Abstract
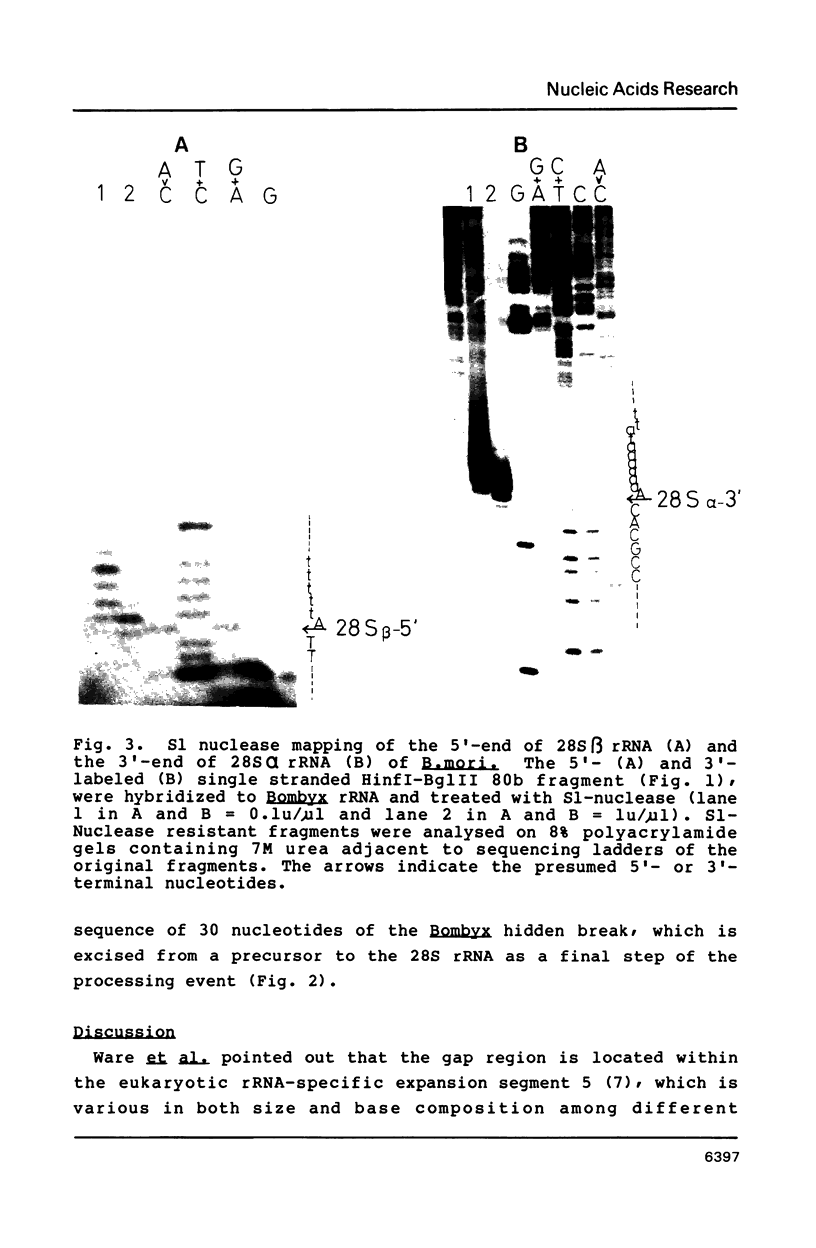
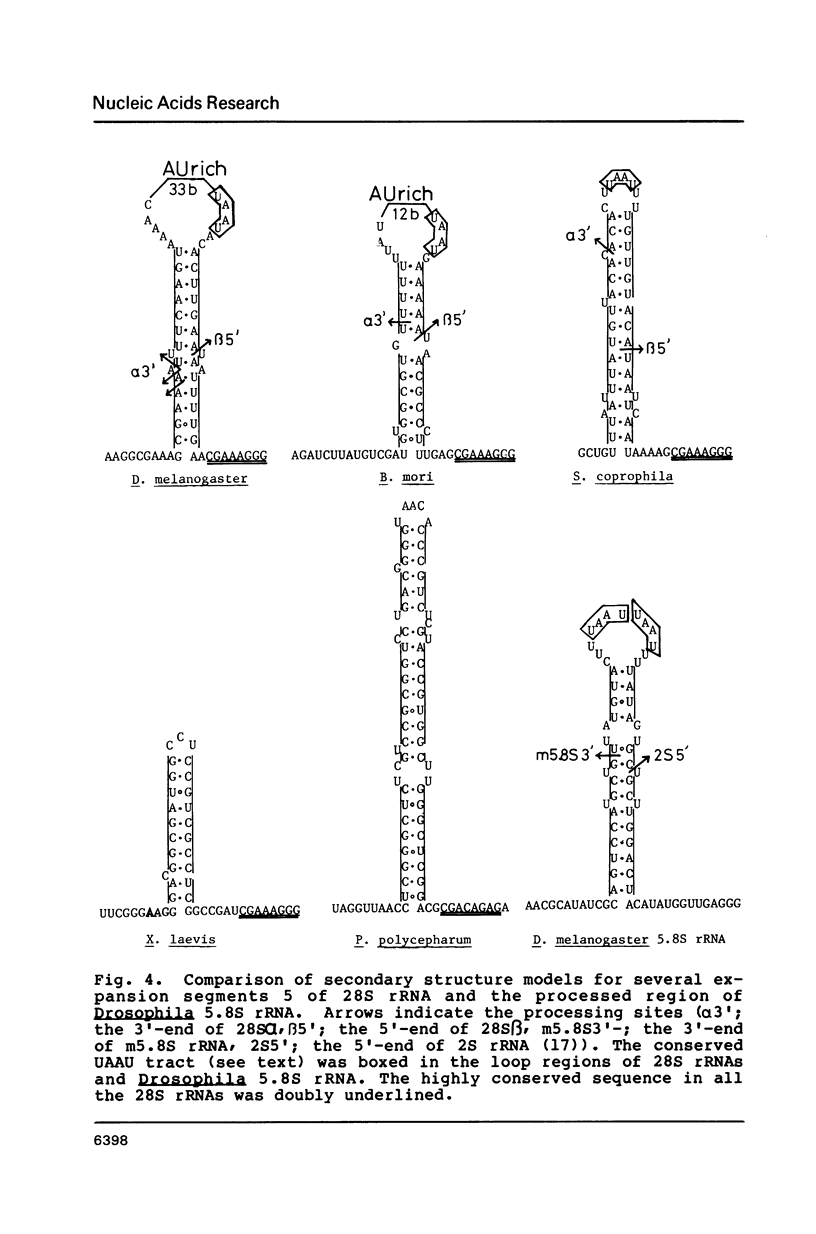
We determined the structure around the gap region between 28S alpha and 28S beta rRNA of Bombyx mori, a lepidopteran insect, to know the introduction mechanism of the hidden break, an interruption of the phosphodiester bond specific to the 28S rRNA of protostomes. Sequence analysis and S1 nuclease mapping suggested that a stretch of 30 nucleotides is excised from the mid region of the 28S rRNA to generate the hidden break. The length of excluded stretch was very various among three insects so far studied. However, the gap responsible for the hidden break was located in a fixed position in the 28S rRNA irrespective of insect species. It was suggested that extremely AU-rich sequence including the specific UAAU tract forming a loop can be a signal for the introduction of the hidden break. The same signal seemed also involved in splitting the dipteran 5.8S rRNA.
Full text
PDF

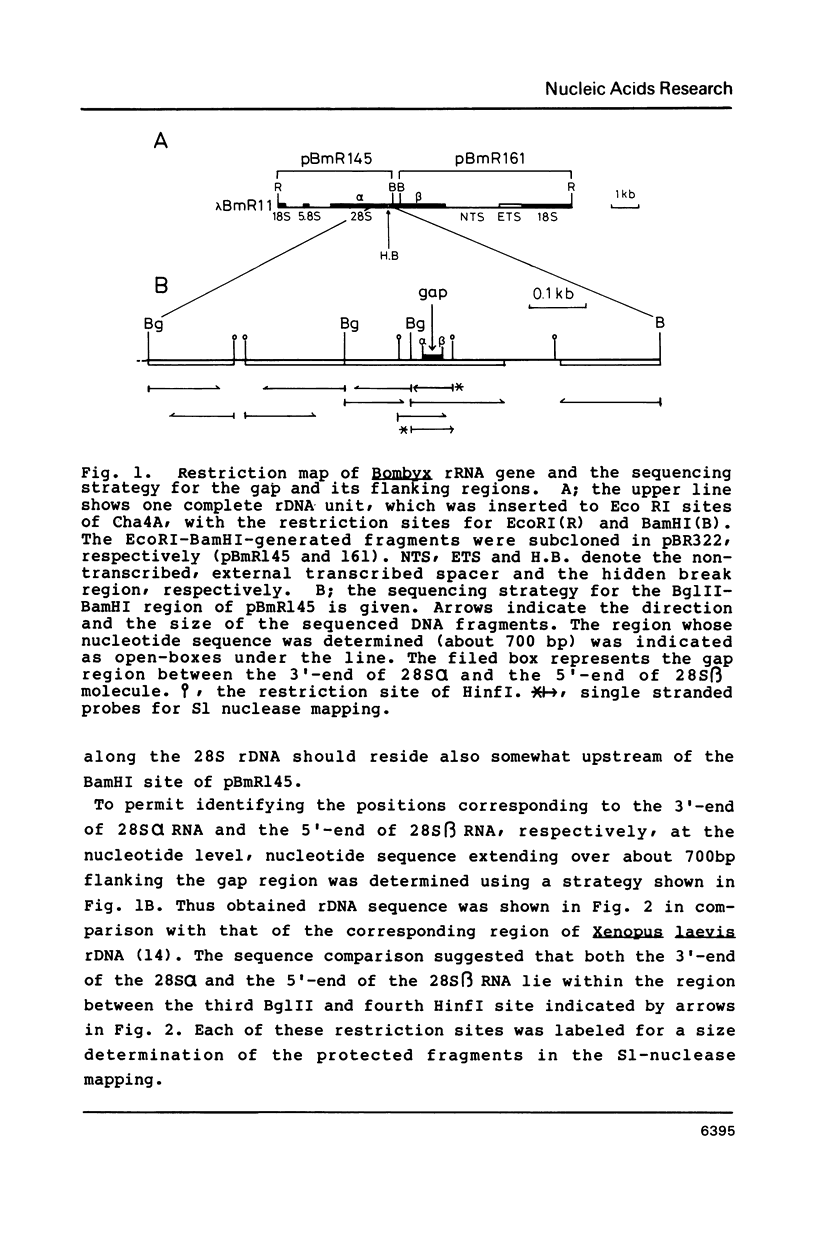




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. G., Tague B. W., Ware V. C., Gerbi S. A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984 Aug 10;12(15):6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanversin G., Jacq B. Séquence de la région de la coupure centrale du précurseur de l'ARN ribosomique 26S de Drosophile. C R Seances Acad Sci III. 1983;296(22):1041–1044. [PubMed] [Google Scholar]
- Fujiwara H., Ishikawa H. Primary and secondary structures of Tetrahymena and aphid 5.8S rRNAs: structural features of 5.8S rRNA which interacts with the 28S rRNA containing the hidden break. Nucleic Acids Res. 1982 Sep 11;10(17):5173–5182. doi: 10.1093/nar/10.17.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Ogura T., Takada N., Miyajima N., Ishikawa H., Maekawa H. Introns and their flanking sequences of Bombyx mori rDNA. Nucleic Acids Res. 1984 Sep 11;12(17):6861–6869. doi: 10.1093/nar/12.17.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Newburgh R. W. Studies of the thermal conversion of 28 S RNA of Galleria mellonella (L.) to an 18 S product. J Mol Biol. 1972 Feb 28;64(1):135–144. doi: 10.1016/0022-2836(72)90325-7. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3565–3573. doi: 10.1093/nar/8.16.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening U. E. Molecular weights of ribosomal RNA in relation to evolution. J Mol Biol. 1968 Dec;38(3):355–365. doi: 10.1016/0022-2836(68)90391-4. [DOI] [PubMed] [Google Scholar]
- Manning R. F., Samols D. R., Gage L. P. The genes for 18S, 5.8S and 28S ribosomal RNA of Bombyx mori are organized into tandem repeats of uniform length. Gene. 1978 Oct;4(2):153–166. doi: 10.1016/0378-1119(78)90027-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982 Oct;30(3):669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Ware V. C., Renkawitz R., Gerbi S. A. rRNA processing: removal of only nineteen bases at the gap between 28S alpha and 28S beta rRNAs in Sciara coprophila. Nucleic Acids Res. 1985 May 24;13(10):3581–3597. doi: 10.1093/nar/13.10.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]