfADF1 remained in the recovered supernatant across all filament concentrations tested (Fig. S4), thus precluding determination of the Kd but suggesting that PfADF1 has a very low affinity for actin filaments. Control measurement with PfADF2, which has a more canonical F-loop and C-terminal helix (Fig. S5), although still low, showed a Kd of 64.6 μM (Fig. 4A and Fig. S4).
Fig. 4.
Filament disassembly by PfADF1 and its binding affinity for filaments. (A) Graph of bound/sites versus free protein for PfADF1 and PfADF2. Curve is the best fit to the data, with a Kd of 64.6 μM (n = 3) for PfADF2. A Kd for PfADF1 was not determinable. (B) Ultracentrifugation of PfADFs with preformed actin filaments. P, pellet; S, supernatant. (C) Quantification of proportion of actin in respective fractions (from Fig. 4B). Data shown represent mean ± SEM for n = 3; 0 vs. 16 μM PfADF2, P = 0.023, t test; 0 vs. 16 μM PfADF1, P = 0.035. (D) Electron microscopy of rabbit actin filaments ± PfADF1 and PfADF1.K72A mutant. (E) Quantification of actin filament length in presence of PfADF1 and PfADF1.K72A. Data presented are mean ± SEM. (F–H) TIRF microscopy observation of real-time filament severing by PfADF1. (F) Representative time-lapse micrographs of the indicated reactions (with addition of buffer alone, 250 nM PfADF1, or 250 nM SpCOF at time 0). See also Movie S1. Numbers indicate time in seconds. (G) Plot of the severing rate (breaks μm−1 s−1) in the presence of different concentrations of PfADF1. (H) Plot of the severing rate (breaks μm−1 s−1) in the presence of indicated proteins (n = 10 filaments per condition) at 100 nM. Values are mean ± SD.
Despite showing low affinity for actin filaments, sedimentation analysis in which PfADF1 was varied with respect to a fixed concentration of preformed filaments resulted in concentration-dependent increase of actin in the supernatant (P < 0.05, t test, 0 vs. 16 μM of PfADF1; Fig. 4 B and C). This result, in contrast to the previous report (10), unexpectedly demonstrates that PfADF1 can facilitate actin filament severing and/or depolymerization despite possessing a substantially divergent F-actin binding motif. This result is in line with recent observations from its homolog in T. gondii, TgADF1 (28). PfADF2 similarly was able to mediate filament disassembly, whereas GST alone had no effect (Fig. 4 B and C). To examine whether PfADF1 can disassemble filaments composed of native P. falciparum actin1, recombinant PfAct1 was expressed and purified from E. coli (Fig. S6), verified for structural integrity by circular dichroism spectroscopy (Fig. S6 A, B, and E), and functionally analyzed by sedimentation under polymerizing conditions (Fig. S6F). As with rabbit actin, PfADF1 was able to induce concentration-dependent disassembly of preformed PfAct1 filaments (P < 0.05, t test, 0 vs.16 μM of PfADF1; Fig. S6 G and H).
To explore the nature of PfADF1-mediated filament disassembly, we visualized preformed actin filaments in the presence of PfADF1 by electron microscopy (Fig. 4D). Coincubation of actin filaments in the presence of 30 or 250 nM of PfADF1 was strongly associated with shorter lengths of preformed actin filaments (P < 0.05, t test, control vs. 30 or 250 nM PfADF1; Fig. 4E), suggesting severing. To verify severing, direct observation of actin filament kinetics was undertaken with total internal reflection fluorescence (TIRF) microscopy (29). Although no increased rate of depolymerization was seen, PfADF1 efficiently severed filaments at a rate comparable to that of Schizosaccharomyces pombe cofilin (SpCOF) and human cofilin 1 (COF1) (Fig. 4 F–H and Movie S1) (16), demonstrating maximal severing rates between 10 and 100 nM. Combined, these results conclusively demonstrate filament severing by PfADF1. They establish that severing can occur despite a substantial reduction in the extended F-loop and C-terminal α4 helix. Furthermore, they demonstrate that severing does not require a high-affinity interaction between an AC protein and an actin filament, consistent with previous findings in which the optimal severing concentration is significantly lower than the Kd between an AC protein and an actin filament (16).
Minimum of One Solvent-Exposed Basic Residue in the F-Loop Is Essential for AC-Mediated Filament Severing.
To investigate a universal mechanism of AC-mediated filament severing, we explored whether Lys72, the only conserved solvent-exposed basic residue retained in the F-loop of PfADF1 in comparison with other AC proteins (Fig. 3B and Figs. S2 and S5), is the minimal requirement for severing of actin filaments. A mutant PfADF1, PfADF1.K72A, was generated in which the exposed basic residue was replaced by a nonpolar alanine. Circular dichroism spectroscopy confirmed the folding of PfADF1.K72A to be identical to that of the wild-type protein (Fig. S6 C, D, and E). Electron and TIRF microscopy combined with actin sedimentation assays showed conclusively that the PfADF1.K72A mutant is not able to mediate filament disassembly or severing (Fig. 4 C–E and H). This result establishes that Lys72, the only solvent-exposed basic residue in the F-loop, is the essential requirement for PfADF1-dependent filament-severing activity. We explored the universality of this mechanism by generating an HsCOF mutant in which the native F-loop was replaced by that of PfADF1, along with comparable elimination of its α4 helix. This HsCOF-Pf mutant was able to mediate severing (Fig. 4H, P < 0.05, t test, HsCOF-Pf vs. buffer control). As a control, a reciprocal PfADF1 with the F-loop and α4 helix of HsCOF was generated (PfADF1-Hs), which retained a similar degree of severing (Fig. 4H, P < 0.05, t test, PfADF1-Hs vs. buffer control). The reduction of activity compared with native ADFs for both mutants probably was the result of their chimeric nature. Collectively, these data establish that a single solvent-exposed basic residue in the F-loop is the minimal requirement for AC protein-mediated actin filament severing.
Discussion
Cell motility and host cell invasion in malaria parasites is critically dependent on the regulated assembly and disassembly of actin filaments (5, 6). However, despite its centrality to parasite development, malaria and related parasites from the phylum Apicomplexa possess a markedly reduced repertoire of known eukaryotic actin regulators (3). The core repertoire retained includes two formins, profilin, a cyclase-associated protein (CAP)/Srv2 homolog capping protein subunits, coronin, and two members of the AC family of proteins (reviewed in refs. 3, 9). Of these, CAP/Srv2 and one of the capping protein subunits have been shown to be nonessential for asexual development (9). The importance of actin dynamics in asexual development and in invasion in particular (5) suggests that the roles of the remaining actin regulators probably are critical. To further their detailed characterization, we present here the crystal structure of an apicomplexan AC protein, PfADF1, which reveals a substantial divergence in motifs traditionally associated with F-actin binding. Despite these differences in structure and apparent low binding affinity for filaments as compared with other AC proteins, PfADF1 is capable of mediating filament disassembly via severing. This result is in line with observations from its homolog in T. gondii, TgADF1 (28). Importantly, PfADF1 is capable of disassembling PfAct1 filaments, which are markedly divergent from those of other eukaryotes (3).
The crystal structure of PfADF1 completes the triad of the core actin monomer-binding proteins in apicomplexan parasites. Structures recently have been published for PfProfilin (30) and CAP/Srv2 from the malaria-related parasite Cryptosporidium parvum (31); each, like PfADF1, possesses features unique to the phylum. Our structural analysis of PfADF1 reveals that it retains the conserved G-actin binding motif, comprised of the N terminus, long α3 helix, and turn connecting the β6 strand and α4 helix. However, structural characteristics associated with filament binding are more divergent. Present understanding has linked F-actin binding and disassembly/severing to charged residues located at the extended F-loop and C-terminal α4 helix as well as the C-terminal tail, which fold together to form a highly conserved F-actin binding motif (19–21). A marked reduction in the size of the F-loop and the near absence of the α4 helix in PfADF1 would suggest, based on the present understanding of the process, that filament disassembly is not a feature of apicomplexan AC proteins. However, we clearly demonstrate that PfADF1 is capable of F-actin severing. This observation is strongly supported by recent biochemical data from the malaria-related parasite T. gondii, in which the direct ortholog of PfADF1, TgADF, also mediated actin filament disassembly and severing (28). The ability to mediate disassembly independent of the implicated filament-binding motifs (19, 21) is inconsistent with expectations. Previous characterizations of AC proteins suggested these properties were dependent on charged residues located at the extended F-loop (Arg80 and Lys82 of ScCOF and Lys95,96 of HsCOF; Fig. S2 and Fig. 3B) and α4 helix (Glu134, Arg135, and Arg138 of ScCOF ; Fig. S2 and Fig. 3A) (19, 21), leading to a model in which molecular interactions between AC proteins and the actin filament could be driven partly by salt-bridge or polar interactions. The structure of PfADF1 shows that these charged residues, with the exception of Lys72 (homologous to Lys82 of ScCOF), are absent. This result suggests either that PfADF1 and its apicomplexan orthologs use a unique mechanism that facilitates actin severing in the absence of conserved motifs or that our understanding of AC-mediated disassembly, in general, requires refinement.
Toward the latter possibility, comparative structural analysis of representative AC proteins and their varying capacity to bind and sever F-actin (Table 1) leads us to suggest a revised model in which filament severing does not require high-affinity binding to filaments but instead depends on the availability of a free basic residue in the F-loop exposed for interaction with F-actin (Fig. S7). Filament binding is observed with several structurally defined AC proteins, including budding yeast ScCOF and S. cerevisiae actin-binding protein 1 (ScABP1), HsCOF, and Acanthamoeba castellani actophorin (AcActophorin) (Table 1). Each has a stabilized F-actin binding fold that is anchored by polar interactions between the F-loop and α4 helix [Lys82 of β5 with the main chain amide of Val136 of α4 in ScCOF (19)] (Fig. 3B). In ScABP1, which does not cause filament disassembly (32), anchoring is achieved via the homologous Lys80 interacting with the main chains of Ile135, Ser136, and Ala138. Indeed, the substitution ofLys80 for an alanine residue significantly reduces the binding of the protein to filaments (32), presumably by disrupting the F-binding fold. Similar phenotypes are observed when this anchoring residue is mutated in ScCOF (Lys82 to alanine in ScCOF) (19). In HsCOF, anchoring is inverted, with interactions mediated between Lys152 of α4 and Asp98 of the F-loop (Fig. 3B). Mouse twinfilin retains an extended F-loop and α4 helix (26) and superficially resembles a canonical AC protein. However, its F-loop contains significantly divergent residues (Fig. S2 and Fig. 3B), potentially explaining twinfilin's inability to bind or disassemble actin filaments (33). In this context, the reduction of the F-loop and α4 helix in PfADF1 and the absence of necessary polar interactions that facilitate an anchored fold are consistent with PfADF1’s apparent low affinity for filaments. This evidence suggests that the anchored conformation is essential for high-affinity binding to actin filaments. Supporting this possibility, PfADF2, which retains a more classical F-loop and C-terminal α4 helix in its predicted structure, showed greater affinity for actin filaments than did PfADF1.
Table 1.
Summary of AC protein actin filament-binding and -severing properties
| ScCOF residue numbering |
|||||||||
| AC protein | PDB ID code | F-loop | α4 helix | F-binding | Severing | K79 | K80 | K82 | Ref. |
| ScCOF* | 1COF | KRSK | Extended | High | +++¶ | K | R exposed | K anchored | 17, 19 |
| ScCOF.R80A.K82A | — | KASA | Extended | —§ | -‖ | K | A | A | 17,19 |
| HsCOF* | 1Q8G | KKED | Extended | High | +++ | K | K exposed | D | 21 |
| HsCOF.K95Q.K96Q | — | QQED | Extended | — | — | Q | Q | D | 21 |
| HsCOF-Pf† | — | AVSK | Truncated | — | ++ | A | V | K ∼exposed | — |
| AcActophorin* | 1AHQ | QRNK | Extended | High | +++ | Q | R exposed | K exposed | 36 |
| PfADF1* | 3Q2B | AVSK | Truncated | Low | +++ | A | V | K exposed | — |
| PfADF1-K72A | — | AVSA | Truncated | Low | — | A | V | A | — |
| PfADF1-Hs | — | KKED | Extended | High | ++ | K | K | D | — |
| ScABP1 | 1HQZ | DVEK | Extended | High | — | D | V | K anchored | 32 |
| MM_TWIN1_C | 3DAW | YLES | Extended | — | — | Y | L | S | 33, 37 |
| TgADF | — | CGNK | Truncated | Low | +++ | C | G | K ∼exposed | 28 |
| TgADF-t‡ | — | CGNK | ∼Extended | High | — | C | G | K ∼anchored | 28 |
∼ indicates predicted structure in the absence of experimental proof.
*Structurally defined AC proteins with demonstrated severing activity.
†HsCOF mutant with HsCOF F-loop replaced by PfADF1 F-loop.
‡A mutant form of TgADF with an α4 helix of SpCOF fused to the C terminus of the protein.
§No binding to filaments.
¶Strong severing activity.
‖Inability to sever.
Each of the structurally characterized AC proteins able to sever actin filaments retains a basic residue in the F-loop that is exposed to the solvent (Table 1). The side chain of this basic residue (Arg80 in ScCOF, Lys96 in HsCOF, and Arg76 in AcActophorin; Fig. 3B), which positions toward the α4 helix (following ScCOF nomenclature), always faces the same direction as the anchoring residue (Lys82 of ScCOF) or the exposed residue (Asp98 in HsCOF and Lys78 of AcActophorin) present in the F-loop and is predicted to be orientated to interact with subdomain 2 of an actin monomer in the filament (Fig. 3B) (21, 27). This exposed basic residue is notably absent in ScABP1, where the sole basic Lys80 of the F-loop is occupied in polar interactions with the α4 helix. However, it is present in PfADF1, despite reduction of the α4 helix and extended F-loop. This exposed Lys72 is the only structurally conserved motif, already implicated in filament binding, which can singularly explain the potential for filament severing in AC proteins. Importantly, we show that substitution of Lys72 to alanine disrupts the ability of PfADF1 to mediate filament disassembly and severing. Furthermore, chimeras in which the native F-loop and α4 helices of PfADF1 and HsCOF are interchanged can still mediate filament severing. Collectively, these data demonstrate that AC-mediated filament severing requires a minimum of one exposed basic residue in the F-loop.
In the recent characterization of TgADF1 (also assumed to lack the anchored fold), the addition of the C-terminal α4 helix of SpCOF reduced the ability of this AC protein to mediate filament disassembly with a reported, although not shown, increase in F-actin binding potential (28). We predict that addition of an α4 helix structurally altered this protein, making it more akin to ScABP1 in which the previously exposed Lys68 is drawn into a polar interaction with the C-terminal helix. The presence of an exposed basic residue in the F-loop therefore would be expected to enable filament severing, irrespective of the existence of an anchored F-loop/α4 helix fold. In HsCOF, Lys95 was reported to be required for filament severing; an HsCOF.K95QK96Q mutant was unable to sever filaments (21) (Table 1). However, the ScCOF.R80AK82A mutant that still possessed Lys79 (homologous to Lys95 of HsCOF) lost severing activity (19) (Table 1). This result rules out Lys95 of HsCOF as a requirement for severing; in accordance with the model we present here, the loss of activity found in HsCOF.K95QK96Q is caused solely by the K96Q substitution. Consistent with this hypothesis, Lys95 of HsCOF and Lys79 of ScCOF both face away from the α4 helix (Fig. 3B). Collectively these observations indicate that for AC-mediated severing to occur, the exposed basic residue must orientate in the direction of the α4 helix.
High-resolution details of the interface formed between AC proteins and the actin filament will be needed to determine the molecular mechanism through which such an exposed residue could mediate severing. Cryo-electron microscopy of AC-decorated actin filaments suggests that the two sites on opposing ends of the AC protein (the F-actin and G-actin binding motifs) mediate binding to subdomains 1 and 2 of the lower actin subunit and to subdomains 1–3 of the upper actin subunit, respectively (21, 27). It also has been suggested that a local twist in the filament structure induced by AC binding is associated with filament severing (27). Our data suggest the the ability to sever must reside in a balance between the presence of functional G-actin binding surfaces and the availability of an exposed basic residue in the F-loop (Fig. S7).
The reduced repertoire of actin regulators in malaria and other apicomplexan parasites highlights the usefulness of these remarkable ancient eukaryotes for investigation of universal mechanisms of actin regulation. Furthermore, the reliance on a minimal set of actin regulators in the malaria parasite may constitute a hitherto unexplored target for therapeutic intervention against this increasingly drug-resistant pathogen of global significance.
Materials and Methods
Parasite Cultures, Imaging, and Immunoblotting.
Wild-type (D10) asexual parasites were maintained in standard culture conditions and synchronized as described (24). Free merozoites were isolated and processed for microscopy following ref. 24. Antibodies and image processing are described in SI Materials and Methods.
Expression and Purification of Recombinant AC Proteins and PfAct1.
Full-length PfADF1 and PfADF2 were PCR amplified from P. falciparum genomic DNA and expressed as GST-fusion proteins using the pGEX4T vector (GE Healthcare). PfAct1 was expressed as a hexa-His fusion protein using the pET28 vector (Novagen). Full details on amplification, expression, and purification of each AC protein and mutants are given in SI Materials and Methods.
Crystallization and X-Ray Data Collection.
Purified PfADF1 (minus the GST tag) in storage buffer [20 mM Tris (pH8), 10 mM NaCl, 5 mM 2-mercaptoethanol, and 0.02% NaN3] was concentrated to 6.6 mg/mL, and crystals were grown at 22 °C by hanging-drop vapor diffusion mixing 1 μL protein solution with 1 μL of reservoir buffer consisting of 1.8 M (NH4)2SO4, 0.2 M KNa tartrate, 0.1 M NaOAc, pH 4.6. Crystals were equilibrated into cryoprotectant consisting of 0.2 M KNa tartrate, 3 M Na malonate, pH 4.6. X-ray diffraction data were collected at the Australian Synchrotron. Data were integrated and scaled with HKL2000 (34). Structure determination and refinement are described in SI Materials and Methods. Data collection and refinement statistics for PfADF1 are presented in Table S1. Coordinate and structure factors are available from the Protein Data Bank (PDB ID code 3Q2B).
Actin Biochemical Assays.
Filament disassembly was assayed by analytical ultracentrifugation at 100,000 × g (TLA100 rotor, Beckman Coulter Optima TL Ultracentrifuge) with rabbit actin (4 μM; Cytoskeleton, Inc.) or PfAct1 (4 μM) essentially as described (28) with varying concentrations of recombinant PfADF1, PfADF2, or control GST added to preformed actin filaments prepared under polymerizing conditions for 1 h in a predominately ADP + Pi-rich form 1 h before sedimentation. Quantification of protein in pellet and supernatant fractions was performed by densitometry analysis using a GS-800 calibrated densitometer (Bio-Rad). PfADF1 and PfADF2 (fixed at 5 μM) were added to varying concentrations of preformed actin filaments (polymerized for 1 h) for 30 min at room temp. A precentrifugation aliquot was removed, and the remainder was centrifuged 100,000 × g for 1 h. An equal amount was analyzed by SDS/PAGE and densitometry pre- and postcentrifugation. The amount of ADF bound to preformed actin filaments was determined as total ADF (5 μM) − free ADF postcentrifugation. The free actin concentration was determined as total actin − bound ADF. Using a fixed concentration of PfADF1 (5 μM), the equilibrium Kd was calculated as bound/sites (5 μM) = free actin/Kd − free actin (35). The data were fit to this equation using nonlinear regression (Prism software, GraphPad). Electron and TIRF microscopy followed established protocols (21, 29) detailed further in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Andrew Holmes, Dejan Bursac, Trevor Lithgow, Tony Hodder, and Wai-Hong Tham for experimental help and reagents and the staff of the CSIRO Bio21 Collaborative Crystallization Centre and Australian Synchrotron for invaluable assistance. Human erythrocytes were kindly provided by the Red Cross Blood Bank (Melbourne). This work was supported by National Health and Medical Research Council Project Grant 516747 (to J.B.). J.B. is supported through a Future Fellowship (FT100100112) from the Australian Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3Q2B).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018927108/-/DCSupplemental.
References
- 1.Baum J, et al. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J Biol Chem. 2006;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- 2.Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat Rev Microbiol. 2006;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- 4.Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 5.Miller LH, Aikawa M, Johnson JG, Shiroishi T. Interaction between cytochalasin B-treated malarial parasites and erythrocytes. Attachment and junction formation. J Exp Med. 1979;149:172–184. doi: 10.1084/jem.149.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuno Y, et al. Effect of jasplakinolide on the growth, invasion, and actin cytoskeleton of Plasmodium falciparum. Parasitol Res. 2002;88:844–848. doi: 10.1007/s00436-002-0666-8. [DOI] [PubMed] [Google Scholar]
- 7.Schüler H, Mueller AK, Matuschewski K. Unusual properties of Plasmodium falciparum actin: New insights into microfilament dynamics of apicomplexan parasites. FEBS Lett. 2005;579:655–660. doi: 10.1016/j.febslet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Wetzel DM, Håkansson S, Hu K, Roos D, Sibley LD. Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol Biol Cell. 2003;14:396–406. doi: 10.1091/mbc.E02-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sattler JM, Ganter M, Hliscs M, Matuschewski K, Schuler H. Actin regulation in the malaria parasite. Eur J Cell Biol. 2011 doi: 10.1016/j.ejcb.2010.11.011. 10.1016/j.ejcb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Schüler H, Mueller AK, Matuschewski K. A Plasmodium actin-depolymerizing factor that binds exclusively to actin monomers. Mol Biol Cell. 2005;16:4013–4023. doi: 10.1091/mbc.E05-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 12.Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- 13.Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 14.van Rheenen J, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J. 2010;98:2327–2336. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC. Structure determination of yeast cofilin. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- 18.Guan JQ, Vorobiev S, Almo SC, Chance MR. Mapping the G-actin binding surface of cofilin using synchrotron protein footprinting. Biochemistry. 2002;41:5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 19.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono S, et al. The C-terminal tail of UNC-60B (actin depolymerizing factor/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- 21.Pope BJ, Gonsior SM, Yeoh S, McGough A, Weeds AG. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J Mol Biol. 2000;298:649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- 22.Doi Y, Shinzawa N, Fukumoto S, Okano H, Kanuka H. ADF2 is required for transformation of the ookinete and sporozoite in malaria parasite development. Biochem Biophys Res Commun. 2010;397:668–672. doi: 10.1016/j.bbrc.2010.05.155. [DOI] [PubMed] [Google Scholar]
- 23.Mehta S, Sibley LD. Actin depolymerizing factor controls actin turnover and gliding motility in Toxoplasma gondii. Mol Biol Cell. 2011;22:1290–1299. doi: 10.1091/mbc.E10-12-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riglar DT, et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe. 2011;9:9–20. doi: 10.1016/j.chom.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 26.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta S, Sibley LD. Toxoplasma gondii actin depolymerizing factor acts primarily to sequester G-actin. J Biol Chem. 2010;285:6835–6847. doi: 10.1074/jbc.M109.068155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kursula I, et al. Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 2008;16:1638–1648. doi: 10.1016/j.str.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Hliscs M, et al. Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector. J Biol Chem. 2010;285:11572–11583. doi: 10.1074/jbc.M109.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintero-Monzon O, Rodal AA, Strokopytov B, Almo SC, Goode BL. Structural and functional dissection of the Abp1 ADFH actin-binding domain reveals versatile in vivo adapter functions. Mol Biol Cell. 2005;16:3128–3139. doi: 10.1091/mbc.E05-01-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paavilainen VO, et al. Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc Natl Acad Sci USA. 2007;104:3113–3118. doi: 10.1073/pnas.0608725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. In: Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology, Macromolecular Crystallography, part A. Carter CWJ, Sweet RM, editors. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 35.Wachsstock DH, Schwartz WH, Pollard TD. Affinity of alpha-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophys J. 1993;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchoin L, Robinson RC, Choe S, Pollard TD. Phosphorylation of Acanthamoeba actophorin (ADF/cofilin) blocks interaction with actin without a change in atomic structure. J Mol Biol. 2000;295:203–211. doi: 10.1006/jmbi.1999.3336. [DOI] [PubMed] [Google Scholar]
- 37.Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J Cell Sci. 2002;115:881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.