Abstract
With the rapidly growing availability of the entire genome sequences of microbial pathogens, there is unmet need for increasingly sensitive systems to monitor the gene-specific markers for diagnosis of bacteremia that enables an earlier detection of causative agent and determination of drug resistance. To address these challenges, a novel FISH-type genomic sequence-based molecular technique is proposed that can identify bacteria and simultaneously detect antibiotic resistance markers for rapid and accurate testing of pathogens. The approach is based on a synergistic combination of advanced Peptide Nucleic Acid (PNA)-based technology and signal-enhancing Rolling Circle Amplification (RCA) reaction to achieve a highly specific and sensitive assay. A specific PNA-DNA construct serves as an exceedingly selective and very effective biomarker, while RCA enhances detection sensitivity and provide with a highly multiplexed assay system. Distinct-color fluorescent decorator probes are used to identify about 20-nucleotide-long signature sequences in bacterial genomic DNA and/or key genetic markers of drug resistance in order to identify and characterize various pathogens. The technique's potential and its utility for clinical diagnostics are illustrated by identification of S. aureus with simultaneous discrimination of methicillin-sensitive (MSSA) versus methicillin-resistant (MRSA) strains. Overall these promising results hint to the adoption of PNA-based rapid sensitive detection for diagnosis of other clinically relevant organisms. Thereby, new assay enables significantly earlier administration of appropriate antimicrobial therapy and may, thus have a positive impact on the outcome of the patient.
Key words: PNA, bacteral detection, drug resistance, S. aureus, RCA
Introduction
Infectious diseases are still a major healthcare problem. Widespread use of antibiotics has resulted in the emergence and spread of drug-resistant bacterial pathogens, increasing the demand for better clinical diagnostic assays. The goal for such assays is to rapidly identify an infectious agent and simultaneously determine its antimicrobial resistance profile in order to facilitate optimal therapeutic management. Despite recent advancement in pathogens' identification, most of the gold-standard diagnostic methods have limitations, including laborious sample preparation, bulky instrumentation and slow data readout.
Since its introduction, fluorescent in situ hybridization (FISH) has become a widely used method for a variety of assays in the clinical diagnostic laboratory. This method makes possible the accurate detection of microbes directly from clinical specimens or after culture enrichment.1–3 Due to its high copy number, ribosomal RNA (rRNA) sequences are commonly used as targets for the fluorescent-labeled probes.4 Peptide nucleic acids (PNA) are the most advanced technology in respect to applications in rRNA-FISH, and the robustness and usefulness of the method is evident in commercially available diagnostic tests such as those developed by AdvanDx Inc., (Woburn, MA USA; www.advandx.com). However, major limitations are inherent in rRNA- targeted FISH. Notably, ribosomal RNA sequences of closely related strains, subspecies or even different species are often identical and therefore cannot always be used as differential markers; also, ribosomal inquiry cannot be used to characterize antimicrobial resistance.5
We have recently proposed and tested on model bacteria a new, more versatile, isothermal approach for bacterial detection in a convenient FISH format that is based on exceedingly specific and sensitive recognition of short signature sites within bacterial genomes.6,7 Here we report our progress in using this approach for detection of several important bacterial pathogens: methicillin-sensitive and methicillin-resistant strains of S. aureus (MSSA and MRSA, respectively) and a common, often multidrug-resistant pathogen P. aeruginosa.
Staphylococcus aureus is a formidable healthcare-associated pathogen.8,9 Hospital-associated staphylococcal infection is associated with significant morbidity and mortality and increased costs to the healthcare system. This is especially true for MRSA infections as compared to those caused by MSSA. In certain nosocomial settings, the rate of MRSA is estimated to be greater than 50% of all S. aureus isolates. In addition, during the last decade community-acquired MRSA infection has emerged as a formidable clinical problem as well.
A particular diagnostic challenge concerns the rapid identification of S. aureus in blood cultures. Clinically significant blood cultures with the pathogen S. aureus must be distinguished from cultures that recover phlebotomy-associated skin contaminants. For staphylococci, this is a particularly vexing problem because of similarities between a common contaminant (coagulase-negative staphylococcal species) and the pathogen S. aureus. About 10% of all blood cultures are considered indicative of true bloodstream infection, compared to 30% or more that represent so-called pseudobacteremias due to non S. aureus contaminants like coagulase-negative staphylocci (CNS). Because empiric therapy is often determined by Gram stain morphology of organisms in a positive blood culture, and because S. aureus and CNS share similar Gram stain morphologies, patients are often unnecessarily treated with broad-spectrum antibiotics until confirmatory subculture is completed. On the other hand, delays in the identification of S. aureus and determination of MSSA or MRSA can be associated with inadequate antimicrobial administration and suboptimal clinical outcomes. Thus, rapid identification and differentiation for staphylococci is a critical task for diagnostic laboratories in order to facilitate timely directed therapy.
Here we show that our new molecular technique is capable of detecting S. aureus and it can reliably distinguish methicillin-sensitive (MSSA) from methicillin-resistant strains (MRSA) and either one from coagulase-negative staphylococci (CNS). Initial matrix experiments were successful when the bacteria were admixed in packed red cells and blood culture broth media, implying the method's potential as a diagnostic test for bacteremia. Data also demonstrate the successful detection of Pseudomonas aeruginosa, another important pathogen (see Detection and discrimination of P. aeruginosa, Sup. Material). With additional optimization, our method has potential to provide diagnostic results with a limit of detection and turn-around-time that is both clinically relevant and competitive with currently available molecular methods.
Results and Discussion
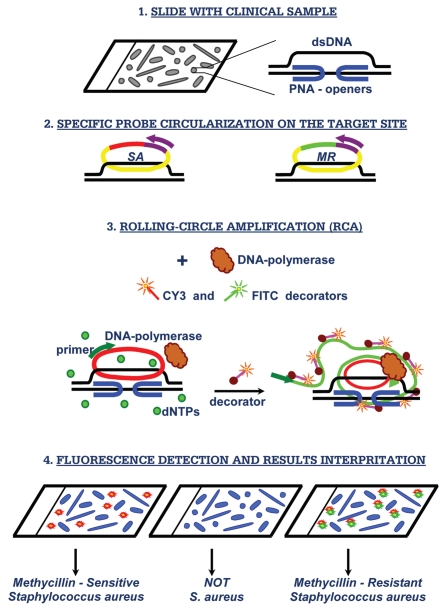
Our method is based on so-called PD-loop structure, which makes it possible to sequence-specifically assemble a small single-stranded DNA circle on dsDNA using peptide nucleic acid (PNA) openers (Fig. 1). PNAs are artificial nucleic acid analogs containing a neutral peptide-like backbone onto which nucleobases are grafted in a designed sequence.10 Most significantly, specially designed bis-PNA molecules consisting of two PNA oligomers connected by a flexible linker, spontaneously invade double-stranded DNA (dsDNA), binding to one of the two dsDNA strands with high affinity and sequence specificity owing to the simultaneous formation of Watson-Crick and Hoogsteen base pairs.11–13 As a result, these synthetic DNA mimics have a unique ability to locally pry open duplex DNA binding to one DNA strand and leaving the other strand accessible for hybridization with synthetic DNA probes.14 To uniquely define a specific fragment in genomic DNA, probes with a length of at least 15–20 nucleotides are required. To this end, we used two nested PNA probes. The binding sites for each of two bis-PNA openers are short (7–10 bp-long) homopurine-homopyrimidine tracts that can be separated by an arbitrary sequence of nucleobases up to 10 bp.14
Figure 1.
PNA-based diagnostic scheme. Major steps of the proposed assay for in situ detection of S. aureus and its methicillin-resistant strains.
Next, a highly specific assembly of the circular probe occurs on the DNA strand displaced due to PNA binding to the other strand. The circularization reaction is performed by the DNA ligase, which provides robust distinction between single-nucleotide variants under standard reaction conditions. The circularization reaction is very specific since it can only occur if both termini of the circularizable probe hybridize correctly to the target sequence, while leaving non-hybridized oligonucleotide probes linear.15 A two-component probe greatly improves selectivity of nucleic acid recognition in comparison to a single one (e.g., primers in PCR); as 15–20 nucleotides length duplex hybrids are too stable to be sensitive to a single-base mispairing. Since only chosen sites are made accessible by PNA openers and the rest of DNA remains predominantly in duplex form and therefore inaccessible for mismatch hybridization, the circular probe assembly is exceedingly sequence specific16,17 (see Sequence specificity of detection, Sup. Material). The region of the circularizable probe that does not participate in target DNA hybridization contains generic primer sequence for amplification and a tag sequence for fluorescent decorator probe. In our design, the circularized probe becomes topologically linked to the target DNA site.12,18
To generate a strong signal from individual circularized probes, the rolling-circle amplification RCA reaction is performed using the circular probe-specific primer and phi29 DNA polymerase. The generated long repetitive single-stranded DNA product is multiply-labeled by fluorophore-tagged decorator probes. The reaction product remains bound to its target site and provides localized fluorescent signal that is easily visualized using a standard fluorescence microscope. The multiplexing is obviously inherent and based on the unique properties of circularizable probes to find their specific targets, circularize and become amplifiable targets for a RCA. Monitoring of individual signals from different circles offers an opportunity for simultaneous detection of different signature sites using spectral multiplexing. The linear amplification process with a single primer, and the low concentration of the padlock probes, diminishes the problem of unwanted oligonucleotide interactions that regular multiplex PCR tend to suffer from. Others have demonstrated that more than 30 circularizable probes can be included in multiplex assay without specificity problem.19 In this report we explore this opportunity by using two different probe sets: one specific for any S. aureus and the other specific to methicillin-resistant strains (see schematics in Fig. 1).
Resistance to methicillin and other β-lactam antibiotics is conferred by the mecA gene. mecA is situated on a mobile genetic element known as the Staphylococcal Cassette Chromosome mec (SCCmec). To date, five SCCmec types have been distinguished, and several variants of these SCCmec types have been described.20 In our proof-of-principle experiments for detection and identification of MRSA versus MSSA we used three arbitrarily chosen S. aureus- specific PD-loop sites to detect S. aureus species (SA-1: AAA GAA AAA GCA ACA AGA GGA A, SA-2: AGA GGA AGC AGA GCG CAA GGG AAA, SA-3: AAA AGA AGA AAG ATT CAG AGG AAG) and two PD-loop sites located within mecA gene for penicillin-binding protein, which is specific for all MRSA strains (MR-1: AAG GAG GAT ATT GAT GAA AAA GA and MR-2: GGA AGA AAA ATA TTA TTT CCA AAG AAA A; PNA binding sites are underlined; see also Sup. Table 1).
In order to evaluate the diagnostic potential of the proposed method, we performed limited matrix experiments that included some components of human peripheral blood cultures. S. aureus cells were resuspended in blood culture media to which was added an aliquot of packed red blood cells. The bacteria used in these experiments were strains commercially available through the American Type Culture Collection (ATCC) and obtained from AdvanDx, Inc., (Woburn, MA).
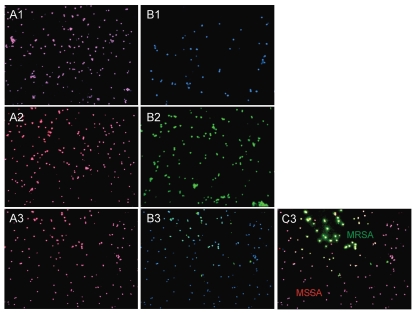
Specifically, two series of probes SA and MR were applied to aliquots of the admixed media that were spiked with (1) methicillin-sensitive strains (MSSA), (2) an Mu3 methicillin-resistant strain (MRSA) and (3) both MSSA and MRSA strains together. Figure 2 demonstrates that both series of probes work with similar efficiency (data with SA-3 and MR-1 are shown) and no signal was observed when probes specific to the mecA gene were applied to the MSSA strain. The obtained data suggest that this general approach can be developed for in situ recognition of drug-resistant pathogens based on short genomic signature sites located in known genetic markers of drug resistance.
Figure 2.
Detection discrimination of methicillin-sensitive (MSSA) versus methicillin-resistant (MRSA) strains. Images of S. aureus bacterial cells observed by fluorescent microscopy in experiments performed with simulated matrix (packed red cells and blood culture media) spiked with S. aureus bacterial cells. A combination of two probes (SA-3 and MR-1) was applied to simulated matrix specimens spiked with: the regular MSS A strain (A1 and B1); the Mu3 MRSA strain (A2 and B2) and their mixture (A3–C3). The fluorescent signals were acquired separately using three filter sets: A1–A3 present a superposition of two separate images, with DAPI and CY3; B1–B3 present a superposition of two separate images, with DAPI and FITC; C3 presents a superposition of three separate images, with DAPI, CY3 and FITC.
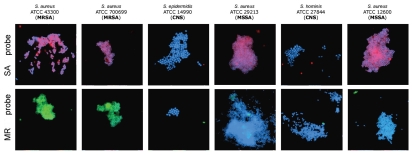
Also, we conducted experimenter-blinded tests using six de-identified, codified panels contained S. aureus (both MSSA and MRSA) and coagulase-negative staphylococcal species (CNS) (i.e., S. epidermidis and S. hominis that are closely related to and have similar microscopic morphology in common with S. aureus). Specimens were evaluated to determine if chosen sets of probes would cross-react with other staphylococcal species. Since the genomic sequences of most of the aforementioned organisms are not yet available, this has been an appropriate method to test for cross-reaction and validation of our approach.
Representative results obtained in these experiments are presented in Figure 3. The proposed method is capable of detecting S. aureus and reliably distinguishing methicillin-sensitive (MSSA) from methicillin-resistant (MRSA) strains in these proof-of-principle studies. The method can also discriminate S. aureus from coagulase-negative staphylococcal species. The data in Figure 3 show that our method is applicable to detection of S. aureus in an experimental matrix of packed red blood cells and blood culture media.
Figure 3.
Experimenter-blinded tests with de-identified, codified bacteria contained S. aureus (both MSSA and MRSA) and coagulase-negative staphylococcal species (CNS). Images of methicillin-sensitive S. aureus (MSSA), coagulase-negative staphylococcus (CNS) and methicillin-resistant S. aureus (MRSA) cells observed by fluorescent microscopy when probes specific to SA (MSSA/MRSA) or the MR (to mec cassette—MRSA only) were applied. The fluorescent signals were acquired separately using three filter sets (DAPI for DNA and Cy3 or FITC for the RCA product). Images are superposition of two images: (SA-probe) DAPI and CY3; (MR-probe) DAPI and FITC. Signals were pseudocolored in blue for DAPI, red for CY3 and green for FITC. SA-probes (PNA binding sites are underlined): SA-1: AAA GAA AAA GCA ACA AGA GGA A, SA-2: AGA GGA AGC AGA GCG CAA GGG AAA, SA-3: AAA AGA AGA AAG ATT CAG AGG AAG; MR-probes (within mecA gene for penicillin-binding protein): MR-1: AAG GAG GAT ATT GAT GAA AAA GA, MR-2: GGA AGA AAA ATA TTA TTT CCA AAG AAA A.
Ideally, our novel approach would be developed for direct assay of a peripheral whole blood sample from a bacteremic patient—if the limit of detection allows. Notably, PCR has been reported to have difficulties with whole blood specimen protocols, despite its inherent promise.21 Staphylococcal bacteremia has been estimated to have a low-end range of 1–30 CFU per ml of blood, this low concentration is sufficient for obtaining growth in blood culture bottles but below the sensitivity of a PCR assay. Therefore, a PCR diagnostic cannot replace blood culture in the microbiological workup of suspected Bloodstream Infections.
Preliminary trials (data not shown) using serial dilutions suggested that the limit of detection (LOD) for our current protocol for S. aureus suspended in packed red cells is 103 CFU per ml. This is by two orders of magnitude better than was achieved with convenient PNA-FISH—105 CFU directly from blood cultures.22 Therefore, as a practical matter, our assay should also be compatible with several proprietary routine blood culture media—to facilitate testing, if needed, after culture amplification of a patient sample. Results to date are encouraging to future larger efforts that should include refined in vitro matrix studies and clinical samples.
We plan to further improve the assay primarily by optimizing the rolling circle amplification step. The RCA has proven to be very powerful technique allowing for signal enhancement by thousands times depending on amplification time and as a result providing with the sensitivity of detection on the level of a single copy of the target DNA site per genome.15,23 Therefore, by increasing the duration of the rolling circle amplification step or by using multiply fluorescence-labeled decorator probes we will be able to achieve a clinically-relevant LOD with our approach.
Finally, in order to demonstrate that our method can be extrapolated to another important bacterial pathogen, we tested it against Pseudomonas aeruginosa.
P. aeruginosa is a major cause of morbidity and mortality due to healthcare-associated infections. It is an important nosocomial bloodstream pathogen, as well as a common cause of ventilator-associated pneumonia in intensive care unit patients. Since the outer membrane of P. aeruginosa is less permeable than that of other Gram-negative bacteria, it is intrinsically resistant to many antimicrobials. This outer membrane presents a potential technical challenge since our protocol depends on PNA and RCA molecules being able to easily enter bacterial cells. Based on genomic sequences of two P. aeruginosa strains that were available in Bacteria Genomes Database we chose three unique target sites specific for different gene regions. We were able to reliably distinguish P. aeruginosa from a mixture of P. aeruginosa and a different (Gram positive) bacillus, B. subtilis (see Detection and discrimination of P. aeruginosa, Sup. Material).
Our method combines a highly specific and sensitive recognition of signature sequences by PNA openers and circularizable probes, in conjunction with efficient local signal amplification by RCA. The method expands both the utility and resolving power of whole-cell FISH for the detection of microbes and the entire process is very compatible with routine clinical diagnostics. In contrast to traditional FISH methods but similar to PCR, this method provides robust distinction between family-, genus- and species-specific targets down to single-nucleotide variants.
To demonstrate this potential, we performed a series of experiments on mismatch discrimination (see Sequence specificity of detection, Sup. Material). Nearly all experiments showed no signal when mismatched probes (with mismatches in PNA or in the circularizable probe's binding site) were used. Only in one experimental design, when a single-nucleotide mismatch was located far from the ligation point at the circularizable probe binding site, we observed a discordant signal from about 20% of cells. Therefore, in case a single mismatch happens at fourth position or farther from the ligation point, discrimination may be insufficient under our standard conditions and a modification of the protocol may be necessary. The current protocol can be still be applied in case the potential signature site is carefully checked to avoid distant single-nucleotide replacements. Another possibility to enhance selectivity of detection consists in simultaneous targeting of two signature sites within the chosen bacterial genome. Furthermore, different sites can be detected using different fluorophores with resolvable emission bands. Such double-coincidence detection will eliminate the possibility of false positives.
Nonetheless, unlike PCR, the output of this assay is in a format familiar to the clinician who will get both the standard microscopy result (cell morphology) to confirm the presence and shape of the pathogen along with the drug-resistant signature determined by the proposed method. This novel method offers the advantages of a high-technology molecular assay in a format that is familiar, requires a minimum of training, and is relatively cost-effective.
We believe that our method presents a unique, user-friendly alternative to other genome-based methods for detection of drug resistant and/or otherwise important microbial pathogens.24–26 While a slide-based assay does not lend itself to high-throughput workflow, additional advantages inherent in the FISH format of genomic interrogation include facile on-demand specimen testing, correlation of morphology with signal detection and preservation of slides for future clinical correlation and quality assurance. Determining the potential use of this method for clinical diagnostics awaits further protocol optimization and the establishment of relevant performance characteristics.
Materials and Methods
Bacterial strains, culture condition and cell fixation.
In this study we used the following strains: MSSA (ATCC29213 and ATCC12600); MRSA (ATCC43300, ATCC700698 and ATCC700699) and CNS (S. epidermidis—ATCC14990 and S. hominis—ATCC27844). Strains were cultured overnight at 37°C in Brain Heart Infusion broth (ATCC#44).
During initial simulated matrix experiments, bacterial detection was performed after a pellet of corresponding bacterial cells was resuspended in blood culture media (REDOX 1® aerobic, 80 ml, Trek Diagnostic System, Cleveland, OH) to which was added one milliliter of expired packed human red blood cells (AB+, courtesy of Dr. Karen Quillen of Transfusion Medicine, Boston Medical Center), to achieve an approximate concentration of 108 colony-forming units/mL of bacteria. Then an aliquot of the experimental admixture was dropped on a clean polylysine microscope slide. The slides were fixed 15 min in freshly-prepared fixative (methanol/glacial acetic acid, 3:1) at room temperature and washed twice with new fixative. After fixation, the slide was washed twice with phosphate-buffered saline (PBS; pH 7.4), then dehydrated through an alcohol series (70%, 90%, 100%) for 2 min and thoroughly dried.
To these preparations, the different combinations of various probes for S. aureus were applied.
PNA openers, circularizable probes, RCA primers and decorator probes.
PNAs, circularizable oligonucleotides (ODNs), primers and decorator probes used in this study are summarized in Supplementary Table 1. All PNA samples were obtained as a gift from Dr. P.E. Nielsen (Copenhagen University) or PANAGENE (South Korea) (eg1, Lys and J denote a bis-PNA linker segment, the amino acid lysine and the nucleobase pseudoisocytosine, respectively). All primers and labeled probes with a phosphate group (p), Cy3 or FITC were supplied by MWG biotech. Circularizable probes carried a phosphate group at the 5′-end and sequences complementary to target DNA sites at the 3′- and 5′-ends. They also carried the sequence complementary to the RCA primer and an arbitrary sequence in the middle, which was designed to hybridize specific decorator probes to the RCA product and to discriminate bacterial strains from one another.
PD-loop assembly, ODN ligation and rolling circle amplification.
We dropped on each slide about 50 µl 10 mM Na phosphate buffer (pH 7.0) with 0.8 µM of each PNA. The slide was covered by 24 × 50 mm slip and sealed with rubber cement. Binding a pair of PNA openers to bacterial chromosomal DNA was carried out at 45°C for 4 hours. After completion of this step, the rubber cement was gently peeled off and the cover slip was removed. The slide was washed twice in 10 mM Na phosphate buffer (pH 7.0) to remove non-bound PNAs.
The circularizable probe was then added and the ligation reaction was performed by dropping on the slide 50 µl of ligation mixture containing 1× T4 ligase buffer with 5 u T4 DNA ligase (USB) and 2 µM of the circularizable probe. The slide was incubated under the cover slip at room temperature for 1 hour. Then the slide was washed twice in 2× SSC buffer to remove excess non-ligated ODN and washed once in buffer A (100 mM TrisHCl, 150 mM NaCl and 0.05% Tween20).
Next, the RCA reaction was performed by dropping on the slide 50 µl of the RCA reaction mixture containing 2 µM of primer, 10 u phi29 DNA polymerase (New England Biolabs), 200 µM dNTPs and 1× phi29 DNA polymerase buffer. In addition, the mixture contained 2 µM of fluorescently labeled decorator probe. Decorator probes carried fluorescent labels on 3′-ends and could not be extended by DNA polymerase. The slide was covered by the slip and sealed with rubber cement. The RCA reaction was performed at 37°C, for 2 hours in a humidifying chamber. Then the slide was washed twice in buffer A and once in 4XT buffer.
After a drop of counterstain DAPI (4,6-diamidino-2-phenylindole) was added, the coverslip was applied and the slide was allowed to stand at room temperature for 5 min, then the cover slip was secured with nail polish.
Signal detection.
Cells on the slides and intracellular amplicon were observed by using a fluorescence microscope Olympus BX-70 equipped with a cooled charge-coupled device camera. Fluorescence signals were captured separately using appropriate filter sets for DAPI, CY3 and FITC, respectively. Images were pseudo-colored, processed and merged using the CytoVision image software (Applied Imaging) and then stored as a digital file.
Acknowledgements
We thank Dr. Peter Nielsen for providing us with PNA oligomers, Mark Fiandaca for S. aureus bacterial strains and Dr. Karen Quillen for expired packed human red blood cells.
This work was supported by the Wallace C. Coulter Foundation to Maxim D. Frank-Kamenetskii and Nancy S. Miller and a NIH research grant (1R21RR025371-01) to Irina Smolina.
Supplementary Material
References
- 1.Mothershed EA, Whitney AM. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clinica Chimica Acta. 2006;363:206–220. doi: 10.1016/j.cccn.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 2.Procop GW. In situ hybridization for the detection of infectious agents. Clinical Microbiology Newsletter. 2002;24:121–125. [Google Scholar]
- 3.Mothershed M, Horn M, Daims H. Fluorescence in situ hybridization for the identification and characterization of prokaryotes. Curr Opinion Microbol. 2003;6:302–309. doi: 10.1016/s1369-5274(03)00054-7. [DOI] [PubMed] [Google Scholar]
- 4.Moter A, Gobel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods. 2000;41:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 5.Zwirglmaier K. Fluorescence in situ hybridisation (FISH)—the next generation. FEMS Microbiol Lett. 2005;246:151–158. doi: 10.1016/j.femsle.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Smolina IV, Lee C, Frank-Kamenetskii MD. Detection of low copy genomic DNA sequences in individual bacterial cells using PNA-assisted rolling circle amplification and fluorescence in situ hybridization. Appl Environ Microbiol. 2007;73:2324–2328. doi: 10.1128/AEM.02038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smolina IV, Kuhn H, Lee C, Frank-Kamenetskii MD. Fluorescence-based detection of short DNA sequences under non-denaturing conditions. Bioorg Med Chem. 2008;16:84–93. doi: 10.1016/j.bmc.2007.04.063. [DOI] [PubMed] [Google Scholar]
- 8.Beekmann SE, Diekema DJ, Chapin KC, Doern GV. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J Clin Microbiol. 2003;41:3119–3125. doi: 10.1128/JCM.41.7.3119-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbarth S, Samore MH. Antimicrobial resistance determinants and future control. Emerg Infect Diseases. 2005;11:794–801. doi: 10.3201/eid1106.050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 11.Egholm M, Christensen L, Dueholm KL, Buchardt O, Coull J, Nielsen PE. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demidov VV, Frank-Kamenetskii MD. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem Sci. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn H, Demidov VV, Frank-Kamenetskii MD, Nielsen PE. Kinetic sequence discrimination of cationic bis-PNAs upon targeting of double-stranded DNA. Nucleic Acids Res. 1998;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukanov NO, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc Natl Acad Sci USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson M. Lock and roll: single-molecule genotyping in situ using padlock probes and rolling-circle amplification. Histochem Cell Biol. 2006;126:159–164. doi: 10.1007/s00418-006-0213-2. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn H, Demidov VV, Frank-Kamenetskii MD. An earring for the double helix: Assembly of topological links comprising duplex DNA and a circular oligodeoxynucleotide. J Biomol Struct Dyn. 2000;11:221–225. doi: 10.1080/07391102.2000.10506625. [DOI] [PubMed] [Google Scholar]
- 17.Demidov VV, Kuhn H, Lavrentieva-Smolina IV, Frank-Kamenetskii MD. Peptide nucleic acid-assisted topological labeling of duplex DNA. Methods. 2001;23:123–131. doi: 10.1006/meth.2000.1113. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn H, Demidov VV, Frank-Kamenetskii MD. Topological links between duplex DNA and a circular DNA single strand. Angewandte Chemie Int Ed. 1999;38:1446–1449. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1446::AID-ANIE1446>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Baner J, Gyarmati P, Yacoub A, Hakhverdyan M, Stenberg J, et al. Microarray-based molecular detection of foot-and-mouth disease, vesicular stomatitis and swine vesicular disease viruses, using padlock probes. J Virol Methods. 2007;143:200–206. doi: 10.1016/j.jviromet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 21.Wellinghausen N, Siegel D, Gebert S, Winter J. Rapid detection of Staphylococcus aureus bacteremia and methicillin resistance by real-time PCR in whole blood samples. Eur J Clin Microbiol Infect Diseases. 2009;28:1001–1005. doi: 10.1007/s10096-009-0723-7. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira K, Procop GW, Wilson D, Coull J, Stender H. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J Clin Microbiol. 2002;40:247–251. doi: 10.1128/JCM.40.1.247-251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvius J, Melin J, Göransson J, Stenberg J, Fredriksson S, Gonzalez-Rey C, et al. Digital quantification using amplified single-molecule detection. Nature Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 24.Sundsfjord A, Simonsen GS, Haldosen BC, Haaheim H, Hjelmevoll SO, et al. Genetic methods for detection of antimicrobial resistance. APMIS. 2004;112:815–837. doi: 10.1111/j.1600-0463.2004.apm11211-1208.x. [DOI] [PubMed] [Google Scholar]
- 25.Mothershed EA, Whitney AM. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clinica Chimica Acta. 2006;363:206–220. doi: 10.1016/j.cccn.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 26.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.