Abstract
Acute exacerbations of chronic obstructive pulmonary disease (COPD) are a major source of morbidity and contribute significantly to healthcare costs. Although bacterial infections are implicated in nearly 50% of exacerbations, only a handful of pathogens have been consistently identified in COPD airways, primarily by culture-based methods, and the bacterial microbiota in acute exacerbations remains largely uncharacterized. The aim of this study was to comprehensively profile airway bacterial communities using a culture-independent microarray, the 16S rRNA PhyloChip, of a cohort of COPD patients requiring ventilatory support and antibiotic therapy for exacerbation-related respiratory failure. PhyloChip analysis revealed the presence of over 1,200 bacterial taxa representing 140 distinct families, many previously undetected in airway diseases; bacterial community composition was strongly influenced by the duration of intubation. A core community of 75 taxa was detected in all patients, many of which are known pathogens. Bacterial community diversity in COPD airways is substantially greater than previously recognized and includes a number of potential pathogens detected in the setting of antibiotic exposure. Comprehensive assessment of the COPD airway microbiota using high-throughput, culture-independent methods may prove key to understanding the relationships between airway bacterial colonization, acute exacerbation, and clinical outcomes in this and other chronic inflammatory airway diseases.
Introduction
Chronic obstructive pulmonary disease (COPD) affects more than 12 million individuals in the United States and is the fourth leading cause of chronic morbidity and mortality (Rabe et al., 2007). A significant proportion of COPD-related healthcare costs are attributable to hospitalization for respiratory exacerbations (Mannino and Braman, 2007), with severe exacerbations having been associated with high mortality rates (Connors et al., 1996; Nseir et al., 2006). Bacterial infections are implicated in approximately 50% of COPD exacerbations (Sethi and Murphy, 2008). However, in most studies to date, bacterial identification has primarily relied on culture-based methods (Papi et al., 2006; Rosell et al., 2005; Soler et al., 2007) or species-specific, targeted PCR approaches (Murphy et al., 2004). Therefore, the true depth of bacterial diversity present in COPD airways is unknown, and the potential role for mixed-species bacterial communities in the pathogenesis of chronic airway colonization and acute exacerbations have been largely overlooked.
It is increasingly recognized that the human host is colonized by diverse, site-specific microbial communities that constitute the human microbiome (Dethlefsen et al., 2007; Eckburg et al., 2005). Growing interest in characterizing these consortia and their interactions with the host, is exemplified by the formation in 2008 of the International Human Microbiome Consortium (IHMC), a collaborative effort to merge data generated through the U.S. NIH Human Microbiome Project (HMP; http://nihroadmap.nih.gov/hmp) and the E.U. Metagenomics of the Human Intestinal Tract (MetaHIT; www.metahit.eu) initiatives. Constant exposure of the respiratory tract with the external environment could conceivably lead to important microbe–microbe and microbe–host interactions, yet the human airway microbiota, particularly that present in the context of pulmonary disease, remains largely uncharacterized. Given the limitations of culture-based methods, culture-independent approaches that identify species, or groups of closely related species, based on sequence polymorphisms in conserved genes, such as the 16S ribosomal RNA (16S rRNA) gene, enable a more comprehensive assessment of microbial members present in a mixed-species population. Such methods have increasingly been applied to identify bacterial consortia in a variety of human niches with demonstration that community composition is related to disease states such as obesity, ventilator-associated pneumonia and cystic fibrosis airway disease (Flanagan et al., 2007; Harris et al., 2007; Rogers et al., 2004; Turnbaugh et al., 2006).
An alternative approach to traditional culture-independent methods of microbial community analysis is the 16S rRNA PhyloChip, a high-density microarray containing 500,000 probes that can detect approximately 8,500 bacterial taxa [taxa are defined as a group of bacteria sharing at least 97% sequence homology within the 16S rRNA gene sequence; (Brodie et al., 2006)] The PhyloChip has previously been shown to detect substantially greater bacterial diversity compared in parallel with traditional clone library sequencing approaches (Brodie et al., 2006; Desantis et al., 2007; Flanagan et al., 2007). Using this microarray, we analyzed airway specimens from eight COPD patients who were being managed for severe respiratory exacerbations to determine if a more diverse bacterial community is present during pulmonary exacerbation in the setting of antibiotic administration.
Materials and Methods
Subject selection and sample collection
Potential subjects for this study were screened from a database of airway specimens collected between August 2004 and April 2006 from mechanically ventilated patients admitted to the intensive care units at Moffitt-Long Hospital (University of California, San Francisco), who were enrolled in a parent study of Pseudomonas aeruginosa in intubated patients (Flanagan et al., 2007). Subjects admitted to the ICU with a primary diagnosis of “COPD exacerbation” were identified for inclusion in our study. Available endotracheal aspirates (ETAs) from eight patients were processed for 16S rRNA PhyloChip analysis, as detailed below. To compare results from PhyloChip analysis with conventional clinical cultures, we obtained results of quantitative clinical laboratory bacterial cultures (blood agar, chocolate agar, and EMB media) performed on minibronchoalveolar lavage (m-BAL) airway samples, collected within 1–5 days of the ETA specimen analyzed by PhyloChip, as previously described (Flanagan et al., 2007). We have previously found m-BALs to possess a similar bacterial community composition to that of ETAs obtained concurrently from the same patient (Flanagan et al., 2007). Clinical data (Table 1) were recorded in a secure database, including whether a diagnosis of pneumonia by conventional clinical and radiologic criteria (Zhuo et al., 2008) was made during the patient's hospitalization and the time frame between diagnosis and collection of airway samples. The Committee on Human Research at UCSF approved all study protocols, and all patients or their surrogates provided written, informed consent.
Table 1.
Clinical Characteristics of Subjects and Samples
| Patient | Age | Gender | Intubation days at sample collection | Antimicrobial therapy received within the past month | Days of active antimicrobial therapy at time of sample collection | Culture resultsa |
|---|---|---|---|---|---|---|
| 1 | 63 | M | 16 | ceftazidime | 16 | PAb* |
| 2 | 69 | F | 6 | vancomycin, tobramycin, levofloxacin | 5 | PAb* |
| 3 | 78 | M | 1 | vancomycin, piperacillin/tazobactam, levofloxacin | 1 | PAb*, KPb*, AF |
| 4 | 78 | M | 21 | piperacillin/tazobactam | 31 | PAb*, SMb* |
| 5 | 86 | F | 17 | levofloxacin | 17 | PAb* |
| 6 | 85 | F | 16 | doxycycline, moxifloxacin, vancomycin | 1 | PAb* |
| 7 | 61 | M | 5 | vancomycin, piperacillin/tazobactam | 7 | PAb*, SAb |
| 8 | 73 | M | 3 | piperacillin/tazobactam | 3 | PAb, EAb* |
mini-BAL, minibronchoalveolar lavage clinical culture. The most recent, available culture data were obtained from within 1–5 days prior to the endotracheal aspirate sample analyzed by PhyloChip.
Detected by PhyloChip; *≥10,000 colony-forming units on quantitative mini-BAL culture.
PA, Pseudomonas aeruginosa; KP, Klebsiella pneumoniae; SA, Staphylococcus aureus; EA, Enterobacter aerogenes; SM, Stenotrophomonas maltophilia; AF, Aspergillus fumigatus.
DNA extraction, 16S rRNA gene amplification, PhyloChip processing
Total DNA was extracted from ETAs (200 μL) using a bead-beating step (5.5 m s−1 for 30 s, FastPrep system) (MP Biomedicals, Cleveland, OH) prior to nucleic acid extraction using the Wizard Genomic DNA Purification kit (Promega, Madison, WI). Twelve, 25-cycle PCR reactions, containing 100 ng of DNA, 2.5 mM each of dNTPs, 1.5 μM each primer (Bact-27F and Bact-1492R) (Lane, 1991) and 0.02 U/μL of ExTaq (Takara Bio, Japan), were performed for each sample across a gradient of annealing temperatures (48–58°C), to maximize the diversity recovered. The resulting PCR products were pooled and gel-purified using the MinElute Gel Extraction kit (Qiagen, Chatsworth, CA). Known concentrations of synthetic 16S rRNA gene fragments and non-16S rRNA gene fragments were spiked into the pooled, purified PCR product, which served as internal standards for normalization. A total of 250 ng of purified PCR product per sample was fragmented, biotin-labeled, and hybridized to the microarray as previously described (Brodie et al., 2006). Washing, staining, and scanning of arrays were conducted according to standard Affymetrix protocols. Background subtraction, noise calculations and scaling were carried out as described previously (Brodie et al., 2006; Desantis et al., 2007).
Analysis of PhyloChip data
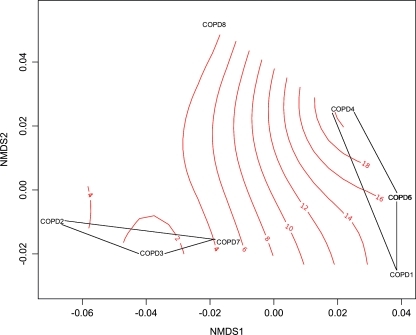
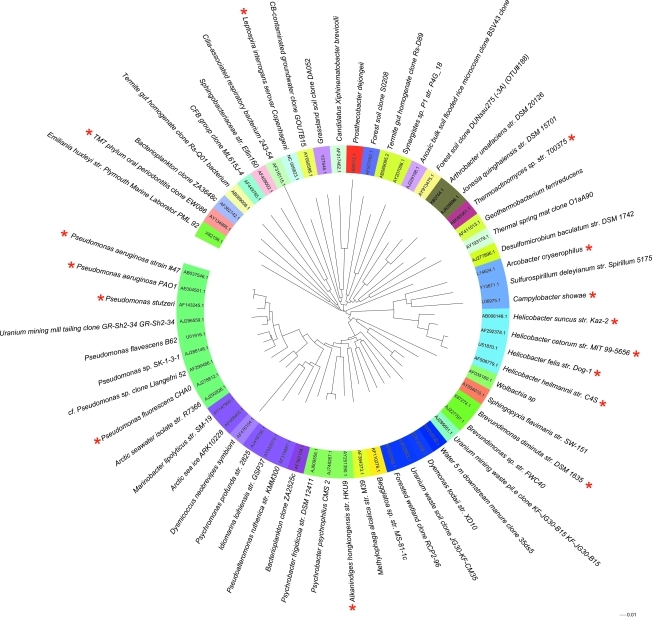
Relatively conservative detection and quantification criteria for each taxon were applied, as previously described (Desantis et al., 2007). Briefly, probe-pairs consisting of a perfectly matched and mismatched cross hybridization control probe (containing a mismatch at the 13th nucleotide) were scored as positive if they met two criteria: (1) the fluorescence intensity of the perfectly matched probe was ≥1.3 times greater than that of the mismatched probe, and (2) the difference in intensity in each probe pair was 130 times greater than the squared noise value for that array. The positive fraction (pf ) of probe sets (minimum of 11, median of 24 probe-pairs per taxon) was calculated, and a taxon was considered “present” if the calculated pf was ≥90%. Statistical analyses were performed in the R environment (www.R-project.org), using the ecological community analysis package vegan (version 1.16-1). Log-transformed fluorescence intensities were used to calculate Bray-Curtis dissimilarity measures of ecological distance. Nonmetric multidimensional scaling (NMDS), a nonparametric ordination method that maps community relatedness, in this case using the Bray-Curtis distance metric, was used to assess variability in bacterial community structure. The function adonis (Anderson, 2001), which conducts a matrix-based nonparametric analysis of variance, was applied to explore relationships between community composition and clinical variables, including age, gender, number of intubation days, presence of pneumonia, time frame between pneumonia diagnosis and sample collection, antibiotic and corticosteroid treatments, and survival to ICU discharge. Between-group differences in taxon abundance were assessed by two-tailed t-testing with significance adjusted for false discovery using q-values, as previously described (Storey and Tibshirani, 2003). Taxa exhibiting q values <0.05, a p-value ≤0.02 and a change of >1,000 fluorescence units (log-fold change in 16S rRNA copy number) were considered statistically and biologically significant. Phylogenetic trees were constructed using representative 16S rRNA sequences from the Greengenes database (Desantis et al., 2006). A neighbor-joining tree with nearest-neighbor interchange was produced using FastTree (Price et al., 2009) and uploaded to the Interactive Tree of Life project (http://itol.embl.de/) for annotation (Letunic and Bork, 2007).
Quantitative polymerase chain reaction (Q-PCR)
To confirm that changes in array fluorescence intensities were reflective of changes in target organism abundance, triplicate, Q-PCR reactions were performed for selected taxa containing species of interest, using a Stratagene MxP3000 real-time system and the QuantiTect SYBR Green PCR kit (Qiagen). Primers for taxa containing selected species of interest were designed based on PhyloChip probes for the target taxon (Table 2). Reaction conditions were similar to those previously described (Brodie et al., 2007) with the exception that 10 ng of DNA extract were used in 40 cycles of reaction using the annealing temperatures listed in Table 2 for each primer set. Regression analyses of inverse cycle threshold values plotted against PhyloChip fluorescence intensities were determined for each targeted taxon.
Table 2.
Primers Used for Q-PCR Validation of Targeted Species
| Species | Primers | Annealing temperature |
|---|---|---|
| P. aeruginosa | 5′-CAGTAAGTTAATACCTTGCTGTGCTG-3′ | 55°C |
| 5′- TGCTGAACCACCTACGCGC-3′ | ||
| S. maltophilia | 5′-GCCGGCTAATACCTGGTTGGGA-3′ | 55°C |
| 5′- CTACCCTCTACCACACTCTAGTCGC-3′ | ||
| H. cetorum | 5′-GCGTTACTCGGAATCACTGGGCGTA-3′ | 48°C |
| 5′- ATGAGTATTCCTCTTGATCTCTACG-3′ | ||
| C. mucosalis | 5′-ATGTGGTTTAATTCGAAGATACGCG-3′ | 52°C |
| 5′- CACGAGCTGACGACAGCCGTGCAGC-3′ |
Results
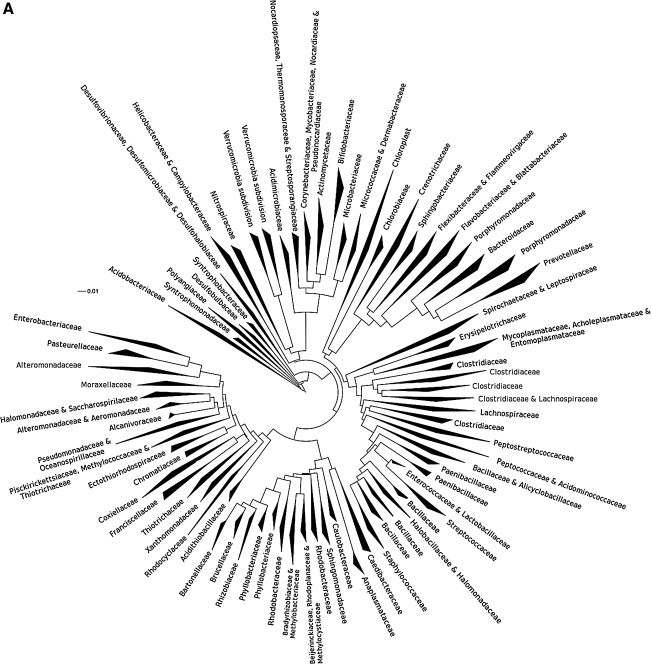
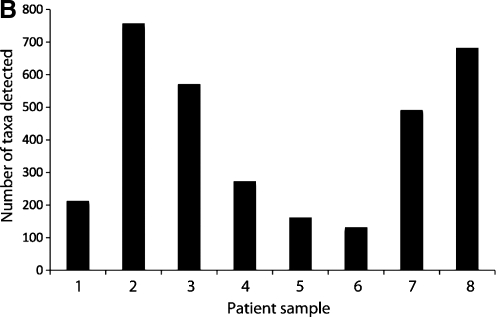
16S rRNA PhyloChip analysis identified a total of 1,213 bacterial taxa present in airway samples from COPD patients obtained during the course of acute exacerbations (complete list is provided in Table 3; see at end of article). Despite recent or ongoing exposure to antibiotics across the group, the mean number of taxa detected in each sample was 411 ± 246 taxa (SD). Identified taxa represented a diverse group of species belonging to 38 bacterial phyla and 140 distinct families (Fig. 1A). Bacterial families detected included members of the Pseudomonadaceae, Pasteurellaceae, Helicobacteraceae, Enterobacteriaceae, Comamonadaceae, Burkholderiaceae, and Alteromonadaceae, among many others. In addition, recently described phyla such as the TM7 subgroup of Gram-positive uncultivable bacteria were also detected in the airways of these patients (Table 3).
Table 3.
All Bacterial Taxa Detected by 16S rRNA Phylochip in Airway Samples of COPD Patients Being Treated for Severe Exacerbations
| Phylum | Class | Order | Family | S-Fa | Taxon IDb | Representative speciesc |
|---|---|---|---|---|---|---|
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 508 | uranium mining waste pile clone JG37-AG-81 sp. |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 541 | uranium mill tailings soil sample clone GuBH2-AG-47 sp. |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 209 | uranium mining waste pile clone JG37-AG-29 sp. |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6425 | Great Artesian Basin clone B27 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6335 | forested wetland clone FW45 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_6 | 6345 | soil sample uranium mining waste pile near town Johanngeorgenstadt clone JG36-TzT-77 bacterium |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6350 | soil isolate Ellin337 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6356 | forested wetland clone FW47 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_6 | 6359 | PCE-contaminated site clone CLi114 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_6 | 6362 | grassland soil clone DA052 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6366 | PCB-polluted soil clone WD228 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6368 | soil clone UA2 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6378 | Acidobacterium capsulatum |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6410 | |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6412 | acid mine drainage clone TRB82 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_16 | 6414 | PCE-contaminated site clone CLs73 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6421 | PCB-polluted soil clone WD217 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_6 | 6423 | coal effluent wetland clone FW92 |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_14 | 6424 | sphagnum peat bog clone K-5b10 |
| Acidobacteria | Unclassified | Unclassified | Unclassified | sf_1 | 572 | forested wetland clone FW144 |
| Acidobacteria | Acidobacteria-4 | Ellin6075/11-25 | Unclassified | sf_1 | 435 | anaerobic VC-degrading enrichment clone VC47 bacterium |
| Acidobacteria | Acidobacteria-5 | Unclassified | Unclassified | sf_1 | 523 | soil metagenomic library clone 17F9 |
| Acidobacteria | Acidobacteria-4 | Ellin6075/11-25 | Unclassified | sf_1 | 790 | soil clone 11-25 |
| Acidobacteria | Acidobacteria-4 | Ellin6075/11-25 | Unclassified | sf_1 | 87 | activated sludge clone 2951 |
| Acidobacteria | Acidobacteria-6 | Unclassified | Unclassified | sf_1 | 350 | Mammoth cave clone CCM15a |
| Acidobacteria | Acidobacteria-6 | Unclassified | Unclassified | sf_1 | 897 | Mammoth cave clone CCM8b |
| Acidobacteria | Acidobacteria-6 | Unclassified | Unclassified | sf_1 | 1049 | soil clone C112 |
| Acidobacteria | Solibacteres | Unclassified | Unclassified | sf_1 | 6426 | Great Artesian Basin clone B11 |
| Acidobacteria | Acidobacteria-4 | Unclassified | Unclassified | sf_1 | 6363 | soil clone 32-11 |
| Acidobacteria | Unclassified | Unclassified | Unclassified | sf_1 | 4222 | forested wetland clone FW105 |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Acidimicrobiaceae | sf_1 | 1090 | |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Acidimicrobiaceae | sf_1 | 1749 | forest soil clone DUNssu275 (-3A) (OTU#188) |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Acidimicrobiaceae | sf_1 | 1856 | forested wetland clone RCP2-105 |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Acidimicrobiaceae | sf_1 | 1360 | forested wetland clone RCP2-103 |
| Actinobacteria | Actinobacteria | Actinomycetales | Acidothermaceae | sf_1 | 1399 | uranium mill tailings clone Gitt-KF-183 |
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | sf_1 | 1684 | Varibaculum cambriense str. CCUG 44998 |
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | sf_1 | 1227 | Actinomyces naeslundii |
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | sf_1 | 1672 | Actinomyces odontolyticus str. CCUG 28084 |
| Actinobacteria | Actinobacteria | Actinomycetales | Actinosynnemataceae | sf_1 | 1463 | Saccharothrix texasensis str. NRRL B-16107T |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1351 | Bifidobacterium psychraerophilum str. T16 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1967 | Bifidobacterium pseudocatenulatum str. JCM1200 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 2040 | Bifidobacterium adolescentis str. E-981074T |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1109 | Bifidobacterium thermacidophilum porcinum subsp. suis str. P3-14 subsp. |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1987 | human subgingival plaque clone CX010 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1444 | Bifidobacteriaceae genomosp. C1 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1835 | Bifidobacterium breve str. KB 92 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | sf_1 | 1875 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Cellulomonadaceae | sf_1 | 1586 | Cellulomonas gelida str. DSM 20111T |
| Actinobacteria | Actinobacteria | Actinomycetales | Cellulomonadaceae | sf_1 | 1748 | Beutenbergia cavernosa str. DSM 12333 |
| Actinobacteria | Actinobacteria | Coriobacteriales | Coriobacteriaceae | sf_1 | 1258 | ground water deep-well injection disposal site radioactive wastes Tomsk-7 clone S15A-MN25 |
| Actinobacteria | Actinobacteria | Coriobacteriales | Coriobacteriaceae | sf_1 | 1800 | ground water deep-well injection disposal site radioactive wastes Tomsk-7 clone S15A-MN100 |
| Actinobacteria | Actinobacteria | Coriobacteriales | Coriobacteriaceae | sf_1 | 1958 | Atopobium vaginae VA14183_00 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1517 | Corynebacterium xerosis str. DSM 20743 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1492 | Corynebacterium tuscaniae str. ISS-5309 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1820 | Corynebacterium jeikeium str. ATCC 43734 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1089 | Corynebacterium mucifaciens National Microbiology Laboratory Special identifier 01-0118 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1374 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1428 | Corynebacterium simulans National Microbiology Laboratory Special identifier 00-0186 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1493 | Corynebacterium tuberculostearicum str. CIP102346 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | sf_1 | 1803 | Corynebacterium spheniscorum str. CCUG 45512 |
| Actinobacteria | Actinobacteria | Actinomycetales | Dermabacteraceae | sf_1 | 2053 | Brachybacterium nesterenkovii str. DSM 9573 |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | sf_1 | 1598 | lichen-dominated Antarctic cryptoendolithic community clone FBP402 |
| Actinobacteria | Actinobacteria | Actinomycetales | Kineosporiaceae | sf_1 | 1961 | Kineococcus aurantiacus str. IFO 15268 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | sf_1 | 1667 | Microbacterium lacticum |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | sf_1 | 1197 | Arctic sea ice ARK10173 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | sf_1 | 1437 | freshwater clone SV1-16 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1266 | Arthrobacter psychrolactophilus |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1557 | Arthrobacter oxydans str. DSM 20119 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1593 | Arthrobacter globiformis |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1610 | Arthrobacter sp str. AC-51 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1966 | TCE-contaminated site clone ccspost2208 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1324 | glacial ice isolate str. CanDirty1 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1494 | Arthrobacter agilis str. DSM 20550 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1573 | Arthrobacter nicotianae str. SB42 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1889 | Citricoccus sp. str. 2216.25.22 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 2019 | Micrococcus luteus str. HN2-11 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1724 | Rothia mucilaginosa str. DSM |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 2020 | Rothia dentocariosa str. ChDC B200 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 2063 | Rothia dentocariosa str. ATCC 17931 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | sf_1 | 1213 | Kocuria roseus |
| Actinobacteria | Actinobacteria | Actinomycetales | Micromonosporaceae | sf_1 | 1876 | Couchioplanes subsp. caeruleus str. IFO13939 |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1175 | Mycobacterium cf. xenopi 'Hymi_Wue Tb_939/99' str. Hymi_Wue Tb_939/99 |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1262 | Mycobacterium holsaticum str. 1406 |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1308 | Mycobacterium pyrenivorans str. DSM 44605 |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1365 | Mycobacterium chelonae str. CIP 104535T |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1650 | Mycobacterium tuberculosis str. NCTC 7416 H37Rv |
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | sf_1 | 1726 | Mycobacterium terrae str. ATCC 15755 |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiaceae | sf_1 | 1834 | Nocardia transvalensis str. DSM 43405 |
| Actinobacteria | Actinobacteria | Actinomycetales | Nocardiopsaceae | sf_1 | 1385 | Streptomonospora salina str. YIM90002 |
| Actinobacteria | Actinobacteria | Actinomycetales | Promicromonosporaceae | sf_1 | 1671 | Cellulosimicrobium cellulans str. NCIMB 11025 |
| Actinobacteria | Actinobacteria | Actinomycetales | Promicromonosporaceae | sf_1 | 1711 | Promicromonospora sukumoe str. DSM 44121 |
| Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | sf_1 | 1863 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | sf_1 | 1343 | Saccharomonospora azurea str. M.Goodfel K161 = NA128 (type st |
| Actinobacteria | Actinobacteria | Rubrobacterales | Rubrobacteraceae | sf_1 | 1551 | soil isolate Ellin301 |
| Actinobacteria | Actinobacteria | Rubrobacterales | Rubrobacteraceae | sf_1 | 1739 | |
| Actinobacteria | Actinobacteria | Rubrobacterales | Rubrobacteraceae | sf_1 | 1843 | uranium mining waste pile soil sample clone JG30-KF-A23 |
| Actinobacteria | Actinobacteria | Actinomycetales | Sporichthyaceae | sf_1 | 1695 | lichen-dominated Antarctic cryptoendolithic community clone FBP417 |
| Actinobacteria | Actinobacteria | Actinomycetales | Streptosporangiaceae | sf_1 | 1190 | Nonomuraea polychroma str. IFO 14345 |
| Actinobacteria | Actinobacteria | Actinomycetales | Thermomonosporaceae | sf_1 | 1741 | Actinomadura pelletieri str. IMSNU 22169T |
| Actinobacteria | Actinobacteria | Actinomycetales | Thermomonosporaceae | sf_1 | 1546 | Actinomadura fulvescens str. DSM 43923T |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | sf_2 | 1233 | |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | sf_1 | 1898 | termite gut homogenate clone Rs-J10 bacterium |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | sf_1 | 1367 | |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | sf_1 | 1370 | forested wetland clone RCP1-37 |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Unclassified | sf_1 | 1666 | |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_4 | 1337 | Sturt arid-zone soil clone #0425-2M17 |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1486 | deep marine sediment clone MB-A2-108 |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | sf_1 | 1676 | |
| Actinobacteria | Actinobacteria | Acidimicrobiales | Unclassified | sf_1 | 1217 | DCP-dechlorinating consortium clone SHA-34 |
| Actinobacteria | BD2-10 group | Unclassified | Unclassified | sf_2 | 1652 | marine sediment clone Bol7 |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 2045 | hypersaline lake clone ML602J-44 |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1130 | Georgenia muralis str. 1A-C |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1687 | Jonesia quinghaiensis str. DSM 15701 |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1243 | termite gut homogenate clone Rs-M95 bacterium |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1577 | termite gut homogenate clone Rs-N91 bacterium |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1405 | Arthrobacter ureafaciens str. DSM 20126 |
| AD3 | Unclassified | Unclassified | Unclassified | sf_1 | 2338 | uranium mining waste pile soil clone JG30-KF-C12 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | sf_12 | 5256 | termite gut homogenate clone Rs-D38 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | sf_12 | 5320 | Bacteroides distasonis |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | sf_12 | 5474 | Bacteroides acidofaciens str.A24 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | sf_12 | 5551 | Bacteroides uniformis |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Bacteroidaceae | sf_12 | 5979 | Bacteroides fragilis str. YCH46 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Blattabacteriaceae | sf_1 | 5828 | Blattabacterium species |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 5334 | autotrophic nitrifying biofilm clone NB-11 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 5619 | anaerobic VC-degrading enrichment clone VC10 bacterium |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 5888 | penguin droppings sediments clone KD9-169 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 6123 | Flexibacter japonensis str. IFO 16041 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 6267 | Cilia-respiratory isolate str. 243-54 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 6249 | Haliscomenobacter hydrossis |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flammeovirgaceae | sf_5 | 6084 | Microscilla arenaria str. IFO 15982 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 6079 | synonym: CFB group clone APe4_42 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5367 | patient's bronchoalveolar lavage isolate str. MDA2507 sp. |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5915 | groundwater deep-well injection disposal site radioactive wastes Tomsk-7 clone S15A-MN27 bacterium |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5997 | Flavobacterium aquatile |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 6274 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5317 | Tenacibaculum maritimum str. IFO 15946 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5991 | Tenacibaculum ovolyticum str. IAM14318 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 6252 | Riftia pachyptila's tube clone R103-B20 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5263 | subgingival plaque clone DZ074 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5401 | Capnocytophaga gingivalis str. ChDC OS45 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5836 | Capnocytophaga granulosa str. LMG 12119; FDC SD4 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5423 | Aequorivita antarctica str. QSSC9-14 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5942 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5955 | Flavobacterium sp. str. V4.MS.29 = MM_2747 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5971 | Cytophaga uliginosa |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5436 | Arctic sea ice ARK10004 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5473 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5267 | bacterioplankton clone AEGEAN_179 |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | sf_1 | 5914 | Psychroserpens burtonensis str. S2-64 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5563 | Cytophaga sp. I-545 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5542 | Cytophaga sp. I-1787 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5307 | Microscilla sericea str. IFO 16561 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5357 | Flexibacter ruber str. IFO 16677 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5372 | |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5566 | Hongiella mannitolivorans str. IMSNU 14012 JC2050 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5667 | penguin droppings sediments clone KD6-118 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 5994 | Hymenobacter sp. str. NS/50 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 6124 | Flexibacter flexilis subsp. pelliculosus str. IFO 16028 subsp. |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_19 | 6297 | EBPR sludge lab scale clone HP1A92 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | sf_20 | 10311 | Cytophaga sp. str. BHI60-57B |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5295 | swine intestine clone p-987-s962-5 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5680 | termite gut clone Rs-106 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5800 | Porphyromonas endodontalis str. ATCC 35406 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5817 | termite gut homogenate clone Rs-N56 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5961 | chlorobenzene-degrading consortium clone IA-16 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5454 | Dysgonomonas wimpennyi str. ANFA2 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5510 | sphagnum peat bog clone 26-4b2 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 6012 | mouse feces clone L11-6 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Porphyromonadaceae | sf_1 | 5460 | mouse feces clone F8 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5718 | Prevotella tannerae str. 29-1 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5437 | cow rumen clone BE1 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5916 | cow rumen clone BE14 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 6011 | rumen clone F24-B03 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 6152 | rumen clone RF37 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 6259 | |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5249 | Prevotella denticola str. ATCC 35308 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5484 | oral periodontitis clone FX046 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5706 | oral cavity clone 3.3 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5769 | Bacteroidaceae str. A42 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5905 | swine intestine clone p-2443-18B5 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5940 | Prevotella sp. str. E7_34 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 5946 | tongue dorsa clone DO027 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 6047 | deep marine sediment clone MB-A2-107 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Prevotellaceae | sf_1 | 6239 | tongue dorsa clone DO033 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Rikenellaceae | sf_5 | 5892 | anoxic bulk soil flooded rice microcosm clone BSV73 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | sf_1 | 5513 | crevicular epithelial cells clone AZ123 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | sf_1 | 5913 | Sphingobacteriaceae str. Ellin160 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5573 | termite gut homogenate clone Rs-D44 bacterium |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Unclassified | sf_6 | 5439 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-40 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5475 | SHA-25 clone |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5544 | marine? clone KD3-17 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5783 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-15 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5874 | Paralvinella palmiformis mucus secretions clone P. palm 53 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5890 | penguin droppings sediments clone KD1-125 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 6046 | chlorobenzene-degrading consortium clone IIIB-1 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5820 | cow rumen clone BF24 |
| Bacteroidetes | Unclassified | Unclassified | Unclassified | sf_1 | 5745 | |
| Bacteroidetes | Flavobacteria | Flavobacteriales | Unclassified | sf_3 | 5248 | Delaware River estuary clone 1G12 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5355 | DCP-dechlorinating consortium clone SHA-5 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5481 | marine sediment above hydrate ridge clone Hyd89-72 bacterium |
| Bacteroidetes | Unclassified | Unclassified | Unclassified | sf_4 | 5703 | |
| Bacteroidetes | Unclassified | Unclassified | Unclassified | sf_4 | 5785 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-56 |
| Bacteroidetes | Unclassified | Unclassified | Unclassified | sf_4 | 5787 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-1 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 5957 | Paralvinella palmiformis mucus secretions clone P. palm C/20 bacterium |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | sf_15 | 6324 | temperate estuarine mud clone KM02 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Unclassified | sf_3 | 6168 | Toolik Lake main station at 3 m depth clone TLM11/TLMdgge04 |
| Bacteroidetes | KSA1 | Unclassified | Unclassified | sf_1 | 5951 | CFB group clone ML615J-4 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Unclassified | sf_3 | 6298 | travertine hot spring clone SM1C04 |
| BRC1 | Unclassified | Unclassified | Unclassified | sf_2 | 118 | penguin droppings sediments clone KD1-1 |
| BRC1 | Unclassified | Unclassified | Unclassified | sf_1 | 5051 | soil clone PBS-III-24 |
| BRC1 | Unclassified | Unclassified | Unclassified | sf_1 | 5143 | soil clone PBS-II-1 |
| Caldithrix | Unclassified | Caldithrales | Caldithraceae | sf_2 | 91 | benzoate-degrading consortium clone BA059 |
| Caldithrix | Unclassified | Caldithrales | Caldithraceae | sf_1 | 2384 | saltmarsh clone LCP-89 |
| Chlamydiae | Chlamydiae | Chlamydiales | Chlamydiaceae | sf_1 | 4820 | Chlamydophila pneumoniae str. AR39 |
| Chlamydiae | Chlamydiae | Chlamydiales | Parachlamydiaceae | sf_1 | 4964 | neutral pH mine biofilm clone 44a-B1-34 |
| Chlorobi | Chlorobia | Chlorobiales | Chlorobiaceae | sf_1 | 262 | Chlorobium ferrooxidans DSM 13031 str. KofoX |
| Chlorobi | Chlorobia | Chlorobiales | Chlorobiaceae | sf_1 | 859 | Chlorobium phaeovibrioides str. 2631 |
| Chlorobi | Chlorobia | Chlorobiales | Chlorobiaceae | sf_1 | 995 | Chlorobium limicola str. M1 |
| Chlorobi | Unclassified | Unclassified | Unclassified | sf_8 | 5822 | Saltmarsh mud clone K-790 |
| Chlorobi | Unclassified | Unclassified | Unclassified | sf_6 | 5294 | Mammoth cave clone CCM9b |
| Chlorobi | Unclassified | Unclassified | Unclassified | sf_9 | 6146 | sludge clone A12b |
| Chlorobi | Unclassified | Unclassified | Unclassified | sf_8 | 549 | benzene-degrading nitrate-reducing consortium clone Cart-N2 bacterium |
| Chlorobi | Unclassified | Unclassified | Unclassified | sf_8 | 636 | benzene-degrading nitrate-reducing consortium clone Cart-N3 bacterium |
| Chloroflexi | Thermomicrobia | Unclassified | Unclassified | sf_1 | 1041 | Antarctic cryptoendolith clone FBP471 |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_2 | 818 | |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_5 | 1051 | forest soil clone DUNssu055 (-2B) (OTU#087) |
| Chloroflexi | Anaerolineae | Chloroflexi-1a | Unclassified | sf_1 | 927 | Paralvinella palmiformis mucus secretions clone P. palm C 37 bacterium |
| Chloroflexi | Thermomicrobia | Unclassified | Unclassified | sf_2 | 652 | uranium mining waste pile soil sample clone JG30-KF-CM45 |
| Chloroflexi | Anaerolineae | Chloroflexi-1a | Unclassified | sf_1 | 106 | DCP-dechlorinating consortium clone SHD-231 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 375 | forest soil clone C043 |
| Chloroflexi | Anaerolineae | Chloroflexi-1a | Unclassified | sf_1 | 487 | thermophilic UASB granular sludge isolate str. IMO-1 bacterium |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 576 | DCP-dechlorinating consortium clone SHA-36 |
| Chloroflexi | Anaerolineae | Chloroflexi-1a | Unclassified | sf_1 | 583 | anaerobic bioreactor clone SHD-238 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 72 | sediments collected at Charon's Cascade near Echo River October 2000 clone CCD21 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 727 | forest soil clone S0208 |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_7 | 757 | DCP-dechlorinating consortium clone SHA-8 |
| Chloroflexi | Anaerolineae | Chloroflexi-1a | Unclassified | sf_1 | 76 | DCP-dechlorinating consortium clone SHA-147 |
| Chloroflexi | Anaerolineae | Chloroflexi-1b | Unclassified | sf_2 | 789 | travertine hot spring clone SM1D10 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 946 | temperate estuarine mud clone KM87 |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2339 | uranium mill tailings soil sample clone Sh765B-TzT-20 bacterium |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2367 | deep marine sediment clone MB-B2-113 |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2438 | deep marine sediment clone MB-A2-110 |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2445 | deep marine sediment clone MB-A2-103 |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2485 | |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | sf_1 | 2497 | forested wetland clone FW60 |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_12 | 2523 | sponge clone TK10 |
| Chloroflexi | Chloroflexi-4 | Unclassified | Unclassified | sf_2 | 2344 | forest soil clone C083 |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_1 | 2534 | forest soil clone S085 |
| Coprothermobacteria | Unclassified | Unclassified | Unclassified | sf_1 | 751 | Coprothermobacter sp. str. Dex80-3 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 4967 | Toolik Lake main station at 3 m depth clone TLM14 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5147 | Emiliania huxleyi str. Plymouth Marine Laborator PML 92 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5112 | Cyanidium caldarium str. 14-1-1 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5006 | |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_11 | 5098 | Euglena tripteris str. UW OB |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_11 | 5123 | Lepocinclis fusiformis str. ACOI 1025 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 4966 | Adiantum pedatum |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 4976 | Calypogeia muelleriana |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_13 | 5000 | Mitrastema yamamotoi |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5040 | Solanum nigrum |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5182 | Epifagus virginiana—chloroplast |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5183 | Pisum sativum—chloroplast |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5192 | Cycas revoluta |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_5 | 4998 | |
| Cyanobacteria | Cyanobacteria | Thermosynechococcus | Unclassified | sf_1 | 5012 | Synechococcus sp. str. PCC 6312 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_9 | 5038 | Rumen isolate str. YS2 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_9 | 5164 | termite gut homogenate clone Rs-H34 |
| Cyanobacteria | Cyanobacteria | Oscillatoriales | Unclassified | sf_1 | 5189 | Oscillatoria sancta str. PCC 7515 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_5 | 5015 | Chlorogloeopsis fritschii str. PCC 6912 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_5 | 5030 | Hapalosiphon welwitschii |
| Cyanobacteria | Cyanobacteria | Nostocales | Unclassified | sf_1 | 5057 | Nodularia sphaerocarpa str. UTEX B 2093 |
| Cyanobacteria | Cyanobacteria | Oscillatoriales | Unclassified | sf_1 | 5049 | Oscillatoria spongeliae str. 520 bg |
| Cyanobacteria | Cyanobacteria | Plectonema | Unclassified | sf_1 | 5190 | Plectonema sp. str. F3 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_8 | 5206 | |
| Deinococcus-Thermus | Unclassified | Unclassified | Unclassified | sf_1 | 178 | Thermus sp. str. C4 |
| Deinococcus-Thermus | Unclassified | Unclassified | Unclassified | sf_1 | 563 | Vulcanithermus mediatlanticus str. TR |
| Deinococcus-Thermus | Unclassified | Unclassified | Unclassified | sf_2 | 637 | hypersaline pond clone LA7-B27N |
| Deinococcus-Thermus | Unclassified | Unclassified | Unclassified | sf_3 | 920 | |
| DSS1 | Unclassified | Unclassified | Unclassified | sf_2 | 38 | DCP-dechlorinating consortium clone SHA-109 |
| DSS1 | Unclassified | Unclassified | Unclassified | sf_1 | 4405 | benzoate-degrading consortium clone BA143 |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 3955 | Weeping tea tree witches' broom phytoplasma tree |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 3961 | Clover yellow edge mycoplasma-like organism |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 3975 | Black raspberry witches' broom phytoplasma str. BRWB witches' broom room |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 3976 | |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 4044 | |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 4045 | Chinaberry yellows phytoplasma |
| Firmicutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | sf_1 | 4046 | Pigeon pea witches' broom mycoplasma-like organism |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3386 | feedlot manure clone B87 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3522 | Aerococcus viridans |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3631 | Abiotrophia defectiva str. GIFU12707 (ATCC49176) |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3870 | Abiotrophia para-adiacens str. TKT1 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3323 | Trichococcus flocculiformis str. DSM 2094 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3326 | Nostocoida limicola I str. Ben206 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3504 | Marinilactibacillus psychrotolerans str. O21 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3553 | Desemzia incerta str. DSM 20581 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3833 | Carnobacterium alterfunditum |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | sf_1 | 3840 | Trichococcus pasteurii str. KoTa2 |
| Firmicutes | Bacilli | Bacillales | Alicyclobacillaceae | sf_1 | 3368 | geothermal site isolate str. G1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3612 | Bacillus schlegelii str. ATCC 43741T |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3419 | Bacillus algicola str. KMM 3737 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3424 | uranium mill tailings clone Gitt-KF-76 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3661 | Bacillus sp. str. 2216.25.2 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3688 | Bacillus sp. str. SAFN-006 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3926 | Lake Bogoria isolate 64B4 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 234 | Bacillus vulcani str. 3S-1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 283 | Geobacillus thermocatenulatus str. DSM 730 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 305 | Bacillus thermoleovorans |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3460 | Geobacillus jurassicus str. DS1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3540 | Geobacillus thermoleovorans str. B23 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3763 | Geobacillus stearothermophilus |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3836 | Geobacillus stearothermophilus str. 46 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 385 | Geobacillus stearothermophilus str. T10 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 462 | Geobacillus thermodenitrificans str. DSM 466 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 571 | Bacillus caldotenax str. DSM 406 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 829 | Geobacillus sp. str. YMTC1049 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3635 | Bacillus aeolius str. 4-1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3827 | Bacillus acidogenesis str. 105-2 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3845 | hot synthetic compost clone pPD15 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3895 | Bacillus sporothermodurans str. M215 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3283 | Bacillus niacini str. IFO15566 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3439 | Bacillus siralis str. 171544 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3589 | Bacillus senegalensis str. RS8; CIP 106 669 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3650 | |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 1050 | Bacillus firmus CV93b |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 246 | Bacillus sp. 6160m-C1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3550 | Bacillus megaterium str. QM B1551 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3345 | Bacillus pumilus str. S9 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3328 | Pseudobacillus carolinae |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3370 | Bacillus sp. str. TGS437 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3492 | Bacillus subtilis str. IAM 12118T |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3579 | Bacillus sp. str. TGS750 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3675 | Bacillus mojavensis str. M-1 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3706 | Bacillus sonorensis str. NRRL B-23155 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3831 | Bacillus licheniformis str. KL-068 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3900 | Bacillus licheniformis str. DSM 13 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3909 | Bacillus subtilis subsp. Marburg str. 168 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3918 | Bacillus subtilis |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3467 | Bacillus luciferensis str. LMG 18422 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3489 | Bacillus silvestris str. SAFN-010 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3482 | garbage compost isolate str. M32 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3383 | |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | sf_1 | 3517 | Planococcus maritimus str. TF-9 |
| Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | sf_1 | 3536 | |
| Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | sf_1 | 3792 | Carnobacterium sp. str. D35 |
| Firmicutes | Bacilli | Bacillales | Caryophanaceae | sf_1 | 3285 | Caryophanon latum str. DSM 14151 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 2764 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 3021 | Clostridium caminithermale str. DVird3 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 2915 | Tepidibacter thalassicus str. SC 562 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 3049 | Clostridium paradoxum str. DSM 7308T |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 3077 | Clostridium glycolicum str. DSM 1288 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4156 | MCB-contaminated groundwater-treating reactor clone RA9C1 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4173 | termite gut homogenate clone Rs-D81 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4187 | Clostridiales oral clone P4PB_122 P3 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4278 | granular sludge clone R1p16 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4297 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4300 | termite gut clone Rs-060 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4310 | termite gut clone Rs-056 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4364 | oral endodontic infection clone MCE3_9 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4406 | termite gut homogenate clone Rs-J39 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4477 | termite gut homogenate clone Rs-N85 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4502 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4584 | Clostridium papyrosolvens str. DSM 2782 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4614 | Clostridium sp. str. JC3 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4622 | termite gut clone Rs-L36 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4638 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4554 | termite gut clone Rs-068 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4180 | termite gut homogenate clone Rs-M23 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4225 | termite gut clone Rs-116 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4265 | termite gut homogenate clone Rs-N70 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4266 | termite gut homogenate clone Rs-M86 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4272 | termite gut homogenate clone Rs-M34 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4321 | termite gut homogenate clone Rs-C76 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4357 | Lachnospiraceae bacterium 19gly4 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4359 | termite gut homogenate clone Rs-C69 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4369 | termite gut homogenate clone Rs-N73 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4415 | termite gut homogenate clone Rs-K32 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4418 | termite gut homogenate clone Rs-H18 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4475 | termite gut homogenate clone Rs-N02 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4507 | termite gut homogenate clone Rs-N21 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4524 | termite gut clone Rs-093 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4550 | swine intestine clone p-320-a3 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4559 | cow rumen clone BF30 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4566 | swine intestine clone p-2657-65A5 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4582 | swine intestine clone p-2600-9F5 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4627 | termite gut homogenate clone Rs-A13 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4306 | UASB reactor granular sludge clone PD-UASB-4 bacterium |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4607 | Clostridium novyi str. NCTC538 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4229 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4551 | Clostridium acetobutylicum str. ATCC 824 (T) |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4339 | Clostridium chauvoei str. ATCC 10092T |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4598 | Clostridium sardiniense str. DSM 600 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | sf_12 | 4169 | |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3261 | Enterococcus mundtii str. LMG 10748 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3288 | Isolation and identification hyper-ammonia producing swine storage pits manure |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3298 | Enterococcus saccharolyticus str. LMG 11427 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3318 | Enterococcus ratti str. ATCC 700914 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3382 | |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3392 | Vagococcus lutrae str. m1134/97/1; CCUG 39187 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3433 | Tetragenococcus muriaticus |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3598 | Enterococcus solitarius str. DSM 5634 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3680 | Melissococcus plutonius str. NCDO 2440 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3713 | Enterococcus cecorum str. ATCC43198 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | sf_1 | 3881 | Enterococcus dispar str. LMG 13521 |
| Firmicutes | Mollicutes | Entomoplasmatales | Entomoplasmataceae | sf_1 | 4074 | swine intestine clone p-2013-s959-5 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | sf_3 | 3952 | Erysipelothrix rhusiopathiae str. Pecs 56 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | sf_3 | 3965 | TCE-contaminated site clone ccslm238 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | sf_3 | 3981 | phototrophic sludge clone PSB-M-3 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | sf_3 | 768 | |
| Firmicutes | Clostridia | Clostridiales | Eubacteriaceae | sf_1 | 28 | termite gut homogenate clone Rs-H81 bacterium |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3633 | Bacillus clausii str. GMBAE 42 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3344 | Halobacillus yeomjeoni str. MSS-402 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3488 | Halobacillus salinus str. HSL-3 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3702 | Amphibacillus xylanus str. DSM 6626 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3756 | Salibacillus sp. str. YIM-kkny16 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | sf_1 | 3769 | Gracilibacillus sp. str. YIM-kkny13 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2698 | termite gut homogenate clone Rs-B88 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2804 | Clostridium amygdalinum str. BR-10 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2961 | termite gut homogenate clone Rs-F92 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3042 | swine intestine clone p-2876-6C5 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3036 | termite gut homogenate clone Rs-F27 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2668 | termite gut homogenate clone Rs-G40 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3017 | termite gut homogenate clone Rs-D48 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3076 | Clostridium nexile |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2825 | Butyrivibrio fibrisolvens str. LP1265 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2834 | Butyrivibrio fibrisolvens str. OB156 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2844 | Pseudobutyrivibrio ruminis str. pC-XS2 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3059 | Butyrivibrio fibrisolvens str. NCDO 2249 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2994 | termite gut clone Rs-L15 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3038 | swine intestine clone p-1594-c5 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3171 | Lachnospira pectinoschiza |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2931 | termite gut homogenate clone Rs-G77 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3060 | termite gut homogenate clone Rs-B14 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 3218 | termite gut homogenate clone Rs-N53 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 2681 | termite gut homogenate clone Rs-K41 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4212 | termite gut clone Rs-061 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4273 | termite gut homogenate clone Rs-M14 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4281 | granular sludge clone UASB_brew_B86 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4315 | termite gut homogenate clone Rs-N94 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4331 | granular sludge clone UASB_brew_B84 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4335 | termite gut homogenate clone Rs-N86 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4434 | termite gut homogenate clone Rs-K11 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4510 | termite gut homogenate clone Rs-Q53 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4511 | ckncm314-B7-17 clone |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4512 | granular sludge clone UASB_brew_B25 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4514 | termite gut homogenate clone Rs-B34 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4533 | termite gut homogenate clone Rs-N06 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4535 | ckncm297-B1-1 clone |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4539 | termite gut homogenate clone Rs-C61 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4540 | termite gut homogenate clone Rs-M18 bacterium |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4567 | human colonic clone HuCB5 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4571 | Faecalibacterium prausnitzii str. ATCC 27766 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4613 | rumen clone 3C0d-3 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4623 | human colonic clone HuCA1 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | sf_5 | 4525 | termite gut homogenate clone Rs-Q18 bacterium |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3330 | Lactobacillus kitasatonis str. KM9212 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3342 | Lactobacillus crispatus str. DSM 20584 T |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3478 | Lactobacillus crispatus str. ATCC33197 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3490 | Lactobacillus suntoryeus str. LH |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3618 | Lactobacillus jensenii str. KC36b |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3696 | Lactobacillus kalixensis str. Kx127A2; LMG 22115T; DSM 16043T; CCUG 48459T |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3395 | Lactobacillus reuteri str. DSM 20016 T |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3547 | Lactobacillus frumenti str. TMW 1.666 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3566 | Lactobacillus pontis str. LTH 2587 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3798 | Lactobacillus fermentum str. MD-9 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3521 | Pediococcus inopinatus str. DSM 20285 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3885 | Pediococcus pentosaceus |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3634 | Lactobacillus letivazi str. JCL3994 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3767 | Lactobacillus suebicus str. CECT 5917T |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3810 | Lactobacillus brevis |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3829 | Lactobacillus paralimentarius str. DSM 13238 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3366 | Lactobacillus saerimneri str. GDA154 LMG 22087 DSM 16049 (T); CCUG 48462 (T) |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3418 | Lactobacillus subsp. aviarius |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3703 | Lactobacillus salivarius str. RA2115 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3914 | Lactobacillus cypricasei str. LMK3 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3821 | Lactobacillus casei |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3768 | Lactobacillus perolens str. L532 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | sf_1 | 3526 | Lactobacillus sakei |
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | sf_1 | 3497 | Weissella koreensis S-5673 |
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | sf_1 | 3573 | Leuconostoc ficulneum str. FS-1 |
| Firmicutes | Mollicutes | Mycoplasmatales | Mycoplasmataceae | sf_1 | 3929 | Mycoplasma gypsbengalensis str. Gb-V33 |
| Firmicutes | Mollicutes | Mycoplasmatales | Mycoplasmataceae | sf_1 | 3997 | Mycoplasma salivarium str. PG20(T) |
| Firmicutes | Mollicutes | Mycoplasmatales | Mycoplasmataceae | sf_1 | 4014 | Mycoplasma pulmonis str. UAB CTIP |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 3415 | Paenibacillus nematophilus str. NEM1b |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 3630 | |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 3595 | Paenibacillus sp. str. MB 2039 |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 3299 | Brevibacillus borstelensis str. LMG 15536 |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 3641 | Brevibacillus sp. MN 47.2a |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 319 | Ammoniphilus oxalaticus str. RAOx-FF |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | sf_1 | 625 | Ammoniphilus oxalivorans str. RAOx-FS |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 304 | Selenomonas ruminantium str.JCM6582 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 709 | Selenomonas ruminantium str.S20 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 710 | Centipeda periodontii str. HB-2 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 131 | pig feces clone |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 181 | Allisonella histaminiformans str. MR2 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 59 | swine intestine clone p-1941-s962-3 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 940 | Veillonella dispar str. DSM 20735 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 1036 | Great Artesian Basin clone G07 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 428 | chlorobenzene-degrading consortium clone IIIA-1 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 534 | chlorobenzene-degrading consortium clone IIA-26 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 992 | anoxic bulk soil flooded rice microcosm clone BSV43 clone |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 242 | Desulfosporosinus orientis str. DSMZ 7493 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 300 | benzene-contaminated groundwater clone ZZ12C8 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 39 | forested wetland clone RCP2-71 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2721 | termite gut homogenate clone Rs-N71 bacterium |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2729 | DCP-dechlorinating consortium clone SHA-58 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2679 | termite gut homogenate clone BCf9-13 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2694 | oral periodontitis clone FX028 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2714 | termite gut homogenate clone Rs-N27 bacterium |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2913 | termite gut homogenate clone Rs-N82 bacterium |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 3080 | termite gut homogenate clone Rs-F43 bacterium |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 3112 | Evry municipal wastewater treatment plant clone 012C11_B_SD_P15 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 3182 | termite gut homogenate clone Rs-Q64 bacterium |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2993 | oral clone P2PB_46 P3 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2738 | Mogibacterium neglectum str. ATCC 700924 (= P9a-h) |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2805 | oral periodontitis clone FX033 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 2797 | Isolation and identification hyper-ammonia producing swine storage pits manure |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 619 | TCE-dechlorinating microbial community clone 1G |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 224 | Finegoldia magna str. ATCC 29328 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 58 | Peptostreptococcus sp. str. E3_32 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 1037 | Finegoldia magna |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 616 | Peptoniphilus lacrimalis str. CCUG 31350 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | sf_5 | 393 | Anaerococcus vaginalis str. CCUG 31349 |
| Firmicutes | Bacilli | Bacillales | Sporolactobacillaceae | sf_1 | 3365 | Bacillus sp. clone ML615J-19 |
| Firmicutes | Bacilli | Bacillales | Sporolactobacillaceae | sf_1 | 3747 | Bacillus sp. str. C-59-2 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3258 | Staphylococcus auricularis str. MAFF911484 ATCC33753T |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3284 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3545 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3569 | Staphylococcus saprophyticus |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3585 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3592 | Staphylococcus caprae str. DSM 20608 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3605 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3628 | Staphylococcus haemolyticus str. CCM2737 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3638 | Staphylococcus sp str. AG-30 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3654 | Staphylococcus pettenkoferi str. B3117 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3684 | Staphylococcus sciuri |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3794 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3822 | Staphylococcus succinus str. SB72 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3494 | Micrococcus luteus B-P 26 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3865 | Macrococcus lamae str. CCM 4815 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | sf_1 | 3432 | deep-sea sediment isolate str. P_wp0225 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3722 | Lactococcus Il1403 subsp. lactis str. IL1403 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3869 | Streptococcus equi subsp. zooepidemicus str. Tokyo1291 subsp. |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3699 | Streptococcus agalactiae str. 2603V/R |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3907 | aortic heart valve patient with endocarditis clone v6 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3250 | Streptococcus bovis str. B315 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3253 | derived cheese sample clone 32CR |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3313 | Streptococcus salivarius str. ATCC 7073 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3397 | Streptococcus macedonicus str. ACA-DC 206 LAB617 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3422 | Streptococcus thermophilus str. DSM 20617 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3543 | |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3588 | Streptococcus downei str. ATCC 33748 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3906 | Streptococcus bovis str.ATCC 43143 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3499 | Streptococcus constellatus str. ATCC27823 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3446 | Streptococcus bovis str. HJ50 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3560 | Streptococcus gallinaceus str. CCUG 42692 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3753 | Streptococcus suis str. 8074 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3251 | Streptococcus cristatus str. ATCC 51100 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3287 | tongue dorsum scrapings clone FP015 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3290 | Streptococcus mitis str. Sm91 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3629 | Streptococcus mutans str. UA96 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | sf_1 | 3685 | Streptococcus gordonii str. ATCC 10558 |
| Firmicutes | Clostridia | Clostridiales | Syntrophomonadaceae | sf_5 | 2456 | granular sludge clone R4b14 |
| Firmicutes | Bacilli | Bacillales | Thermoactinomycetaceae | sf_1 | 3301 | Thermoactinomyces sp. str. 700375 |
| Firmicutes | Unclassified | Unclassified | Unclassified | sf_8 | 546 | Ferribacter thermoautotrophicus |
| Firmicutes | Clostridia | Clostridiales | Unclassified | sf_17 | 2324 | |
| Firmicutes | Desulfotomaculum | Unclassified | Unclassified | sf_1 | 2351 | Desulfotomaculum thermobenzoicum str. DSM 6193 |
| Firmicutes | Desulfotomaculum | Unclassified | Unclassified | sf_1 | 2359 | UASB granular sludge clone JP |
| Firmicutes | Clostridia | Unclassified | Unclassified | sf_3 | 2373 | |
| Firmicutes | Symbiobacteria | Symbiobacterales | Unclassified | sf_1 | 2388 | G+C Gram-positive clone YNPRH70A |
| Firmicutes | Unclassified | Unclassified | Unclassified | sf_8 | 2433 | Ferribacter thermoautotrophicus str. JW/JH-Fiji-2 |
| Firmicutes | Desulfotomaculum | Unclassified | Unclassified | sf_1 | 2443 | Desulfotomaculum thermoacetoxidans str. DSM 5813 |
| Firmicutes | Desulfotomaculum | Unclassified | Unclassified | sf_1 | 2490 | Desulfotomaculum solfataricum str. V21 |
| Firmicutes | Clostridia | Unclassified | Unclassified | sf_4 | 2398 | deep marine sediment clone MB-C2-106 |
| Firmicutes | Symbiobacteria | Symbiobacterales | Unclassified | sf_1 | 77 | thermal soil clone YNPFFP9 |
| Firmicutes | Clostridia | Clostridiales | Unclassified | sf_17 | 926 | |
| Firmicutes | Desulfotomaculum | Unclassified | Unclassified | sf_1 | 198 | Pelotomaculum sp. str. JT |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_4 | 2716 | termite gut homogenate clone Rs-F76 bacterium |
| Firmicutes | Clostridia | Clostridiales | Unclassified | sf_17 | 3476 | |
| Firmicutes | Bacilli | Lactobacillales | Unclassified | sf_1 | 3289 | Isobaculum melis CCUG 37660T |
| Firmicutes | Bacilli | Lactobacillales | Unclassified | sf_1 | 3481 | |
| Firmicutes | Clostridia | Clostridiales | Unclassified | sf_17 | 4168 | |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_4 | 4503 | termite gut homogenate clone Rs-H83 bacterium |
| Firmicutes | Unclassified | Unclassified | Unclassified | sf_8 | 4536 | Mono Lake at depth 35m station 6 July 2000 clone ML635J-14 G+C |
| Firmicutes | Clostridia | Unclassified | Unclassified | sf_7 | 4216 | |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_1 | 4261 | termite gut homogenate clone Rs-G04 bacterium |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_1 | 4293 | termite gut homogenate clone Rs-Q01 bacterium |
| Firmicutes | gut clone group | Unclassified | Unclassified | sf_1 | 4298 | human mouth clone P4PA_66 |
| Firmicutes | Clostridia | Clostridiales | Unclassified | sf_17 | 4307 | |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_4 | 4526 | TCE-contaminated site clone ccslm210 |
| Firmicutes | gut clone group | Unclassified | Unclassified | sf_1 | 4616 | rumen clone F23-C12 |
| Fusobacteria | Fusobacteria | Fusobacterales | Fusobacteriaceae | sf_3 | 367 | Leptotrichia amnionii str. AMN-1 |
| Fusobacteria | Fusobacteria | Fusobacterales | Fusobacteriaceae | sf_3 | 558 | Sneathia sanguinegens str. CCUG 41628T |
| Fusobacteria | Fusobacteria | Fusobacterales | Fusobacteriaceae | sf_1 | 488 | Fusobacterium nucleatum subsp. vincentii str. ATCC 49256 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 442 | forest soil clone S0134 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 227 | uranium mining waste pile clone JG37-AG-36 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 9464 | lodgepole pine rhizosphere soil British Columbia Ministry Forests Long-Term Soil Productivity |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 10112 | forest soil clone NOS7.157WL |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 317 | penguin droppings sediments clone KD8-87 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 1127 | uranium mining waste pile near Johanngeorgenstadt soil clone JG37-AG-21 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 1565 | uranium mining waste pile clone JG34-KF-418 |
| Gemmatimonadetes | Unclassified | Unclassified | Unclassified | sf_5 | 2047 | soil clone #0319-7G21 |
| LD1PA group | Unclassified | Unclassified | Unclassified | sf_1 | 10118 | anoxic marine sediment clone LD1-PA38 |
| Lentisphaerae | Unclassified | Unclassified | Unclassified | sf_5 | 10027 | Cytophaga sp. str. Dex80-43 |
| Lentisphaerae | Unclassified | Unclassified | Unclassified | sf_5 | 10330 | Mono lake clone ML635J-58 |
| Lentisphaerae | Unclassified | Unclassified | Unclassified | sf_5 | 9704 | Cytophaga sp. str. Dex80-64 |
| marine group A | mgA-2 | Unclassified | Unclassified | sf_1 | 6344 | bacterioplankton clone ZA3648c |
| marine group A | mgA-1 | Unclassified | Unclassified | sf_1 | 6408 | Sargasso Sea |
| marine group A | mgA-1 | Unclassified | Unclassified | sf_1 | 6454 | marine clone SAR406 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | sf_1 | 769 | fjord ikaite column clone un-c23 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | sf_1 | 2437 | Mono Lake at depth 23m station 6 July 2000 clone ML623J-19 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | sf_1 | 3570 | Bacillus sp. clone ML1228J-1 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | sf_1 | 3745 | Mono Lake at depth 35m station 6 July 2000 clone ML635J-45 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | sf_1 | 4377 | Mono Lake at depth 35m station 6 July 2000 clone ML635J-65 G+C |
| NC10 | NC10-1 | Unclassified | Unclassified | sf_1 | 452 | vadose clone 5G01 |
| NC10 | NC10-1 | Unclassified | Unclassified | sf_1 | 536 | uranium mill tailings clone GuBH2-AD-8 |
| NC10 | NC10-2 | Unclassified | Unclassified | sf_1 | 10254 | uranium mill tailings soil sample clone Sh765B-TzT-35 |
| Nitrospira | Nitrospira | Nitrospirales | Nitrospiraceae | sf_1 | 984 | uranium mining waste pile clone JG37-AG-131 sp. |
| Nitrospira | Nitrospira | Nitrospirales | Nitrospiraceae | sf_2 | 542 | forested wetland clone FW19 |
| Nitrospira | Nitrospira | Nitrospirales | Nitrospiraceae | sf_2 | 544 | forested wetland clone FW5 |
| Nitrospira | Nitrospira | Nitrospirales | Nitrospiraceae | sf_2 | 697 | forested wetland clone FW118 |
| OP10 | CH21 cluster | Unclassified | Unclassified | sf_1 | 326 | geothermal clone ST01-SN3H |
| OP10 | Unclassified | Unclassified | Unclassified | sf_4 | 484 | forested wetland clone FW68 |
| OP10 | CH21 cluster | Unclassified | Unclassified | sf_1 | 514 | sludge clone SBRA136 |
| OP10 | Unclassified | Unclassified | Unclassified | sf_5 | 9782 | Rocky Mountain alpine soil clone S1a-1H |
| OP3 | Unclassified | Unclassified | Unclassified | sf_4 | 628 | CB-contaminated groundwater clone GOUTB15 |
| OP3 | Unclassified | Unclassified | Unclassified | sf_2 | 349 | soil clone PBS-25 |
| OP9/JS1 | OP9 | Unclassified | Unclassified | sf_1 | 726 | hot spring clone OPB72 |
| OP9/JS1 | OP9 | Unclassified | Unclassified | sf_1 | 969 | DCP-dechlorinating consortium clone SHA-1 |
| phylum_tax | class_tax | order_tax | family_tax | subfamily | otu_id | rep_prokMSAname |
| Planctomycetes | Planctomycetacia | Planctomycetales | Anammoxales | sf_2 | 4683 | anoxic basin clone CY0ARA028B09 |
| Planctomycetes | Planctomycetacia | Planctomycetales | Anammoxales | sf_4 | 4694 | USA: Colorado Fort collins Horsetooth Reservoir clone HT2F11 |
| Planctomycetes | Planctomycetacia | Planctomycetales | Anammoxales | sf_4 | 9662 | Great Artesian Basin clone B83 |
| Planctomycetes | Planctomycetacia | Planctomycetales | Pirellulae | sf_3 | 4670 | |
| Planctomycetes | Planctomycetacia | Planctomycetales | Pirellulae | sf_3 | 4677 | aerobic basin clone CY0ARA032A03 |
| Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | sf_3 | 4652 | anoxic basin clone CY0ARA028C04 |
| Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | sf_3 | 4948 | anoxic basin clone CY0ARA027D01 |
| Proteobacteria | Alphaproteobacteria | Acetobacterales | Acetobacteraceae | sf_1 | 7529 | Gluconacetobacter europaeus str. ZIM B028 V3 |
| Proteobacteria | Gammaproteobacteria | Acidithiobacillales | Acidithiobacillaceae | sf_1 | 8320 | acid mine drainage clone BA11 |
| Proteobacteria | Gammaproteobacteria | Acidithiobacillales | Acidithiobacillaceae | sf_1 | 8552 | Acidithiobacillus ferrooxidans str. D2 |
| Proteobacteria | Gammaproteobacteria | Acidithiobacillales | Acidithiobacillaceae | sf_1 | 9224 | Acidithiobacillus albertensis str. DSM 14366 |
| Proteobacteria | Gammaproteobacteria | Acidithiobacillales | Acidithiobacillaceae | sf_1 | 9497 | Acidithiobacillus ferrooxidans str. ATCC 19859 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 9294 | Arctic deep sea Isolation common chemoorganotrophic oxygen-respiring polar current d 1210 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 8340 | Aeromonas ichthiosmia |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 8364 | Aeromonas allosaccharophila str. CECT 4199 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 8621 | Aeromonas sp. PAR2A |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 9000 | Aeromonas culicicola str. MTCC 3249 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 9026 | Haemophilus piscium str. NCIMB 1952 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 9440 | Aeromonas sobria str. NCIMB 12065 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | sf_1 | 9494 | Aeromonas molluscorum str. 849T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7737 | atrazine-catabolizing microbial presence methanol clone KRA30+06A |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7768 | swine intestine clone p-861-a5 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7788 | atrazine-catabolizing microbial absence methanol clone KRA30-58 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7838 | Alcaligenes defragrans str. PD-19 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7902 | Alcaligenes faecalis str. M3A |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7932 | Achromobacter subsp. denitrificans str. DSM 30026 (T) |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7984 | Waste-gas biofilter clone BIfciii38 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 7992 | Alcaligenes faecalis 5659-H |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 8062 | Brackiella oedipodis str. LMG 1945 R8846 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | sf_1 | 8094 | Alcaligenes sp. str. VKM B-2263 dcm6 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Alcanivoraceae | sf_1 | 8335 | Alcanivorax sp. str. K3-3 (MBIC 4323) |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Alcanivoraceae | sf_1 | 9658 | Alcanivorax sp. str. Haw1 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9035 | Microbulbifer sp. str. JAMB-A94 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8348 | Arctic sea ice ARK10038 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8484 | Alteromonadaceae isolate str. LA50 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8503 | Arctic sea ice ARK10244 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8578 | Marinobacter lipolyticus str. SM-19 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8594 | Marinobacter sp. str. SBS |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9239 | Arctic sea ice ARK10228 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8196 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8222 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8600 | Colwellia piezophila str. Y223G |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8753 | Idiomarina loihiensis str. GSP37 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8174 | attached marine recovered surface clone 17 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8318 | Aestuariibacter salexigens str. JC2042 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8374 | Agarivorans albus str. MKT 89 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8533 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8695 | Arctic pack ice; northern Fram Strait; 80 31.1 N; 01 deg 59.7 min E clone ARKIA-34 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8863 | Alteromonas marina str. SW-47 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8970 | Arctic seawater isolate str. R9879 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8978 | Arctic sea ice ARK10108 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9230 | Antarctic pack ice Lasarev Sea Southern Ocean clone ANTXI/4_14-62 sea |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9236 | attached marine recovered surface clone 18 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9288 | Alteromonas stellipolaris str. LMG 21861 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9292 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9501 | sea water isolate str. BP-PH |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9562 | Alteromonadaceae clone PH-B55N |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8172 | Pseudoalteromonas sp. str. Bdeep-1 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8336 | Alteromonas sp. str. MS23 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8580 | Arctic seawater isolate str. R7076 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8932 | Pseudoalteromonas antarctica str. N-1 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8975 | Alteromonas sp. str. NIBH P1M3 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9058 | Pseudoalteromonas carrageenovora str. ATCC 12662T |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9111 | Pseudoalteromonas sp. str. E36 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9143 | Pseudoalteromonas agarivorans str. KMM 255 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9205 | marine clone Arctic96B-17 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9218 | Pseudoalteromonas haloplanktis str. ATCC 14393 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9324 | Pseudoalteromonas ruthenica str. KMM300 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9386 | Alteromonas sp. str. NIBH P2M11 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9640 | exposed to diatom detritus isolate str. Tw-10 Tw-10 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8643 | Pseudoalteromonas porphyrae str. S2-65 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9369 | Pseudoalteromonas luteoviolacea str. NCIMB 1893T |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9222 | Shewanella hanedai str. CIP 103207T |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9384 | Moritella viscosa str. NVI 88/478T |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8916 | Shewanella algae str. 43940 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9067 | Shewanella algae str. ACM 4733 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9416 | marine isolate str. R8 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9586 | Shewanella gaetbuli str. TF-27 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8579 | Psychromonas profunda str. 2825 |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 6628 | Wolbachia pipientis |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 6648 | Wolbachia sp |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 6803 | Wolbachia sp. Dlem16SWol |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 6908 | Rhinocyllus conicus endosymbiont |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 7481 | Wolbachia pipientis |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Bartonellaceae | sf_1 | 7056 | Bartonella schoenbuchensis str. R1 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Bartonellaceae | sf_1 | 7384 | aortic heart valve patient with endocarditis clone v9 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Bartonellaceae | sf_1 | 7415 | Bartonella quintana str. Toulouse |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Bartonellaceae | sf_1 | 7634 | Bartonella henselae str. Houston-1 |
| Proteobacteria | Deltaproteobacteria | Bdellovibrionales | Bdellovibrionaceae | sf_1 | 10010 | uranium mining waste pile clone JG37-AG-139 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 7401 | Scrippsiella trochoidea NEPCC 15 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 6651 | Beijerinckia indica |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 7275 | Mammoth cave clone CCU18 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 7219 | Methylosinus sporium |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 7640 | Methylosinus trichosporium |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 6762 | acidic forest soil clone UP8 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Beijerinck/Rhodoplan/Methylocyst | sf_3 | 7153 | Methylocella tundrae str. Y1 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Bradyrhizobiaceae | sf_1 | 7029 | |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7403 | Oligotropha carboxidovorans str. S23 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6927 | Nitrobacter hamburgensis str. X14 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6768 | Rhodopseudomonas palustris str. GH |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6799 | Rhodopseudomonas palustris str. ATCC 17001 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7316 | |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7333 | Afipia genosp. 4 str. G3644 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6941 | Rhodopseudomonas rhenobacensis str. Klemme Rb |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7087 | Bradyrhizobium japonicum HA1 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7398 | Bradyrhizobium japonicum str. USDA 38 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6636 | Bradyrhizobium elkanii str. USDA 76 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6867 | heavy metal-contaminated soil clone a13131 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6887 | Bradyrhizobium str. YB2 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7044 | Afipia genosp. 2 str. G4438 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7126 | ground water deep-well injection disposal site radioactive wastes Tomsk-7 clone S15A-MN96 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7390 | Afipia genosp. 10 str. G8996 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7477 | Bradyrhizobium elkanii str. SEMIA 6028 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7522 | Bradyrhizobium sp. str. KKI14 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6878 | Bradyrhizobium japonicum SD5 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 6917 | Bradyrhizobium japonicum str. IAM 12608 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Bradyrhizobiaceae | sf_1 | 7353 | temperate estuarine mud clone HC65 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Brucellaceae | sf_1 | 6757 | Ochrobactrum anthropi str. ESC1 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Brucellaceae | sf_1 | 6981 | Ochrobactrum gallinifaecis str. Iso 196 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Brucellaceae | sf_1 | 6995 | |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7720 | penguin droppings sediments clone KD1-79 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7771 | Burkholderia glathei str. ATCC 29195T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7782 | Burkholderia hospita str. LMG 20598T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7969 | Burkholderia sp. |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 8059 | Burkholderia caribensis str. MWAP71 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 8068 | Burkholderia caryophylli str. ATCC 25418 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7747 | |
| Proteobacteria | Alphaproteobacteria | Consistiales | Caedibacteraceae | sf_4 | 7157 | acid mine drainage clone ASL45 |
| Proteobacteria | Alphaproteobacteria | Consistiales | Caedibacteraceae | sf_5 | 6947 | termite gut homogenate clone Rs-B60 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10446 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10461 | deepest cold-seep area Japan Trench clone JTB360 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10523 | Riftia pachyptila's tube clone R103-B70 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10538 | Arcobacter cryaerophilus |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10447 | Sulfurospirillum deleyianum str. Spirillum 5175 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10464 | Campylobacter sp. str. NO2B |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10434 | Campylobacter gracilis |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10456 | Campylobacter showae |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10463 | Campylobacter subsp. fetus |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10484 | Campylobacter helveticus |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10540 | Campylobacter showae str. LMG 12636 |
| Proteobacteria | Gammaproteobacteria | Cardiobacteriales | Cardiobacteriaceae | sf_1 | 8536 | Cardiobacterium hominis |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 7486 | Asticcacaulis excentricus str. ATCC15261 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 6781 | Brevundimonas intermedia str. MBIC2712 ATCC15262 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 6904 | Brevundimonas vesicularis str. IAM 12105T |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 6909 | Brevundimonas diminuta str. DSM 1635 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 6968 | Brevundimonas diminuta str. IAM 12691T |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 7359 | Brevundimonas bacteroides str. CB7 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 7366 | Brevundimonas subvibrioides str. CB81 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 7436 | Brevundimonas sp. str. FWC40 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 9048 | Allochromatium sp. AT2202 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 8546 | Thiocapsa litoralis |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 8527 | |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 8697 | Thiococcus sp. AT2204 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 9054 | |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 9052 | |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 9356 | |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Chromatiaceae | sf_1 | 9370 | isolate str. HTB019 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8112 | Comamonas testosteroni str. SMCC B329 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7704 | freshwater clone PRD01b009B |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7705 | penguin droppings sediments clone KD4-7 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7801 | Toolik Lake main station at 3 m depth clone TLM05/TLMdgge10 proteobacterium |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7829 | Xylophilus ampelinus str. ATCC 33914 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7928 | penguin droppings sediments clone KD5-43 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7941 | MCB-contaminated groundwater-treating reactor clone RB9C10 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7986 | Arctic sea ice ARK10281 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8138 | Pseudomonas lanceolata str. ATCC 14669T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8139 | Delftia tsuruhatensis str. AD9 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7856 | Variovorax paradoxus |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7964 | napthalene-contaminated sediment clone 76 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7888 | Hydrogenophaga flava str. DSM 619T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7919 | strain isolate str. rM4 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7987 | Acidovorax sp. str. OS-6 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8012 | Acidovorax konjaci str. DSM 7481 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8018 | Acidovorax delafieldii str. ATCC 17505 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8021 | Acidovorax facilis str. CCUG 2113 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8022 | Acidovorax avenae subsp. cattleyae str. NCPPB 961 subsp. |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8031 | strain isolate str. rJ10 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8046 | Acidovorax defluvii str. BSB411 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 8152 | nephridia Octolasion lacteum clone Ol2-2 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7807 | Aquaspirillum metamorphum str. DSM 1837 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7884 | Germany:Elbe River clone Elb37 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | sf_1 | 7965 | Anoxobacterium dechloraticum |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 7893 | agricultural soil clone SC-I-71 |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 8457 | 5' clone CHAB-XI-27 |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 9198 | uranium mining waste pile clone KF-JG30-B15 KF-JG30-B15 |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 8969 | uranium mining waste pile soil sample clone JG30-KF-C15 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 9444 | forested wetland clone FW23 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfoarculaceae | sf_2 | 10227 | marine sediment clone Bol11 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 9666 | marine sediment above hydrate ridge clone Hyd89-13 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 9875 | hydrothermal sediment clone AF420354 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 9800 | forested wetland clone FW57 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10268 | |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10046 | Desulfobacterium cetonicum str. DSM 7267 oil recovery water |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10239 | |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10319 | sulfate-reducing habitat clone SLM-CP-116 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10031 | Antarctic sediment clone SB1_49 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 10083 | Desulfobacter curvatus str. DSM 3379 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | sf_5 | 9940 | Antarctic sediment clone SB2_56 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobulbaceae | sf_1 | 10047 | epibiontic clone C11-D3 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobulbaceae | sf_1 | 10187 | Mono Lake at depth 23 m station 6 July 2000 clone ML623J-57 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobulbaceae | sf_1 | 9734 | Riftia pachyptila's tube clone R103-B13 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobulbaceae | sf_1 | 9739 | gas hydrate clone Hyd89-51 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfohalobiaceae | sf_1 | 9894 | Desulfonauticus submarinus str. 6N |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfomicrobiaceae | sf_1 | 10079 | Desulfomicrobium baculatum str. DSM 1742 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 10262 | Desulfovibrio sp. str. Ac5.2 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 10248 | Desulfovibrio giganteus str. DSM 4370 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 10016 | termite gut homogenate clone Rs-N35 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 9826 | termite gut homogenate clone Rs-M72 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 10071 | Desulfovibrio desulfuricans |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 10212 | |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | sf_1 | 9709 | termite gut homogenate clone Rs-N31 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Desulfuromonaceae | sf_1 | 10020 | uranium mill tailings soil sample clone GuBH2-AG-114 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Ectothiorhodospiraceae | sf_1 | 9450 | Halorhodospira neutrophila str. SG 3304 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Ectothiorhodospiraceae | sf_1 | 9598 | Mono Lake at depth 2 m station 6 July 2000 clone ML602J-47 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_6 | 433 | coal effluent wetland clone RCP2-6 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_6 | 646 | Opitutus sp. str. SA-9 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9309 | Buchnera sp |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8742 | USA:New York isolate str. KN4 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_6 | 8783 | Alterococcus agarolyticus str. ADT3; CCRC17102 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9135 | intestine Zophobas mori clone |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9358 | Salmonella subsp. enterica serovar Waycross str. Swy1 subsp. |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9496 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8886 | Salmonella typhimurium LT2 str. SGSC1412 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8740 | Erwinia chrysanthemi str. 573 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9651 | Pectobacterium subsp. atrosepticum str. GSPB 1710 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8379 | Erwinia amylovora EA G-5 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9142 | Erwinia amylovora str. DSM 30165 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9252 | Pantoea cedenensis str. A34 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9345 | Erwinia amylovora str. BC199(= Ea528) |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8554 | Kluyvera ascorbata 69 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8885 | Morganella morganii str. AP28 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9363 | Citrobacter freundii str. CDC 621-64 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9594 | Morganella morganii str. ATCC35200 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8758 | Pectobacterium cypripedii str. ATCC 29267 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8282 | Antonina pretiosa symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8693 | Pantoea agglomerans str. A40 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8700 | Baumannia cicadellinicola |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9302 | Pantoea subsp. stewartii str. GSPB 2626 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8236 | Vryburgia amaryllidis symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8504 | Dysmicoccus neobrevipes symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8603 | Melanococcus albizziae symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8607 | Amonostherium lichtensioides symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8624 | Erium globosum symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9290 | Baumannia cicadellinicola |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9293 | USA clone 14/7 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9420 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8934 | Pectobacterium subsp. carotovorum str. E155 subsp. |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9266 | Parasite BEV of E.variegatus |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8467 | Serratia marcescens subsp. sakuensis str. KRED subsp. |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9348 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8505 | Buttiauxella warmboldiae str. DSM 9404 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8528 | Enterobacter cloacae Nr. 3 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8530 | Enterobacteriaceae CF01Ent-1 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8640 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8936 | Klebsiella oxytoca str. ChDC OS31 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9060 | Enterobacter ludwigii str. EN-119 = DSMZ 16688 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9274 | Enterobacter sp. CC1 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9361 | Enterobacter intermedius str. JCM1238 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9390 | Enterobacter nimipressuralis str. LMG 10245-T |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8251 | Nitrogen-fixing isolate str. CANF3 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8529 | Raoultella planticola 7 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8627 | Australicoccus grevilleae symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8770 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8890 | Raoultella planticola str. DR3 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8362 | Klebsiella pneumoniae str. ASR1 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8510 | Klebsiella pneumoniae str. DSM 30104 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8773 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8286 | Cyphonococcus alpinus symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8711 | Serratia odorifera str. DSM 4582 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8712 | Serratia proteamaculans str. DSM 4543 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8739 | Serratia entomophila str. DSM 12358 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8892 | Aranicola proteolyticus |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9151 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9417 | Serratia fonticola str. DSM 4576 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8631 | Planococcus ficus symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8283 | Heteropsylla texana symbiont |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8173 | Photorhabdus asymbiotica str. ATCC 43949 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8225 | |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8642 | Erwinia chrysanthemi str. 580 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9029 | Photorhabdus asymbiotica subsp. australis str. MB |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8473 | Hafnia alvei |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9265 | Rahnella aquatilis k 8 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9337 | Rahnella genosp. 3 str. DSM 30078 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8564 | Rahnella aquatilis str. ATCC 33989 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9157 | Secondary symbiont type-U Acyrthosiphon pisum (rrs) clone 5B type-U |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 9262 | Yersinia aldovae str. A125 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 1206 | Dermacentor variabilis symbiont |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Francisellaceae | sf_1 | 9554 | Tilapia parasite TPT-541 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Francisellaceae | sf_1 | 8949 | Caedibacter taeniospiralis |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Geobacteraceae | sf_1 | 482 | trichloroethene-contaminated site clone FTLM205 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Geobacteraceae | sf_1 | 10171 | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 8514 | Chromohalobacter israelensis str. ATCC 43985 T |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 8562 | Halomonas sp. str. TNB I20 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 8576 | Halomonas sp. Ko502 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 8598 | Halomonas desiderata str. FB2 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 8854 | Halomonas variabilis str. ANT9112 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 9471 | Boston Harbor surface water isolate str. UMB18C UMB18C |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | sf_1 | 9141 | Halomonas sp. SK1 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10385 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10428 | Flexispira rappini FH 9702248 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10430 | Helicobacter heilmannii str. MM2 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10436 | Helicobacter aurati str. MIT 97-5075c |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10442 | Helicobacter cetorum str. MIT 99-5656 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10444 | Helicobacter suncus str. Kaz-2 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10448 | Helicobacter felis str. Dog-1 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10451 | Helicobacter heilmannii str. C4S |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10454 | Helicobacter pullorum str. NCTC 12826 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10462 | Helicobacter rodentium str. MIT 96-1312 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10518 | Helicobacter pylori str. ATCC 49396T |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10520 | Helicobacter sp. blood isolate 964 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10548 | Helicobacter rappini W.Tee-Bat |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10552 | Helicobacter winghamensis str. NLEP 97-1611 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10562 | Helicobacter rappini W.Tee-Yu |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10425 | Sulfurimonas autotrophica str. OK5 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10411 | termite gut homogenate clone Rs-P71 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10432 | Riftia pachyptila's tube clone R76-B51 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10438 | hydrocarbon seep clone GCA014 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10590 | termite gut homogenate clone Rs-H40 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10614 | strain isolate str. BHI80-49 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10417 | temperate estuarine mud clone KM61 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10467 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10507 | termite gut homogenate clone Rs-M59 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Hyphomicrobiaceae | sf_1 | 7646 | Hyphomicrobium aestuarii str. DSM 1564 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | sf_1 | 7392 | |
| Proteobacteria | Gammaproteobacteria | Legionellales | Legionellaceae | sf_1 | 8865 | Arctic pack ice; northern Fram Strait; 80 31.1 N; 01 deg 59.7 min E clone ARKCH2Br2-23 |
| Proteobacteria | Alphaproteobacteria | Azospirillales | Magnetospirillaceae | sf_1 | 6922 | Dechlorospirillum sp. str. SN1 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Methylobacteriaceae | sf_1 | 7585 | Methylobacterium thiocyanatum str. ALL/SCN-P |
| Proteobacteria | Gammaproteobacteria | Methylococcales | Methylococcaceae | sf_1 | 8243 | isolate str. IR |
| Proteobacteria | Gammaproteobacteria | Methylococcales | Methylococcaceae | sf_1 | 8821 | Methylobacter psychrophilus str. Z-0021 |
| Proteobacteria | Gammaproteobacteria | Methylococcales | Methylococcaceae | sf_1 | 9438 | marine sediment above hydrate ridge clone Hyd24-01 proteobacterium |
| Proteobacteria | Betaproteobacteria | Methylophilales | Methylophilaceae | sf_1 | 8137 | freshwater clone PRD01a011B |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8366 | Psychrobacter frigidicola str. DSM 12411 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8604 | Moraxella oblonga str. IAM 14971 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8838 | Psychrobacter psychrophilus CMS 28 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8727 | Alkanindiges hongkongensis str. HKU9 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 9359 | Acinetobacter junii str. S33 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 9428 | hydrocarbon-degrading consortium clone AF2-1D |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 9466 | Acinetobacter tandoii str. 4N13 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 9641 | Acinetobacter haemolyticus |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Myxococcaceae | sf_1 | 10358 | Myxococcus fulvus str. Mx f2 |
| Proteobacteria | Epsilonproteobacteria | Nautiliales | Nautiliaceae | sf_1 | 10477 | S17sBac5 complete clone |
| Proteobacteria | Betaproteobacteria | Neisseriales | Neisseriaceae | sf_1 | 7945 | Aquaspirillum serpens str. IAM 13944 |
| Proteobacteria | Betaproteobacteria | Neisseriales | Neisseriaceae | sf_1 | 7675 | Neisseria sp. str. CCUG 46910 |
| Proteobacteria | Betaproteobacteria | Neisseriales | Neisseriaceae | sf_1 | 7662 | Mars Odyssey Orbiter and encapsulation facility clone T5-1 sp. |
| Proteobacteria | Betaproteobacteria | Nitrosomonadales | Nitrosomonadaceae | sf_1 | 7789 | |
| Proteobacteria | Betaproteobacteria | Nitrosomonadales | Nitrosomonadaceae | sf_1 | 7976 | Nitrosomonas sp. str. Nm86 |
| Proteobacteria | Betaproteobacteria | Nitrosomonadales | Nitrosomonadaceae | sf_1 | 7770 | Nitrosomonas europaea str. ATCC 19718 |
| Proteobacteria | Betaproteobacteria | Nitrosomonadales | Nitrosomonadaceae | sf_1 | 8145 | Nitrosomonas eutropha str. Nm57 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Nitrospinaceae | sf_2 | 594 | uranium mining mill tailing clone GR-296.II.52 GR-296.I.52 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Oceanospirillaceae | sf_1 | 9351 | bacterioplankton clone ZA2333c |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7743 | Herbaspirillum sp. str. NAH4 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7843 | Massilia timonae timone |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7845 | Diaphorina citri symbiont |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7866 | Paucimonas lemoignei str. ATCC 17989T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7878 | napthalene-contaminated sediment clone 29 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7921 | Collimonas fungivorans str. Ter331 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 7968 | Oxalobacter formigenes str. OXB ovinen rumen |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 8013 | isolate str. A1020 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 8032 | Aquaspirillum arcticum str. IAM 14963 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 8034 | Janthinobacterium agaricidamnosum str. W1r3T |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | sf_1 | 8058 | Herbaspirillum seropedicae str. DSM 6445 ATCC 35892 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9360 | Pasteurella multocida subsp. gallicida str. MCCM 00021 subsp. |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9349 | Pasteurella sp. str. 91985 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8195 | Haemophilus influenzae str. R2866 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8555 | Haemophilus influenzae str. M9741 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9213 | Haemophilus quentini str. MCCM 02026 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9477 | Haemophilus influenzae str. M11105 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8228 | Actinobacillus indolicus str. H1419 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8861 | Haemophilus parasuis 427 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8614 | Acidithiobacillus thiooxidans str. KCTC 8928P |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8952 | Actinobacillus lignieresii |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9263 | Actinobacillus capsulatus |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8876 | Mannheimia sp. R19.2 str. R19.2; CCUG 38463 R19.2 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9237 | |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8409 | human colonic mucosal biopsy clone ABLCf1 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8432 | |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 8848 | str. 86355 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9533 | Haemophilus segnis str. MCCM 00337 |
| Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | sf_1 | 9628 | Histophilus somni str. CCUG 12839 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 6857 | Mesorhizobium mediterraneum str. PECA20 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 6692 | Phyllobacterium trifolii str. PETP02 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 6916 | lake microbial mat isolate str. R-9219 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 6966 | Mesorhizobium tianshanense str. -1BS; USDA 3592 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7009 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7216 | Ahrensia kielensis str. IAM12618 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7379 | Phyllobacterium myrsinacearum HM35 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7381 | Aminobacter aminovorans str. DSM7048T |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7497 | Pseudaminobacter salicylatoxidans str. KTC001 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | sf_1 | 7300 | marine isolate JP57 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 8664 | Thiomicrospira sp. str. Milos-T2 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 9027 | Thiomicrospira crunogena str. XCL-2 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 9557 | Riftia pachyptila's tube clone R76-B23 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 9291 | Methylophaga alcalica str. M39 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 9392 | Methylophaga sp. str. V4.ME.29 = MM_2343 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 10249 | soil sample uranium mining waste pile near town Johanngeorgenstadt clone JG36-TzT-168 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 10298 | marine tidal mat clone BTM36 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 10353 | sludge clone A9 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 9671 | hydrothermal sediment clone AF420357 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 9735 | uranium mining waste pile clone JG37-AG-15 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 9755 | bacterioplankton clone ZA3704c |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 9874 | uranium mining waste pile clone JG34-KF-243 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 9900 | bioreactor clone mle1-27 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_3 | 10082 | uranium mining waste pile clone JG37-AG-33 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | sf_4 | 9733 | bacterioplankton clone ZA3735c |
| Proteobacteria | Betaproteobacteria | Procabacteriales | Procabacteriaceae | sf_1 | 8136 | Acanthamoeba sp. UWC6 symbiont |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Pseudoalteromonadaceae | sf_1 | 9627 | Pseudoalteromonas sp |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Pseudoalteromonadaceae | sf_1 | 9339 | Pseudoalteromonas sp. str. 05 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8813 | Lyrodus pedicellatus symbiont |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9300 | Lyrodus pedicellatus symbiont |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8487 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8508 | Pseudomonas citronellolis str. TERIDB26 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8691 | Pseudomonas aeruginosa str. PAO1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8754 | Pseudomonas sp. str. P400Y-1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9002 | Paederus fuscipes endosymbiont |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9056 | Pseudomonas aeruginosa str. #47 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9588 | Pseudomonas citronellolis str. TERIDB18 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8288 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8777 | Pseudomonas sp. str. KNA6-5 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8852 | Pseudomonas stutzeri str. KC |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9068 | Pseudomonas stutzeri str. A1501 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9228 | Pseudomonas stutzeri HY-105 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9295 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8344 | Anabaena circinalis AWQC118C isolate str. UNSW3 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8553 | Pseudomonas fulva str. IAM 1587 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8725 | Pseudomonas sp. str. 2N1-1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8850 | Agrobacterium agile str. IAM12615 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9238 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9005 | Pseudomonas sp. str. KY |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9613 | Pseudomonas flavescens str. B62 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8474 | ground water deep-well injection disposal site radioactive wastes Tomsk-7 clone S15A-MN7 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8513 | Pseudomonas monteilii str. CIP 104883 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9049 | uranium mining mill tailing clone GR-Sh2-34 GR-Sh2-34 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9219 | Pseudomonas cf. monteilii 9 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9343 | Cellvibrio subsp. mixtus str. ACM 2601 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9469 | cf. Pseudomonas sp. clone Llangefni 52 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9493 | Pseudomonas sp. str. dcm7B |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8209 | uranium mining waste pile clone JG37-AG-122 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8433 | Pseudomonas syringae pv. broussonetiae str. KOZ 8101 pv. |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8635 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8853 | Pseudomonas cichorii str. ATCC 10857T |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9028 | Pseudomonas koreensis str. Ps 9-14 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9240 | Pseudomonas fluorescens str. CHA0 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9267 | Pseudomonas syringae pv. theae str. PT1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9310 | Pseudomonas sp. str. AC-167 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8338 | Pseudomonas synxantha str. DSM 13080 G |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8561 | Pseudomonas sp. B65 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8601 | Pseudomonas marginalis str. ATCC 10844T |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8687 | Pseudomonas putida str. ATCC 17472 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8708 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9175 | Pseudomonas extremorientalis str. KMM3447 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9221 | Pseudomonas fulgida str. DSM 14938 = LMG 2146 P 515/12 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9243 | Pseudomonas tolaasii str. LMG 2342T () |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9366 | Arctic seawater isolate str. R7366 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8755 | Pseudomonas sp. SK-1-3-1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9172 | Pseudomonas psychrophila str. E-3 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Ralstoniaceae | sf_1 | 7823 | Wautersia basilensis str. DSM 11853 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Ralstoniaceae | sf_1 | 8110 | Wautersia paucula str. LMG 3413 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Ralstoniaceae | sf_1 | 8128 | Cupriavidus necator |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Ralstoniaceae | sf_1 | 7761 | Ralstonia detusculanense str. APF11 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Ralstoniaceae | sf_1 | 7778 | Ralstonia insidiosa str. CCUG 46388 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 7051 | Mycoplana dimorpha str. IAM 13154 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6683 | Sinorhizobium fredii str. ATCC35423 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6725 | Sinorhizobium meliloti str. 1021 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6972 | Ensifer adhaerens str. LMG 20582 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6974 | India: Himalayas Kaza Spiti Valley Cold Desert isolate str. Kaza-35 Kaza-35 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6770 | Rhizobium tropici str. LMG 9517 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6871 | Rhizobium mongolense str. USDA 1832 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 7135 | Rhizobium gallicum str. FL27 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 7568 | Rhizobium etli str. USDA 2667 ATCC 14483 SEMIA 043 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6798 | Agrobacterium tumefaciens TG14 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6804 | Rhizobium sp. str. SH19312 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 6964 | Agrobacterium tumefaciens str. C58 Cereon |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 7334 | Agrobacterium tumefaciens C4 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | sf_1 | 7041 | Rhizobium huautlense str. SO2 () |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 6701 | Roseobacter clone NAC11-3 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 6980 | Loktanella vestfoldensis str. LMG 22003 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7433 | Scrippsiella trochoidea NEPCC 15 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7453 | Sulfitobacter sp. BIO-11 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 6888 | hydrothermal vent strain str. TB66 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7026 | Leisingera methylohalidivorans str. MB2 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7263 | |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7040 | Paracoccus alcaliphilus str. JCM 7364 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7508 | lichen-dominated Antarctic cryptoendolithic community clone FBP492 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 6991 | Rhodobacter sphaeroides str. 2.4.1 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | sf_1 | 7084 | Scrippsiella trochoidea NEPCC 15 |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7800 | sample taken upstream landfill clone BVC77 landfill |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7817 | TCE-contaminated site clone ccs265 |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7956 | |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 8127 | Zoogloea resiniphila str. PIV-3A2y |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 8131 | |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7907 | Thauera aromatica str. LG356 |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7925 | Thauera selenatis str. ATCC 55363T |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 8156 | industrial-phenol-degrading community clone MM1 sp. |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7824 | termite gut homogenate clone Rs-B77 proteobacterium |
| Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | sf_1 | 7762 | Elbe River snow isolate Iso18 Iso18_1411 |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Rickettsiaceae | sf_1 | 7556 | Rickettsia bellii str. strains 369-C and G2D42 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Saccharospirillaceae | sf_1 | 8889 | hypersaline Mono Lake clone ML110J-5 |
| Proteobacteria | Alphaproteobacteria | Consistiales | SAR11 | sf_2 | 7043 | marine clone Arctic95D-8 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Shewanellaceae | sf_1 | 8581 | Shewanella benthica str. DB21MT-2 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Shewanellaceae | sf_1 | 8641 | Moritella abyssi str. 2693 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Shewanellaceae | sf_1 | 9081 | Shewanella sp. str. MTW-1 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Shewanellaceae | sf_1 | 8662 | |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7440 | Sphingobium chungbukense str. DJ77 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7528 | Sphingobium yanoikuyae str. GIFU9882 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7548 | Afipia genosp. 13 str. G8991 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 6650 | Sphingomonas phyllosphaerae str. FA1 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7016 | Sphingomonas sp. str. SAFR-027 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7535 | Sphingomonas paucimobilis str. GIFU2395 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_15 | 7035 | Sphingomonas asaccharolytica str. IFO 10564-T |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7215 | travertine hot spring clone SM2B06 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 6663 | Sphingopyxis flavimaris str. SW-151 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7100 | Novosphingobium capsulatum str. GIFU11526 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 7036 | Lutibacterium anuloederans str. LC8 |
| Proteobacteria | Gammaproteobacteria | Aeromonadales | Succinivibrionaceae | sf_1 | 8822 | Anaerobiospirillum sp. str. 3J102 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophaceae | sf_3 | 10067 | benzoate-degrading consortium clone BA044 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 9864 | uranium mining waste pile clone JG37-AG-133 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 10013 | hydrothermal sediment clone AF420341 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 10021 | uranium mill tailings soil sample clone Sh765B-TzT-29 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 9731 | uranium mining waste pile clone JG37-AG-90 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 9845 | uranium mining waste pile clone JG37-AG-128 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 10184 | granular sludge clone R1p32 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 10221 | granular sludge clone R3p4 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 10294 | Desulfacinum hydrothermale str. MT-96 |
| Proteobacteria | Deltaproteobacteria | Syntrophobacterales | Syntrophobacteraceae | sf_1 | 9661 | DCP-dechlorinating consortium clone SHD-1 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 8321 | Wadden Sea sediment clone Dangast A9 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 8741 | marine sediment clone Limfjorden L10 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 8752 | Beggiatoa sp. str. MS-81-1c |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 9015 | Beggiatoa alba str. B18LD; ATCC 33555 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 9321 | marine sediment clone Tokyo Bay D |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 8703 | Beggiatoa sp. str. AA5A |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Unclassified | sf_3 | 468 | marine sediment clone Sva0515 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7377 | Rocky Mountain alpine soil clone W2b-8C |
| Proteobacteria | Alphaproteobacteria | Verorhodospirilla | Unclassified | sf_1 | 7109 | diesel-polluted Bohai Gulf isolate str. M-5 M-5 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7340 | uranium mining waste pile soil sample clone JG30-KF-AS50 |
| Proteobacteria | Alphaproteobacteria | Azospirillales | Unclassified | sf_1 | 7400 | sphagnum peat bog clone K-5b5 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6694 | forested wetland clone RCP2-92 |
| Proteobacteria | Alphaproteobacteria | Azospirillales | Unclassified | sf_1 | 6732 | Anabaena circinalis AWQC118C isolate str. UNSW7 |
| Proteobacteria | Alphaproteobacteria | Acetobacterales | Unclassified | sf_1 | 7028 | |
| Proteobacteria | Alphaproteobacteria | Ellin314/wr0007 | Unclassified | sf_1 | 7123 | uranium mining waste pile near Johanngeorgenstadt soil clone JG37-AG-102 |
| Proteobacteria | Alphaproteobacteria | Ellin314/wr0007 | Unclassified | sf_1 | 7222 | Great Artesian Basin clone B79 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7575 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Unclassified | sf_1 | 6726 | |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6920 | Pseudovibrio denitrificans str. DN34 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6954 | |
| Proteobacteria | Alphaproteobacteria | Ellin329/Riz1046 | Unclassified | sf_1 | 6945 | Rhizobiales str. A48 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Unclassified | sf_1 | 7067 | Blastochloris sulfoviridis str. GN1 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Unclassified | sf_1 | 7264 | Bosea thiooxidans TJ1 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Unclassified | sf_1 | 7339 | |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6898 | heavy metal-contaminated soil clone a13113 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Unclassified | sf_1 | 7199 | uranium mill tailings clone Gitt-KF-194 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Unclassified | sf_1 | 6899 | |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6665 | hydrocarbon-degrading consortium clone 4-Org2-22 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7312 | |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Unclassified | sf_1 | 6789 | Shinella zoogloeoides str. ATCC 19623 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_2 | 6697 | termite gut homogenate clone Rs-D84 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_2 | 7188 | termite gut homogenate clone Rs-B50 proteobacterium |
| Proteobacteria | Alphaproteobacteria | Consistiales | Unclassified | sf_4 | 7105 | Mariana trough hydrothermal vent water 0.2micro-m filterable fraction clone MT-NB25 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Unclassified | sf_5 | 7471 | sponge clone TK03 |
| Proteobacteria | Alphaproteobacteria | Consistiales | Unclassified | sf_5 | 6735 | Candidatus Pelagibacter ubique str. HTCC1002 |
| Proteobacteria | Unclassified | Unclassified | Unclassified | sf_20 | 6763 | |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Unclassified | sf_2 | 6639 | |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Unclassified | sf_1 | 7156 | termite gut homogenate clone Rs-M62 proteobacterium |
| Proteobacteria | Deltaproteobacteria | AMD clone group | Unclassified | sf_1 | 6830 | coal effluent wetland clone RCP124 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Unclassified | sf_1 | 6653 | Kaistobacter koreensis str. PB229 |
| Proteobacteria | Deltaproteobacteria | Bdellovibrionales | Unclassified | sf_1 | 7382 | marine clone Arctic95C-5 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 6987 | |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7572 | |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Unclassified | sf_1 | 8035 | |
| Proteobacteria | Betaproteobacteria | MND1 clone group | Unclassified | sf_1 | 7808 | Mammoth cave clone CCU25 |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 8007 | |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 8036 | Uranium mill tailings soil sample clone Sh765B-TzT-132 proteobacterium |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 7974 | |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 8114 | |
| Proteobacteria | Betaproteobacteria | MND1 clone group | Unclassified | sf_1 | 8023 | ferromanganous micronodule clone MND1 |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 8045 | |
| Proteobacteria | Betaproteobacteria | MND1 clone group | Unclassified | sf_1 | 7818 | soil sample uranium mining waste pile near town Johanngeorgenstadt clone JG36-TzT-215 proteobacterium |
| Proteobacteria | Betaproteobacteria | Neisseriales | Unclassified | sf_1 | 8037 | Chitinimonas taiwanensis str. cf |
| Proteobacteria | Betaproteobacteria | Unclassified | Unclassified | sf_3 | 7997 | |
| Proteobacteria | Gammaproteobacteria | uranium waste clones | Unclassified | sf_1 | 8747 | uranium waste soil clone JG30-KF-CM35 |
| Proteobacteria | Gammaproteobacteria | GAO cluster | Unclassified | sf_1 | 9059 | activated sludge clone SBRH10 |
| Proteobacteria | Gammaproteobacteria | aquatic clone group | Unclassified | sf_1 | 9246 | Mammoth Cave sediment clone CCD24 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9498 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9568 | forested wetland clone RCP2-96 |
| Proteobacteria | Gammaproteobacteria | Chromatiales | Unclassified | sf_1 | 9282 | |
| Proteobacteria | Gammaproteobacteria | Legionellales | Unclassified | sf_1 | 9418 | uranium mining waste pile clone JG37-AG-14 proteobacterium |
| Proteobacteria | Deltaproteobacteria | EB1021 group | Unclassified | sf_4 | 8169 | forested wetland clone RCP2-54 |
| Proteobacteria | Gammaproteobacteria | Symbionts | Unclassified | sf_1 | 8403 | Selenate-reducing isolate str. KE4OH1 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8488 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8646 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8676 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8926 | inactive deep-sea hydrothermal vent chimneys clone IheB2-13 |
| Proteobacteria | Gammaproteobacteria | aquatic clone group | Unclassified | sf_1 | 8957 | marine clone Arctic97C-5 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9105 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9124 | 10e-6 dilution marine samples Weser estuary clone DC8-80-1 proteobacterium |
| Proteobacteria | Gammaproteobacteria | Symbionts | Unclassified | sf_1 | 9128 | Lucina nassula gill symbiont |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9394 | |
| Proteobacteria | Gammaproteobacteria | Symbionts | Unclassified | sf_1 | 9556 | Seepiophila jonesi symbiont |
| Proteobacteria | Gammaproteobacteria | SUP05 | Unclassified | sf_1 | 8605 | bacterioplankton clone ZA2525c |
| Proteobacteria | Gammaproteobacteria | SUP05 | Unclassified | sf_1 | 8654 | inactive deep-sea hydrothermal vent chimneys clone IheB2-31 |
| Proteobacteria | Gammaproteobacteria | SUP05 | Unclassified | sf_1 | 8965 | Bathymodiolus thermophilus gill symbiont |
| Proteobacteria | Gammaproteobacteria | uranium waste clones | Unclassified | sf_1 | 8231 | uranium waste soil clone JG30a-KF-21 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8339 | water 5 m downstream manure clone 35ds5 |
| Proteobacteria | Gammaproteobacteria | Ellin307/WD2124 | Unclassified | sf_1 | 8532 | |
| Proteobacteria | Gammaproteobacteria | Ellin307/WD2124 | Unclassified | sf_1 | 9458 | uranium mining waste pile clone JG37-AG-94 proteobacterium |
| Proteobacteria | Gammaproteobacteria | SAR86 | Unclassified | sf_1 | 8962 | bacterioplankton clone AEGEAN_234 |
| Proteobacteria | Gammaproteobacteria | Legionellales | Unclassified | sf_3 | 8587 | Mars Odyssey Orbiter and encapsulation facility clone T5-3 |
| Proteobacteria | Gammaproteobacteria | GAO cluster | Unclassified | sf_1 | 9468 | activated sludge clone SBRL2_40 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_4 | 8855 | |
| Proteobacteria | Unclassified | Unclassified | Unclassified | sf_8 | 9558 | |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Unclassified | sf_3 | 8230 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8245 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8883 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9044 | hydrothermal sediment clone AF420370 |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Unclassified | sf_1 | 8323 | hydrothermal sediment clone AF420363 |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 8780 | uranium mining mill tailing clone GR-296.II.89 GR-296.II.89 |
| Proteobacteria | Gammaproteobacteria | Oceanospirillales | Unclassified | sf_3 | 8327 | Arctic sea ice ARK10148 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8606 | |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8714 | Marinobacter hydrocarbonoclasticus str. ATCC 27132T |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8959 | bacterioplankton clone AEGEAN_133 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Unclassified | sf_1 | 8483 | Rheinheimera baltica str. OS140 Baltic # 166 |
| Proteobacteria | Gammaproteobacteria | Shewanella | Unclassified | sf_1 | 9344 | Shewanella algae str. ATCC 51192 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9367 | USA: Pacific Ocean seawater Naha Vents Hawaii isolate str. PV-4 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9473 | Arctic pack ice; northern Fram Strait; 80 31.1 N; 01 deg 59.7 min E clone ARKDMS-58 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Unclassified | sf_1 | 8430 | Salmonella bongori str. JEO 4162 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Unclassified | sf_1 | 9828 | termite gut homogenate clone Rs-M89 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Unclassified | sf_1 | 10092 | heavy metal-contaminated soil clone a13134 |
| Proteobacteria | Deltaproteobacteria | Myxococcales | Unclassified | sf_1 | 10259 | |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_7 | 10048 | |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 10049 | DCP-dechlorinating consortium clone SHA-72 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9760 | deep marine sediment clone MB-A2-137 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9784 | Antarctic sediment clone LH5_30 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9798 | uranium mill tailings soil sample clone GuBH2-AD/TzT-67 proteobacterium |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9876 | deep marine sediment clone MB-B2-106 |
| Proteobacteria | Deltaproteobacteria | EB1021 group | Unclassified | sf_4 | 9884 | forested wetland clone RCP2-62 |
| Proteobacteria | Deltaproteobacteria | AMD clone group | Unclassified | sf_1 | 10084 | acid mine drainage clone AS6 |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Unclassified | sf_1 | 10076 | Great Artesian Basin clone G13 |
| Proteobacteria | Deltaproteobacteria | dechlorinating clone group | Unclassified | sf_1 | 9959 | forested wetland clone FW110 |
| Proteobacteria | Deltaproteobacteria | EB1021 group | Unclassified | sf_4 | 10024 | hydrothermal sediment clone AF420338 |
| Proteobacteria | Deltaproteobacteria | AMD clone group | Unclassified | sf_1 | 9678 | coal effluent wetland clone RCP185 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Unclassified | sf_4 | 9951 | forested wetland clone FW13 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9738 | marine methane seep clone 1513 |
| Proteobacteria | Deltaproteobacteria | AMD clone group | Unclassified | sf_1 | 9945 | acid mine drainage clone BA18 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Unclassified | sf_3 | 9813 | hydrothermal sediment clone AF420340 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 9890 | termite gut homogenate clone Rs-K70 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10543 | hydrothermal vent clone PVB_10 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10427 | hydrothermal vent 9 degrees North East Rise Pacific Ocean clone CH3_17_BAC_16SrRNA_9N_EPR |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10475 | hydrothermal sediment clone AF420359 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10480 | Paralvinella palmiformis mucus secretions clone P. palm C 84 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10489 | S17sBac16 complete clone |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10497 | UASB reactor granular sludge clone PD-UASB-2 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10530 | hydrothermal vent 9 degrees North East Rise Pacific Ocean clone CH5_6_BAC_16SrRNA_9N_EPR |
| Proteobacteria | Unclassified | Unclassified | Unclassified | sf_20 | 2520 | |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | sf_9 | 244 | deep marine sediment clone MB-C2-152 |
| Proteobacteria | Deltaproteobacteria | AMD clone group | Unclassified | sf_1 | 3084 | coal effluent wetland clone RCP216 |
| Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | sf_1 | 8999 | Photobacterium leiognathi str. LN101 |
| Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | sf_1 | 8665 | Vibrio gallicus str. CIP 107867; HT 3-3 |
| Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | sf_1 | 8267 | Vibrio pomeroyi str. LMG 20537 |
| Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | sf_1 | 8798 | Vibrio aestuarianus str. KT0901 |
| Proteobacteria | Gammaproteobacteria | Vibrionales | Vibrionaceae | sf_1 | 8888 | Vibrio aestuarianus str. 01/151 |
| Proteobacteria | Alphaproteobacteria | Bradyrhizobiales | Xanthobacteraceae | sf_1 | 6660 | Azorhizobium caulinodans str. ORS 571 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9167 | pea aphid symbiont clone APe4_38 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8689 | Dyemonas todaii str. XD10 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9332 | wetland ecosystem constructed to remediate mine drainage isolate str. WJ2 WJ2 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8392 | penguin droppings sediments clone KD2-14 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8983 | Iron oxidizing strain ES-1 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9031 | municipal wastewater treatment bioreactor clone LB-P bacterium |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9320 | Waste-gas biofilter clone BIyi3 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8577 | Xanthomonas axonopodis pv. citri str. MA |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9569 | |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8538 | |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8563 | Pseudoxanthomonas mexicana str. AMX 26B |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9270 | Stenotrophomonas rhizophila str. e-p10 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 9286 | Stenotrophomonas maltophilia str. LMG 11104 |
| SPAM | Unclassified | Unclassified | Unclassified | sf_1 | 705 | uranium tailings soil clone Sh765B-AG-45 |
| SPAM | Unclassified | Unclassified | Unclassified | sf_1 | 738 | uranium mining waste clone JG34-KF-252 |
| Spirochaetes | Spirochaetes | Spirochaetales | Leptospiraceae | sf_3 | 6496 | Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6459 | Spirochaeta sp. str. BHI80-158 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_3 | 6558 | Spironema culicis str. BR91 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6526 | Treponema sp. str. 7CPL208 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6479 | Treponema sp |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6580 | Treponema sp. str. III:C:BA213 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6458 | termite gut clone NkS34 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6494 | termite gut homogenate clone Rs-C47 sp. |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6562 | forested wetland clone RCP1-96 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6507 | termite gut clone NkS-Ste2 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6476 | termite gut clone NkS50 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6488 | Treponema primitia str. ZAS-1 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6490 | termite gut homogenate clone BCf4-14 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6491 | termite gut homogenate clone BCf8-03 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6506 | termite gut homogenate clone Rs-J58 sp. |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6508 | termite hindgut clone mpsp2 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6523 | termite gut homogenate clone Rs-J64 sp. |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6565 | termite gut clone NkS-Oxy25 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | sf_1 | 6571 | Mixotricha paradoxa is flagellate hindgut Mastotermes darwiniensis clone mp4 of |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 117 | termite gut homogenate clone Rs-D89 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 353 | UASB reactor granular sludge clone PD-UASB-13 G+C |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 60 | Flexistipes sp. str. E3_33 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 601 | terephthalate-degrading consortium clone TA19 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 719 | Synergistes sp. P1 str. P4G_18 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 740 | swine intestine clone p-4292-4Wa3 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 808 | oral cavity clone BH017 |
| Termite group 1 | Unclassified | Unclassified | Unclassified | sf_2 | 437 | termite gut homogenate clone Rs-D43 group |
| Thermodesulfobacteria | Thermodesulfobacteria | Thermodesulfobacteriales | Thermodesulfobacteriaceae | sf_1 | 667 | Geothermobacterium ferrireducens |
| Thermotogae | Thermotogae | Thermotogales | Thermotogaceae | sf_4 | 51 | Thermosipho sp. str. MV1063 |
| TM6 | Unclassified | Unclassified | Unclassified | sf_1 | 9803 | forest soil clone S1204 |
| TM7 | Unclassified | Unclassified | Unclassified | sf_1 | 5177 | |
| TM7 | TM7-3 | Unclassified | Unclassified | sf_1 | 8155 | oral periodontitis clone EW086 |
| TM7 | TM7-3 | Unclassified | Unclassified | sf_1 | 2697 | midgut homogenate Pachnoda ephippiata larva clone PeM47 |
| TM7 | Unclassified | Unclassified | Unclassified | sf_1 | 3025 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_93 | 925 | 4MB-degrading consortium clone UASB_TL26 |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_106 | 243 | hot spring clone OPB25 |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 485 | thermal spring mat clone O1aA90 |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 226 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 333 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 651 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 6430 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 6456 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 6360 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_140 | 6355 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 7444 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 7767 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 10012 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_95 | 2545 | anaerobic sludge isolate str. JE |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 2488 | |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_156 | 4291 | Mono Lake at depth 35m station 6 July 2000 clone ML635J-21 G+C |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 4410 | |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Unclassified | sf_4 | 169 | anoxic marine sediment clone LD1-PA26 |
| Verrucomicrobia | Unclassified | Unclassified | Unclassified | sf_3 | 40 | Elbe river clone DEV055 |
| Verrucomicrobia | Unclassified | Unclassified | Unclassified | sf_3 | 486 | Elbe river clone DEV045 |
| Verrucomicrobia | Unclassified | Unclassified | Unclassified | sf_5 | 686 | hydrothermal vent sediment clone a2b018 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Unclassified | sf_3 | 11 | sludge clone H2 |
| Verrucomicrobia | Unclassified | Unclassified | Unclassified | sf_4 | 288 | Prosthecobacter dejongeii |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Unclassified | sf_3 | 792 | termite gut homogenate clone Rs-P07 bacterium |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 5 | sf_1 | 530 | anoxic marine sediment clone LD1-PB20 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 5 | sf_1 | 533 | anoxic marine sediment clone LD1-PB12 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 5 | sf_1 | 547 | anoxic marine sediment clone LD1-PB1 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 5 | sf_1 | 629 | anoxic marine sediment clone LD1-PA50 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 7 | sf_1 | 446 | anoxic marine sediment clone LD1-PA34 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 7 | sf_1 | 559 | anoxic marine sediment clone LD1-PA20 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobia SD 7 | sf_1 | 760 | Mono lake clone ML316M-1 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | sf_7 | 29 | Fucophilus fucoidanolyticus str. SI-1234 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | sf_6 | 871 | |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Xiphinematobacteraceae | sf_3 | 888 | Candidatus Xiphinematobacter brevicolli |
| WS3 | Unclassified | Unclassified | Unclassified | sf_3 | 95 | marine sediment above hydrate ridge clone Hyd24-32 |
| WS3 | Unclassified | Unclassified | Unclassified | sf_1 | 2537 | anoxic marine sediment clone LD1-PA39 |
| WS5 | Unclassified | Unclassified | Unclassified | sf_2 | 8119 | hydrothermal vent sediment clone a2b013 |
S-F, Subfamily identification; bTaxon ID, PhyloChip Taxon identification number; cRepresentative species, Taxon bacterial species identifier.
FIG. 1.
(A) Phylogenetic tree exhibiting family level bacterial diversity detected in COPD airways despite antimicrobial administration. (B) Bacterial richness detected in individual patient samples.
Interpersonal variation in bacterial richness (number of taxa detected) was noted across the patient samples (Fig. 1B). Four subjects (patients 1, 4, 5, and 6) exhibited communities with significantly fewer taxa (p < 0.002) compared with the other four subjects. Patients in which fewer bacterial taxa were detected tended to possess more members of the Pseudomonadaceae. In contrast, members of the Clostridiaceae, Lachnospiraceae, Bacillaceae, and Peptostreptococcaceae were detected more commonly in those patients with richer communities (patients 2, 3, 7). Patient 8 was observed to possess a large proportion of taxa belonging to the Enterobacteriaceae family, which have been associated with more advanced COPD lung disease (Sethi and Murphy, 2008). Interestingly, this patient had radiographic evidence of coexisting bronchiectasis, which was not present in the other patients.
Given the variation in bacterial richness among samples, which suggested differences in bacterial community composition, NMDS was used to assess variation in bacterial community structure (based on Bray-Curtis dissimilarity measures) across the sample cohort. This revealed two distinct groups of patient samples and confirmed that patient 8 represented a structurally distinct airway community (Fig. 2). Although the cohort was small, given this separation of subjects based on differing bacterial community structures, the influence of available clinical parameters on community composition was explored. Matrix-based, nonparametric multivariate analysis of variance (Anderson, 2001) revealed that across the cohort, the number of elapsed intubation days was significantly associated with bacterial community composition and structure, accounting for the greatest percentage of the observed variability (44%, p < 0.03; Fig. 2). Group 1 patient samples (patients 2, 3, and 7) were characterized by a shorter intubation duration prior to ETA sample collection (≤6 days), while those in Group 2 were intubated for significantly longer periods of time (p < 0.0007; patients 1, 4, 5, and 6; ≥16 days) and exhibited a significantly less rich community composition compared to that of Group 1 (p < 0.025). Given the community variation between Group 1 and Group 2, differences in the relative abundance of all detected taxa were assessed between the groups, which identified 153 taxa with significantly different relative abundances (Table 4; see at end of article). All of these significant taxa were present in higher abundance in Group 1, the majority of which (77%) belonged to the phylum Firmicutes. These included species such as Lactobacillus kitasatonis, L. perolens, L. sakei, and Bacillus clausii, as well as known pathogenic species such as Streptococcus constellatus, which is a member of the Streptoccocus milleri group (SMG; Table 4). No other clinical variable [including diagnosis of pneumonia (n = 6; p < 0.4) or the number of days between pneumonia diagnosis and sample collection (range: 3–52 days; p < 0.6)] demonstrated a significant association with bacterial community composition in this cohort.
FIG. 2.
NMDS analysis demonstrates that bacterial community composition is highly influenced by the duration of intubation (red isotherms). Subjects COPD 5 and COPD 6 are superimposed on the right side of the figure, indicative of highly similar bacterial community composition.
Table 4.
Bacterial Taxa with Significant Differences in Relative Abundance between COPD Patient Group 1 (≤6 Intubation Days) and Group 2 (≥16 Intubation Days)
| Phylum | Class | Order | Family | S-Fa | Taxon IDb | Representative speciesc | p-value | q-value | Fluorescence difference (Group 1 – Group 2) |
|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | Symbiobacteria | Symbiobacterales | Unclassified | 1 | 77 | thermal soil clone YNPFFP9 | <0.001 | <0.01 | 1264 |
| Proteobacteria | Deltaproteobacteria | Unclassified | Unclassified | 9 | 244 | deep marine sediment clone MB-C2-152 | <0.02 | <0.05 | 1048 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | 9 | 375 | forest soil clone C043 | <0.02 | <0.05 | 1873 |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Geobacteraceae | 1 | 482 | trichloroethene-contaminated site clone FTLM205 proteobacterium | ≤0.01 | <0.05 | 1092 |
| OP10 | CH21 cluster | Unclassified | Unclassified | 1 | 514 | sludge clone SBRA136 | <0.01 | <0.05 | 1081 |
| Chlorobi | Unclassified | Unclassified | Unclassified | 8 | 636 | benzene-degrading nitrate-reducing consortium clone Cart-N3 bacterium | <0.01 | <0.05 | 1475 |
| Unclassified | Unclassified | Unclassified | Unclassified | 160 | 651 | <0.01 | <0.05 | 1750 | |
| Chloroflexi | Unclassified | Unclassified | Unclassified | 7 | 757 | DCP-dechlorinating consortium clone SHA-8 | <0.001 | <0.01 | 1619 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | 1 | 769 | fjord ikaite column clone un-c23 | <0.001 | <0.01 | 1305 |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | 11 | 940 | Veillonella dispar str. DSM 20735 | <0.01 | <0.05 | 1150 |
| OP9/JS1 | OP9 | Unclassified | Unclassified | 1 | 969 | DCP-dechlorinating consortium clone SHA-1 | <0.02 | <0.05 | 1190 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 1050 | Bacillus firmus CV93b | ≤0.001 | <0.05 | 1746 |
| Actinobacteria | Actinobacteria | Unclassified | Unclassified | 1 | 1898 | termite gut homogenate clone Rs-J10 bacterium | <0.01 | <0.05 | 1906 |
| AD3 | Unclassified | Unclassified | Unclassified | 1 | 2338 | uranium mining waste pile soil clone JG30-KF-C12 | <0.001 | <0.01 | 1148 |
| Chloroflexi | Dehalococcoidetes | Unclassified | Unclassified | 1 | 2339 | uranium mill tailings soil sample clone Sh765B-TzT-20 bacterium | <0.02 | <0.05 | 1532 |
| Chloroflexi | Unclassified | Unclassified | Unclassified | 1 | 2534 | forest soil clone S085 | <0.001 | <0.01 | 1193 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 2668 | termite gut homogenate clone Rs-G40 bacterium | <0.01 | <0.05 | 2395 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 2694 | oral periodontitis clone FX028 | <0.02 | <0.05 | 1338 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 2714 | termite gut homogenate clone Rs-N27 bacterium | <0.01 | <0.05 | 2061 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 2729 | DCP-dechlorinating consortium clone SHA-58 | <0.01 | <0.05 | 1402 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 2797 | Isolation and identification hyper-ammonia producing swine storage pits manure | <0.01 | <0.05 | 1611 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 2805 | oral periodontitis clone FX033 | <0.02 | <0.05 | 1625 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 2834 | Butyrivibrio fibrisolvens str. OB156 | <0.01 | <0.05 | 1005 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 2994 | termite gut clone Rs-L15 | <0.001 | <0.01 | 3929 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 3021 | Clostridium caminithermale str. DVird3 | <0.01 | <0.05 | 1944 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 3038 | swine intestine clone p-1594-c5 | <0.01 | <0.05 | 1363 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 3059 | Butyrivibrio fibrisolvens str. NCDO 2249 | <0.01 | <0.05 | 1069 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 3060 | termite gut homogenate clone Rs-B14 bacterium | <0.001 | <0.01 | 3703 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 3076 | Clostridium nexile | <0.001 | <0.01 | 1395 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 3077 | Clostridium glycolicum str. DSM 1288 | <0.01 | <0.05 | 1953 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 3171 | Lachnospira pectinoschiza | <0.01 | <0.05 | 1398 |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 5 | 3182 | termite gut homogenate clone Rs-Q64 bacterium | ≤0.01 | <0.05 | 1107 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3250 | Streptococcus bovis str. B315 | <0.001 | <0.01 | 4284 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3251 | Streptococcus cristatus str. ATCC 51100 | <0.01 | <0.05 | 3986 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3253 | derived cheese sample clone 32CR | <0.02 | <0.05 | 2680 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3258 | Staphylococcus auricularis str. MAFF911484 ATCC33753T | <0.01 | <0.05 | 1525 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | 1 | 3261 | Enterococcus mundtii str. LMG 10748 | <0.02 | <0.05 | 2560 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3283 | Bacillus niacini str. IFO15566 | <0.01 | <0.05 | 1201 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3284 | <0.01 | <0.05 | 1347 | |
| Firmicutes | Bacilli | Bacillales | Caryophanaceae | 1 | 3285 | Caryophanon latum str. DSM 14151 | <0.01 | <0.05 | 1499 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3287 | tongue dorsum scrapings clone FP015 | <0.01 | <0.05 | 3582 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | 1 | 3288 | Isolation and identification hyper-ammonia producing swine storage pits manure | <0.01 | <0.05 | 2528 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3290 | Streptococcus mitis str. Sm91 | ≤0.01 | <0.05 | 3971 |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | 1 | 3299 | Brevibacillus borstelensis str. LMG 15536 | <0.02 | <0.05 | 1035 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3313 | Streptococcus salivarius str. ATCC 7073 | <0.001 | <0.01 | 3189 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | 1 | 3318 | Enterococcus ratti str. ATCC 700914 | <0.02 | <0.05 | 2272 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3323 | Trichococcus flocculiformis str. DSM 2094 | <0.01 | <0.05 | 1431 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3326 | Nostocoida limicola I str. Ben206 | <0.01 | <0.05 | 2363 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3328 | Pseudobacillus carolinae | <0.001 | <0.01 | 2370 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3330 | Lactobacillus kitasatonis str. KM9212 | <0.02 | <0.05 | 1389 |
| Firmicutes | Bacilli | Bacillales | Sporolactobacillaceae | 1 | 3365 | Bacillus sp. clone ML615J-19 | <0.001 | <0.01 | 1757 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3386 | feedlot manure clone B87 | <0.01 | <0.05 | 2321 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | 1 | 3392 | Vagococcus lutrae str. m1134/97/1; CCUG 39187 | <0.001 | ≤0.01 | 1976 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3397 | Streptococcus macedonicus str. ACA-DC 206 LAB617 | <0.01 | <0.05 | 4011 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3418 | Lactobacillus subsp. aviarius | <0.01 | <0.05 | 3036 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3419 | Bacillus algicola str. KMM 3737 | <0.01 | <0.05 | 1590 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3422 | Streptococcus thermophilus str. DSM 20617 | <0.01 | <0.05 | 3243 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | 1 | 3433 | Tetragenococcus muriaticus | <0.01 | <0.05 | 2715 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3446 | Streptococcus bovis str. HJ50 | <0.01 | <0.05 | 3846 |
| Firmicutes | Bacilli | Lactobacillales | Unclassified | 1 | 3481 | <0.01 | <0.05 | 2102 | |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3489 | Bacillus silvestris str. SAFN-010 | <0.001 | <0.01 | 1206 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3492 | Bacillus subtilis str. IAM 12118T | <0.01 | <0.05 | 1320 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3494 | Micrococcus luteus B-P 26 | ≤0.01 | <0.05 | 1334 |
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | 1 | 3497 | Weissella koreensis S-5673 | <0.02 | <0.05 | 1457 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3499 | Streptococcus constellatus str. ATCC27823 | <0.01 | <0.05 | 4476 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3504 | Marinilactibacillus psychrotolerans str. O21 | <0.01 | <0.05 | 1505 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3517 | Planococcus maritimus str. TF-9 | <0.01 | <0.05 | 1358 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3521 | Pediococcus inopinatus str. DSM 20285 | <0.001 | <0.05 | 1122 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3526 | Lactobacillus sakei | <0.02 | <0.05 | 1609 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3545 | <0.01 | <0.05 | 1372 | |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3547 | Lactobacillus frumenti str. TMW 1.666 | <0.01 | <0.05 | 1491 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3550 | Bacillus megaterium str. QM B1551 | ≤0.001 | <0.05 | 1620 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3553 | Desemzia incerta str. DSM 20581 | <0.001 | <0.01 | 1553 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3560 | Streptococcus gallinaceus str. CCUG 42692 | <0.001 | <0.01 | 2835 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3566 | Lactobacillus pontis str. LTH 2587 | <0.01 | <0.05 | 2320 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3569 | Staphylococcus saprophyticus | <0.01 | <0.05 | 1391 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3588 | Streptococcus downei str. ATCC 33748 | <0.01 | <0.05 | 2440 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3589 | Bacillus senegalensis str. RS8; CIP 106 669 | ≤0.02 | <0.05 | 1198 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3592 | Staphylococcus caprae str. DSM 20608 | ≤0.01 | <0.05 | 1322 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3605 | <0.01 | <0.05 | 1472 | |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3612 | Bacillus schlegelii str. ATCC 43741T | <0.01 | <0.05 | 1224 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3628 | Staphylococcus haemolyticus str. CCM2737 | <0.01 | <0.05 | 1572 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3629 | Streptococcus mutans str. UA96 | <0.01 | <0.05 | 1466 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | 1 | 3633 | Bacillus clausii str. GMBAE 42 | <0.001 | <0.01 | 2363 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3634 | Lactobacillus letivazi str. JCL3994 | <0.01 | <0.05 | 1586 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3638 | Staphylococcus sp str. AG-30 | <0.01 | <0.05 | 1359 |
| Firmicutes | Bacilli | Bacillales | Paenibacillaceae | 1 | 3641 | Brevibacillus sp. MN 47.2a | <0.02 | <0.05 | 1735 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3654 | Staphylococcus pettenkoferi str. B3117 | <0.01 | <0.05 | 1310 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3661 | Bacillus sp. str. 2216.25.2 | <0.01 | <0.05 | 1593 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3675 | Bacillus mojavensis str. M-1 | <0.01 | <0.05 | 1535 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3684 | Staphylococcus sciuri | <0.02 | <0.05 | 1324 |
| Firmicutes | Bacilli | Bacillales | Halobacillaceae | 1 | 3702 | Amphibacillus xylanus str. DSM 6626 | ≤0.01 | <0.05 | 1523 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3703 | Lactobacillus salivarius str. RA2115 | <0.01 | <0.05 | 1636 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3706 | Bacillus sonorensis str. NRRL B-23155 | <0.02 | <0.05 | 1324 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3722 | Lactococcus Il1403 subsp. lactis str. IL1403 | ≤0.001 | <0.05 | 2673 |
| Firmicutes | Bacilli | Bacillales | Sporolactobacillaceae | 1 | 3747 | Bacillus sp. str. C-59-2 | <0.001 | <0.01 | 1959 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3753 | Streptococcus suis str. 8074 | <0.02 | <0.05 | 3463 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3767 | Lactobacillus suebicus str. CECT 5917T | <0.01 | <0.05 | 2031 |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | 1 | 3768 | Lactobacillus perolens str. L532 | <0.001 | <0.001 | 1593 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3794 | <0.01 | <0.05 | 1324 | |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | 1 | 3822 | Staphylococcus succinus str. SB72 | <0.01 | <0.05 | 1358 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3827 | Bacillus acidogenesis str. 105-2 | <0.01 | <0.05 | 1996 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3831 | Bacillus licheniformis str. KL-068 | <0.01 | <0.05 | 2057 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3833 | Carnobacterium alterfunditum | <0.001 | <0.01 | 2781 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | 1 | 3840 | Trichococcus pasteurii str. KoTa2 | <0.001 | <0.01 | 2656 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3869 | Streptococcus equi subsp. zooepidemicus str. Tokyo1291 subsp. | ≤0.01 | <0.05 | 1766 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3900 | Bacillus licheniformis str. DSM 13 | <0.01 | <0.05 | 1261 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | 1 | 3906 | Streptococcus bovis str.ATCC 43143 | <0.001 | <0.01 | 4284 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3909 | Bacillus subtilis subsp. Marburg str. 168 | <0.01 | <0.05 | 1367 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | 1 | 3918 | Bacillus subtilis | <0.001 | <0.01 | 1486 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | 3 | 3965 | TCE-contaminated site clone ccslm238 | ≤0.01 | <0.05 | 1844 |
| Firmicutes | Mollicutes | Anaeroplasmatales | Erysipelotrichaceae | 3 | 3981 | phototrophic sludge clone PSB-M-3 | <0.01 | <0.05 | 1361 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4180 | termite gut homogenate clone Rs-M23 bacterium | <0.01 | <0.05 | 1383 |
| Firmicutes | Clostridia | Unclassified | Unclassified | 7 | 4216 | <0.001 | <0.01 | 2447 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4266 | termite gut homogenate clone Rs-M86 bacterium | <0.01 | <0.05 | 1302 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4281 | granular sludge clone UASB_brew_B86 | ≤0.001 | <0.05 | 1333 |
| Firmicutes | gut clone group | Unclassified | Unclassified | 1 | 4298 | human mouth clone P4PA_66 | <0.01 | <0.05 | 1991 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4306 | UASB reactor granular sludge clone PD-UASB-4 bacterium | <0.01 | <0.05 | 1483 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4321 | termite gut homogenate clone Rs-C76 bacterium | ≤0.01 | <0.05 | 1362 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4331 | granular sludge clone UASB_brew_B84 | <0.01 | <0.05 | 1091 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4339 | Clostridium chauvoei str. ATCC 10092T | <0.02 | <0.05 | 1542 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4369 | termite gut homogenate clone Rs-N73 bacterium | <0.01 | <0.05 | 1579 |
| Natronoanaerobium | Unclassified | Unclassified | Unclassified | 1 | 4377 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-65 G + C | <0.01 | <0.05 | 1585 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4418 | termite gut homogenate clone Rs-H18 bacterium | <0.02 | <0.05 | 1173 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4434 | termite gut homogenate clone Rs-K11 bacterium | <0.01 | <0.05 | 1298 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4475 | termite gut homogenate clone Rs-N02 bacterium | <0.001 | <0.01 | 1853 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4477 | termite gut homogenate clone Rs-N85 bacterium | 0.01 | <0.05 | 1505 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4507 | termite gut homogenate clone Rs-N21 bacterium | ≤0.01 | <0.05 | 1169 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4510 | termite gut homogenate clone Rs-Q53 bacterium | <0.02 | <0.05 | 1896 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4512 | granular sludge clone UASB_brew_B25 | <0.01 | <0.05 | 1006 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4514 | termite gut homogenate clone Rs-B34 bacterium | ≤0.001 | <0.05 | 1800 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4524 | termite gut clone Rs-093 | <0.02 | <0.05 | 1269 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4533 | termite gut homogenate clone Rs-N06 bacterium | <0.01 | <0.05 | 1671 |
| Firmicutes | Unclassified | Unclassified | Unclassified | 8 | 4536 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-14 G + C | <0.01 | <0.05 | 1005 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4540 | termite gut homogenate clone Rs-M18 bacterium | <0.01 | <0.05 | 1962 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4598 | Clostridium sardiniense str. DSM 600 | <0.02 | <0.05 | 1253 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4607 | Clostridium novyi str. NCTC538 | <0.01 | <0.05 | 1082 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 5 | 4613 | rumen clone 3C0d-3 | <0.02 | <0.05 | 1321 |
| Firmicutes | gut clone group | Unclassified | Unclassified | 1 | 4616 | rumen clone F23-C12 | <0.01 | <0.05 | 2628 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4622 | termite gut clone Rs-L36 | ≤0.01 | <0.05 | 1112 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 12 | 4638 | <0.01 | <0.05 | 2515 | |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | 9 | 5038 | Rumen isolate str. YS2 | <0.001 | <0.01 | 1724 |
| Bacteroidetes | Bacteroidetes | Unclassified | Unclassified | 15 | 5481 | marine sediment above hydrate ridge clone Hyd89-72 bacterium | <0.02 | <0.05 | 2056 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | 19 | 5542 | Cytophaga sp. I-1787 | <0.01 | <0.05 | 2304 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | 15 | 5783 | Mono Lake at depth 35 m station 6 July 2000 clone ML635J-15 bacterium | <0.01 | <0.05 | 1032 |
| Bacteroidetes | Bacteroidetes | Bacteroidales | Unclassified | 15 | 5874 | Paralvinella palmiformis mucus secretions clone P. palm 53 bacterium | <0.02 | <0.05 | 1805 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Flexibacteraceae | 19 | 6124 | Flexibacter flexilis subsp. pelliculosus str. IFO 16028 subsp. | <0.001 | <0.01 | 1387 |
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | 1 | 6459 | Spirochaeta sp. str. BHI80-158 | <0.001 | ≤0.01 | 2558 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Unclassified | 3 | 9813 | hydrothermal sediment clone AF420340 | <0.01 | <0.05 | 1453 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | 5 | 9875 | hydrothermal sediment clone AF420354 | <0.01 | <0.05 | 1147 |
| Proteobacteria | Deltaproteobacteria | Desulfuromonadales | Geobacteraceae | 1 | 10171 | <0.01 | <0.05 | 1375 | |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfoarculaceae | 2 | 10227 | marine sediment clone Bol11 | <0.01 | <0.05 | 1549 |
| Proteobacteria | Deltaproteobacteria | Desulfobacterales | Desulfobacteraceae | 5 | 10319 | sulfate-reducing habitat clone SLM-CP-116 | <0.01 | <0.05 | 1235 |
S-F, Subfamily identification; bTaxon ID, PhyloChip Taxon identification number; cRepresentative species, Taxon bacterial species identifier.
A common “core” of 75 bacterial taxa representing 27 classified bacterial families was identified in all patients analyzed (Fig. 3). This core group included members of the Pseudomonadaceae, Enterobacteriaceae, Campylobacteraceae, and Helicobacteraceae, amongst others. In addition, taxa containing species of pathogenic potential, such as Arcobacter cryaerophilus, Brevundimonas diminuta, Leptospira interrogans, as well as P. aeruginosa, were detected in all patients (a complete list of the core taxa is provided in Table 5; see at end of article). We also analyzed the array data for organisms that have previously been associated with COPD airways (Sethi and Murphy, 2008). Haemophilus influenzae was detected by the array in two subjects (patients 2 and 8), although corresponding m-BAL cultures were negative for this organism. Moraxella catarrhalis was not detected by PhyloChip or culture in any patient sample. However, other phylogenetically related members in the Moraxellaceae family, including Moraxella oblonga, Acinetobacter haemolyticus, and Psychrobacter psychrophilus were identified by the array in 80–100% of subjects (Table 3). Streptococcus pneumoniae was detected in four subjects (patients 2, 3, 7 and 8) despite all m-BALs being culture-negative for this species. Finally, we also examined PhyloChip data for the presence of the atypical bacteria, Mycoplasma pneumoniae and Chlamydophila pneumoniae, which are associated with 3–5% percent of exacerbations (Sethi and Murphy, 2008). Neither was detected by the PhyloChip, although a related species, Mycoplasma pulmonis, was identified in a single individual (patient 3).
FIG. 3.
Phylogenetic tree illustrating core bacterial taxa detected in COPD airways samples in this study. Known pathogens are denoted with an asterix; distinct bacterial families are indicated by different colors.
Table 5.
Core Community of Bacterial Taxa Detected in all COPD Patients during Treatment for Severe Exacerbations (Representative Species with a Proven Role in Mammalian Pathogenesis are Highlighted)
| Phylum | Class | Order | Family | S-Fa | Taxon IDb | Representative speciesc |
|---|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Acidimicrobiales | Acidimicrobiaceae | sf_1 | 1749 | forest soil clone DUNssu275 (-3A) (OTU#188) |
| Acidobacteria | Acidobacteria | Acidobacteriales | Acidobacteriaceae | sf_6 | 6362 | grassland soil clone DA052 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8578 | Marinobacter lipolyticus str. SM-19 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9239 | Arctic sea ice ARK10228 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8222 | |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8753 | Idiomarina loihiensis str. GSP37 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 9324 | Pseudoalteromonas ruthenica str. KMM300 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | sf_1 | 8579 | Psychromonas profunda str. 2825 |
| Proteobacteria | Alphaproteobacteria | Rickettsiales | Anaplasmataceae | sf_3 | 6648 | Wolbachia sp |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | sf_1 | 7747 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10538 | Arcobacter cryaerophilus |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10447 | Sulfurospirillum deleyianum str. Spirillum 5175 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | sf_3 | 10456 | Campylobacter showae |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 6909 | Brevundimonas diminuta str. DSM 1635 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | sf_1 | 7436 | Brevundimonas sp. str. FWC40 |
| Cyanobacteria | Cyanobacteria | Chloroplasts | Chloroplasts | sf_5 | 5147 | Emiliania huxleyi str. Plymouth Marine Laborator PML 92 |
| Proteobacteria | Gammaproteobacteria | Legionellales | Coxiellaceae | sf_3 | 9198 | uranium mining waste pile clone KF-JG30-B15 KF-JG30-B15 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Crenotrichaceae | sf_11 | 6267 | Cilia-respiratory isolate str. 243-54 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfomicrobiaceae | sf_1 | 10079 | Desulfomicrobium baculatum str. DSM 1742 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | sf_1 | 8504 | Dysmicoccus neobrevipes symbiont |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10385 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10442 | Helicobacter cetorum str. MIT 99-5656 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10444 | Helicobacter suncus str. Kaz-2 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10448 | Helicobacter felis str. Dog-1 |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacteraceae | sf_3 | 10451 | Helicobacter heilmannii str. C4S |
| Spirochaetes | Spirochaetes | Spirochaetales | Leptospiraceae | sf_3 | 6496 | Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8366 | Psychrobacter frigidicola str. DSM 12411 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8838 | Psychrobacter psychrophilus CMS 28 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | sf_3 | 8727 | Alkanindiges hongkongensis str. HKU9 |
| Proteobacteria | Betaproteobacteria | Nitrosomonadales | Nitrosomonadaceae | sf_1 | 7789 | |
| Firmicutes | Clostridia | Clostridiales | Peptococc/Acidaminococc | sf_11 | 992 | anoxic bulk soil flooded rice microcosm clone BSV43 clone |
| Planctomycetes | Planctomycetacia | Planctomycetales | Pirellulae | sf_3 | 4670 | |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Piscirickettsiaceae | sf_3 | 9291 | Methylophaga alcalica str. M39 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8691 | Pseudomonas aeruginosa str. PAO1 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9056 | Pseudomonas aeruginosa str. #47 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9068 | Pseudomonas stutzeri str. A1501 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9295 | |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9613 | Pseudomonas flavescens str. B62 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9049 | uranium mining mill tailing clone GR-Sh2-34 GR-Sh2-34 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9469 | cf. Pseudomonas sp. clone Llangefni 52 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9240 | Pseudomonas fluorescens str. CHA0 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 9366 | Arctic seawater isolate str. R7366 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | sf_1 | 8755 | Pseudomonas sp. SK-1-3-1 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | sf_1 | 5913 | Sphingobacteriaceae str. Ellin160 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | sf_1 | 6663 | Sphingopyxis flavimaris str. SW-151 |
| Firmicutes | Bacilli | Bacillales | Thermoactinomycetaceae | sf_1 | 3301 | Thermoactinomyces sp. str. 700375 |
| Thermodesulfobacteria | Thermodesulfobacteria | Thermodesulfobacteriales | Thermodesulfobacteriaceae | sf_1 | 667 | Fjonesia |
| Proteobacteria | Gammaproteobacteria | Thiotrichales | Thiotrichaceae | sf_3 | 8752 | Beggiatoa sp. str. MS-81-1c |
| Chloroflexi | Unclassified | Unclassified | Unclassified | sf_2 | 818 | |
| Verrucomicrobia | Unclassified | Unclassified | Unclassified | sf_4 | 288 | Prosthecobacter dejongeii |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 117 | termite gut homogenate clone Rs-D89 |
| Synergistes | Unclassified | Unclassified | Unclassified | sf_3 | 719 | Synergistes sp. P1 str. P4G_18 |
| OP3 | Unclassified | Unclassified | Unclassified | sf_4 | 628 | CB-contaminated groundwater clone GOUTB15 |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 485 | thermal spring mat clone O1aA90 |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 226 | |
| Bacteroidetes | KSA1 | Unclassified | Unclassified | sf_1 | 5951 | CFB group clone ML615J-4 |
| Chloroflexi | Anaerolineae | Unclassified | Unclassified | sf_9 | 727 | forest soil clone S0208 |
| Cyanobacteria | Unclassified | Unclassified | Unclassified | sf_8 | 5206 | |
| marine group A | mgA-2 | Unclassified | Unclassified | sf_1 | 6344 | bacterioplankton clone ZA3648c |
| Unclassified | Unclassified | Unclassified | Unclassified | sf_160 | 6430 | |
| Proteobacteria | Alphaproteobacteria | Unclassified | Unclassified | sf_6 | 7575 | |
| TM7 | TM7-3 | Unclassified | Unclassified | sf_1 | 8155 | oral periodontitis clone EW086 |
| Proteobacteria | Gammaproteobacteria | uranium waste clones | Unclassified | sf_1 | 8747 | uranium waste soil clone JG30-KF-CM35 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 9568 | forested wetland clone RCP2-96 |
| Proteobacteria | Gammaproteobacteria | SUP05 | Unclassified | sf_1 | 8605 | bacterioplankton clone ZA2525c |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_3 | 8339 | water 5 m downstream manure clone 35ds5 |
| Proteobacteria | Gammaproteobacteria | Unclassified | Unclassified | sf_4 | 8855 | |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10480 | Paralvinella palmiformis mucus secretions cloneP. palm C 84 proteobacterium |
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Unclassified | sf_1 | 10530 | hydrothermal vent 9 degrees North East Rise PacificOcean clone CH5_6_BAC_16SrRNA_9N_EPR |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1687 | Jonesia quinghaiensis str. DSM 15701 |
| Actinobacteria | Actinobacteria | Actinomycetales | Unclassified | sf_3 | 1405 | Arthrobacter ureafaciens str. DSM 20126 |
| Firmicutes | Clostridia | Unclassified | Unclassified | sf_3 | 2373 | |
| Firmicutes | Catabacter | Unclassified | Unclassified | sf_1 | 4293 | termite gut homogenate clone Rs-Q01 bacterium |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | sf_3 | 8689 | Dyemonas todaii str. XD10 |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Xiphinematobacteraceae | sf_3 | 888 | Candidatus Xiphinematobacter brevicolli |
S-F, Subfamily identification; bTaxon ID, PhyloChip Taxon identification number; cRepresentative species, Taxon bacterial species identifier.
Quantitative PCR was performed to validate that changes in reported array fluorescence intensities for targeted taxa correlated with changes in target species copy number for a selection of known airway pathogens (P. aeruginosa and Stenotrophomonas maltophilia) and two characteristic gastrointestinal organisms (Campylobacter mucosalis and Helicobacter cetorum). Regression analysis of species abundance determined by Q-PCR and array fluorescence intensity demonstrated strong concordance between the two independent methods for each target organism (Table 6), confirming their presence in these COPD airway samples and the ability of the array to accurately reflect changes in organism relative abundance.
Table 6.
Correlation Results for Species Abundance by Q-PCR and 16S rRNA PhyloChip
| Target species | R value | p Value |
|---|---|---|
| P. aeruginosa | 0.77 | <0.05 |
| S. maltophilia | 0.80 | <0.05 |
| Campylobacter mucosalis | 0.68 | <0.10 |
| Helicobacter cetorum | 0.79 | <0.05 |
Discussion
Although nearly half of acute COPD exacerbations are associated with bacterial infection, our knowledge of the microbial species associated with these events is limited to a handful of organisms detected primarily by culture-based methods (Papi et al., 2006; Rosell et al., 2005; Soler et al., 2007). The overall aim of this study was to begin to address the overarching question whether previously undetected bacterial species exist in the airways of COPD patients during acute exacerbations. High-resolution, culture-independent analysis using the 16S rRNA PhyloChip revealed a much greater diversity of bacteria than has previously been appreciated in the airways of COPD patients being managed for severe exacerbations, including members of the Pseudomonadaceae, Enterobacteriaceae, and Helicobacteraceae, among others. The potential for a diverse airway bacterial community to play a role in chronic airway colonization and inflammation, a feature of COPD, has not been previously considered.
The identification of a diversity of bacterial communities in respiratory samples from COPD patients experiencing severe exacerbations suggests that the pathogenesis of these events could involve a polymicrobial process. Future studies in a larger cohort of patients, including nonintubated COPD patients with a greater range of exacerbation severity, will be necessary to determine relationships between community composition, structure, and pulmonary health. Only a handful of bacterial species, such as H. influenzae and P. aeruginosa, have previously been associated with COPD exacerbations. The possible role of other bacterial community members with pathogenic potential identified in this study, e.g. A. cryaerophilus, B. diminuta, and L. interrogans, may merit further investigation. Many of these species have been implicated in other pathogenic processes such as endocarditis (Han and Andrade, 2005; Marques da Silva et al., 2006; Paster et al., 2002) and bacteremia (Hsueh et al., 1997; Woo et al., 2001). L. interrogans, the causative agent of human leptospirosis (Gaudie et al., 2008), has recently been shown to induce pulmonary lesions in an experimental animal model of airway infection (Marinho et al., 2009) and pulmonary hemorrhage in severe cases (Dall'Antonia et al., 2008). Their potential for pathogenesis suggests the possibility of a role for these organisms in COPD chronic airway disease. Future studies involving functional and mechanistic analyses will be necessary to further assess this.
Multiple oropharyngeal and gut-associated bacterial species were also identified by PhyloChip analysis, suggesting a potential role in COPD exacerbations. Although contamination by oropharyngeal secretions is always a possibility, samples were collected through an endotracheal tube, diminishing the degree of direct contamination. Ongoing microaspiration during intubation, however, cannot be completely prevented, and it has been suggested that the oral cavity and gastrointestinal tract act as a microbial reservoir for seeding the airways in vulnerable patient populations (Garrouste-Orgeas et al., 1997; Heyland and Mandell, 1992; Orozco-Levi et al., 2003). In our study the relative abundance of targeted gastrointestinal-associated species, H. cetorum and C. mucosalis (Figura et al., 1993; Garcia-Amado et al., 2007), were confirmed by independent Q-PCR in airway samples from these patients. The presence of oropharyngeal or gut-associated bacteria in the lower airways may have significant implications for a disease population with greater risks from pulmonary infections. For example, Duan et al. (2003) demonstrated in a rat model that coinfection of P. aeruginosa with oropharyngeal bacterial species (isolated from cystic fibrosis patient sputa) resulted in enhanced lung damage and upregulation of P. aeruginosa virulence gene expression. In our study, P. aeruginosa and oropharyngeal and gut-associated species were present in the airways of all patients studied, suggesting the potential for enhanced pathogenesis in this patient population.
We recognize that our study numbers are small and represent a severely ill group of COPD patients. Therefore, caution must be exercised in extrapolating these findings to a broader group of COPD patients, especially nonintubated individuals experiencing less severe exacerbations. Although facilitating access to lower respiratory specimens that are otherwise challenging to obtain during severe exacerbations, the intubation status of these patients is an important consideration in weighing these findings. Although protracted intubation was associated with decreased bacterial community richness, the possibility for a more diverse bacterial community to play a role at least at the onset of exacerbation-associated respiratory failure remains. Control samples from nonintubated COPD and non-COPD patients were not available for this study, nor were longitudinal samples from these patients. In a previous study, however, we have found that endotracheal samples obtained from patients briefly intubated for elective surgery produced no detectable 16S rRNA PCR product (Flanagan et al., 2007). The lack of an association between duration of active antimicrobial therapy and bacterial community structure is most likely due to both the small study numbers as well as the administration of previous antibiotic courses up to one month prior to sample collection in the study (e.g., patient 6).
Results of this study highlight the advantages of complementing culture-based methods with higher resolution approaches for bacterial detection to provide a more comprehensive assessment of the airway microbiota present in COPD patients. Culture-independent methods are particularly relevant to identify viable but nonculturable species that produce and exist in biofilms (Rayner et al., 1998), which are characteristic of chronic airway infections (Costerton et al., 1999; Singh et al., 2000) and have recently been implicated in COPD (Martinez-Solano et al., 2008). Because our PhyloChip analysis was based on DNA extracted from airway samples, it does not provide information on the viability of the species detected. However, we have previously noted that within 24 h of antimicrobial administration to cystic fibrosis patients, bacterial richness decreased approximately 10-fold as detected by the PhyloChip (Lynch, unpublished data). This suggests that DNA turnover is relatively rapid in the airways of chronic pulmonary disease patients, and that the taxa detected during antimicrobial administration in our COPD patients largely represent the viable portion of the community. This is supported by the finding that the PhyloChip detected all species that were isolated by concurrent clinical laboratory culture. Independent Q-PCR analysis of selected taxa also demonstrated strong concordance between calculated 16S rRNA copy number and PhyloChip-based fluorescence intensities, validating the abundance of individual species detected by the array. Although both cultures and the microarray demonstrated low detection rates for H. influenzae and M. catarrhalis, two species commonly associated with COPD exacerbation (potentially due to antibiotic-mediated clearance), only the PhyloChip identified other Haemophilus species across the majority of patients in this study, as well as other members of the Moraxellaceae family. Although these molecular methods may identify potentially relevant species undetected by culture, they provide no information on the viability or activity of these species. Hence, the significance of detecting species phylogenetically related to known pathogens in COPD airways is unclear, but may merit further study.
Conclusions
Application of the 16S rRNA PhyloChip to airway secretions from COPD patients during severe respiratory exacerbations requiring ventilatory support and antibiotic administration, demonstrated the presence of diverse bacterial communities, whose structural variation in this cohort was related to the duration of intubation. A core community of bacterial taxa comprised of many known pathogens, some not previously associated with COPD, was common to all patients studied. Given that the disease model of COPD is generally characterized by chronic airway colonization, recurrent infection-related exacerbations, and a persistent state of chronic airway inflammation, these results highlight the need to consider the polymicrobial community present in COPD airways and the potential functional effects of these consortia on immune response and pulmonary health. Identification of relationships that exist between bacterial communities, their collective gene expression, concomitant host response, and clinical outcomes may ultimately lead to improved understanding of the pathogenesis of COPD.
Acknowledgments
The authors thank Yvette Piceno, Todd DeSantis, and Gary Andersen at Lawrence Berkeley National Laboratory for their advice and technical support in this study, and to Homer Boushey, M.D., for his critique of the manuscript. Part of this work was performed at Lawrence Berkeley National Laboratory under the auspices of the University of California under contract number DOE DE-AC02-05CH11231. Research was supported by a Tobacco-Related Disease Research Program (Univ. of California) fellowship to Y.J.H. and an American Lung Association grant to S.V.L. S.V.L. and E.L.B. are funded also by NIH Award (AI075410).
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austr Ecol. 2001;26:32–46. [Google Scholar]
- Brodie E.L. Desantis T.Z. Joyner D.C. Baek S.M. Larsen J.T. Andersen G.L., et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie E.L. Desantis T.Z. Parker J.P. Zubietta I.X. Piceno Y.M. Andersen G.L. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors A.F., Jr. Dawson N.V. Thomas C. Harrell F.E., JR. Desbiens N. Fulkerson W.J., et al. Outcomes following acute exacerbation of severe chronic obstructive lung disease. The SUPPORT investigators (Study to Understand Prognoses and Preferences for Outcomes and Risks of Treatments) Am J Respir Crit Care Med. 1996;154:959–967. doi: 10.1164/ajrccm.154.4.8887592. [DOI] [PubMed] [Google Scholar]
- Costerton J.W. Stewart P.S. Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dall'Antonia M. Sluga G. Whitfield S. Teall A. Wilson P. Krahe D. Leptospirosis pulmonary haemorrhage: a diagnostic challenge. Emerg Med J. 2008;25:51–52. doi: 10.1136/emj.2007.051524. [DOI] [PubMed] [Google Scholar]
- Desantis T.Z. Hugenholtz P. Larsen N. Rojas M. Brodie E.L. Keller K., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis T.Z. Brodie E.L. Moberg J.P. Zubieta I.X. Piceno Y.M. Andersen G.L. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microbial Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L. McFall-Ngai M. Relman D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K. Dammel C. Stein J. Rabin H. Surette M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- Eckburg P.B. Bik E.M. Bernstein C.N. Purdom E. Dethlefsen L. Sargent M., et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figura N. Guglielmetti P. Zanchi A. Partini N. Armellini D. Bayeli P.F., et al. Two cases of Campylobacter mucosalis enteritis in children. J Clin Microbiol. 1993;31:727–728. doi: 10.1128/jcm.31.3.727-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J.L. Brodie E.L. Weng L. Lynch S.V. Garcia O. Brown R., et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Amado M.A. Al-Soud W.A. Borges-Landaez P. Contreras M. Cedeno S. Baez-Ramirez E., et al. Non-pylori Helicobacteraceae in the upper digestive tract of asymptomatic Venezuelan subjects: detection of Helicobacter cetorum-like and Candidatus Wolinella africanus-like DNA. Helicobacter. 2007;12:553–558. doi: 10.1111/j.1523-5378.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- Garrouste-Orgeas M. Chevret S. Arlet G. Marie O. Rouveau M. Popoff N., et al. Oropharyngeal or gastric colonization and nosocomial pneumonia in adult intensive care unit patients. A prospective study based on genomic DNA analysis. Am J Respir Crit Care Med. 1997;156:1647–1655. doi: 10.1164/ajrccm.156.5.96-04076. [DOI] [PubMed] [Google Scholar]
- Gaudie C.M. Featherstone C.A. Phillips W.S. McNaught R. Rhodes P.M. Errington J., et al. Human Leptospira interrogans serogroup icterohaemorrhagiae infection (Weil's disease) acquired from pet rats. Vet Rec. 2008;163:599–601. doi: 10.1136/vr.163.20.599. [DOI] [PubMed] [Google Scholar]
- Han X.Y. Andrade R.A. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J Antimicrob Chemother. 2005;55:853–859. doi: 10.1093/jac/dki139. [DOI] [PubMed] [Google Scholar]
- Harris J.K. de Groote M.A. Sagel S.D. Zemanick E.T. Kapsner R. Penvari C., et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyland D. Mandell L.A. Gastric colonization by Gram-negative bacilli and nosocomial pneumonia in the intensive care unit patient. Evidence for causation. Chest. 1992;101:187–193. doi: 10.1378/chest.101.1.187. [DOI] [PubMed] [Google Scholar]
- Hsueh P.R. Teng L.J. Yang P.C. Wang S.K. Chang S.C. Ho S.W., et al. Bacteremia caused by Arcobacter cryaerophilus 1B. J Clin Microbiol. 1997;35:489–491. doi: 10.1128/jcm.35.2.489-491.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.J. 16S/23S rRNA sequencing. Wiley; Chichester, UK: 1991. [Google Scholar]
- Letunic I. Bork P. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Mannino D.M. Braman S. The epidemiology and economics of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2007;4:502–506. doi: 10.1513/pats.200701-001FM. [DOI] [PubMed] [Google Scholar]
- Marinho M. Oliveira-Junior I.S. Monteiro C.M. Perri S.H. Salomao R. Pulmonary disease in Hamsters infected with Leptospira interrogans: histopathologic findings and cytokine mRNA expressions. Am J Trop Med Hyg. 2009;80:832–836. [PubMed] [Google Scholar]
- Marques da Silva R. Caugant D.A. Eribe E.R. Aas J.A. Lingaas P.S. Geiran O., et al. Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J Vasc Surg. 2006;44:1055–1060. doi: 10.1016/j.jvs.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Martinez-Solano L. Macia M.D. Fajardo A. Oliver A. Martinez J.L. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin Infect Dis. 2008;47:1526–1533. doi: 10.1086/593186. [DOI] [PubMed] [Google Scholar]
- Murphy T.F. Brauer A.L. Schiffmacher A.T. Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- Nseir S. Di Pompeo C. Cavestri B. Jozefowicz E. Nyunga M. Soubrier S., et al. Multiple-drug-resistant bacteria in patients with severe acute exacerbation of chronic obstructive pulmonary disease: prevalence, risk factors, and outcome. Crit Care Med. 2006;34:2959–2966. doi: 10.1097/01.CCM.0000245666.28867.C6. [DOI] [PubMed] [Google Scholar]
- Orozco-Levi M. Felez M. Martinez-Mirallel E. Solsona J.F. Blanco M.L. Broquetas J.M., et al. Gastro-oesophageal reflux in mechanically ventilated patients: effects of an oesophageal balloon. Eur Respir J. 2003;22:348–353. doi: 10.1183/09031936.03.00048902. [DOI] [PubMed] [Google Scholar]
- Papi A. Bellettato C.M. Braccioni F. Romagnoli M. Casolari P. Caramori G., et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173:1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- Paster B.J. Falkler W.A., Jr. Enwonwu C.O. Idigbe E.O. Savage K.O. Levanos V.A., et al. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J Clin Microbiol. 2002;40:2187–2191. doi: 10.1128/JCM.40.6.2187-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N. Dehal P.S. Arkin A.P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe K.F. Hurd S. Anzueto A. Barnes P.J. Buist S.A. Calverley P., et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- Rayner M.G. Zhang Y. Gorry M.C. Chen Y. Post J.C. Ehrlich G.D. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA. 1998;279:296–299. doi: 10.1001/jama.279.4.296. [DOI] [PubMed] [Google Scholar]
- Rogers G.B. Carroll M.P. Serisier D.J. Hockey P.M. Jones G. Bruce K.D. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell A. Monso E. Soler N. Torres F. Angrill J. Riise G., et al. Microbiologic determinants of exacerbation in chronic obstructive pulmonary disease. Arch Intern Med. 2005;165:891–897. doi: 10.1001/archinte.165.8.891. [DOI] [PubMed] [Google Scholar]
- Sethi S. Murphy T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- Singh P.K. Schaefer A.L. Parsek M.R. Moninger T.O. Welsh M.J. Greenberg E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Soler N. Agusti C. Angrill J. Puig de la Bellacasa J. Torres A. Bronchoscopic validation of the significance of sputum purulence in severe exacerbations of chronic obstructive pulmonary disease. Thorax. 2007;62:29–35. doi: 10.1136/thx.2005.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D. Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J. Ley R.E. Mahowald M.A. Magrini V. Mardis E.R. Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Woo P.C. Chong K.T. Leung K. Que T. Yuen K. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteremia by 16S ribosomal RNA gene sequencing. Diagn Microbiol Infect Dis. 2001;40:125–127. doi: 10.1016/s0732-8893(01)00261-9. [DOI] [PubMed] [Google Scholar]
- Zhuo H. Yang K. Lynch S.V. Dotson H. Glidden D.V. Singh G., et al. Increased mortality of ventilated patients with endotracheal Pseudomonas aeruginosa without clinical signs of infection. Crit Care Med. 2008;36:2495–2503. doi: 10.1097/CCM.0b013e318183f3f8. [DOI] [PubMed] [Google Scholar]