Abstract
The traditional view of bacterial cells as non-compartmentalized, which is based on the lack of membrane-engulfed organelles, is currently being reassessed. Many studies in recent years led to the realization that bacteria have an intricate internal organization that is vital for various cellular processes. Specifically, various machineries were shown to localize to the poles of rod-shaped bacteria. We have recently shown that the control center of the PTS system, which governs carbon uptake and metabolism, localizes to the poles of E. coli cells. Notably, the machinery that controls bacterial taxis along chemical gradients (chemotaxis) has a similar localization pattern. The fact that the two systems need to communicate in order to generate an optimal metabolic response suggests that their similar spatial organization is not a coincidence. Rather, due to their special characteristics, the poles may function as hubs for signaling systems to allow for efficient crosstalk between different pathways in order to improve coordination of their actions.
The regulatory mechanisms that underlie the spatial and temporal organization of microbial cells are largely unknown. Thus far, these mechanisms were believed to rely on embedded features of the localized proteins. In another study, we have recently shown that mRNAs are capable of migrating to particular domains in the bacterial cell where their protein products are required. In contrast to the view that transcription and translation are coupled in bacteria, localization of bacterial transcripts may occur in a translation-independent manner. Hence, it seems that the mechanistic basis for separating transcription and translation is more primitive than assumed up until now. We propose that bacteria synthesize proteins either by a transcription-translation coupled mechanism or by transporting mRNAs away from the transcription apparatus. Obviously, eukaryotic cells rely on the latter mechanism. Hence, the capacity of prokaryotic cells to adopt the division between transcription and translation was a crucial step in the evolution of nucleus-containing cells from the prokaryotic origin. Summarily, the line that separates cells with nucleus and cells without is fading, leading to the realization that bacteria are suitable model organisms for studying universal mechanisms that underlie spatial regulation of cellular processes.
Key words: bacterial cell, subcellular organization, protein localization, PTS, mRNA targeting
The bacterial cell was viewed for many years as a non-compartmentalized vessel containing macromolecules that are distributed quite randomly. In recent years, this view is being re-evaluated, mostly due to advances in imaging capabilities that enable better visualization of such tiny cells. Recent studies have documented localization of proteins, DNA and other molecules, such as phospholipids, to specific sub-cellular domains within microbial cells.1 Consequently, it has become clear that bacteria have a highly ordered spatial organization, and that unique functions are being carried out in inner micro-compartments. Moreover, evidence for the dynamic nature of the internal arrangement of various bacterial cells has been obtained.2 Still, the regulatory mechanisms that underlie the spatial and temporal organization of microbial cells are largely unknown. Here, I highlight our two recent papers. The first adds the PTS, a central bacterial system that determines hierarchal utilization of carbon sources, to the list of localized proteins. The results presented in this paper illustrate the spatial basis for the crosstalk between the PTS and other signaling systems, and demonstrate that signal transduction in bacteria involves dynamic localization of proteins. The second paper proposes mRNA targeting as a mechanism for protein localization in bacteria.
The Sub-Cellular Distribution of Bacterial Proteins Is Not Random
Bacterial cells that undergo asymmetric division inevitably possess mechanisms that coordinate non-random distribution of proteins. For example, the ability of Caulobacter to generate dissimilar progeny cells at each division, which have either a polarly placed flagellum or a stalk, can be partially explained by the well-organized deployment of the chromosomal origin complex.3 Still, the complexity and accuracy of the spatial and temporal events that control this process, which involve targeting of global regulators to different cellular domains, are remarkable for an organism with such a small chromosome.2 Another example is highlighted by the intricate spatial and temporal events that allow asymmetric division and spore formation by Bacillus subtilis cells.4
More surprising is the level of spatial organization in bacterial cells that undergo symmetric division, such as E. coli. The cell division mechanism itself, extensively studied in this organism, reveals some strategies that E. coli can employ to orchestrate spatial and temporal aspects of cellular processes. Thus, the cell division ring-forming protein FtsZ, a tubulin-like protein that drives cytokinesis, is guided to midcell by proteins that inhibit its polymerization; due to the rapid pole-to-pole oscillation of the inhibitory proteins, the cell center has the lowest inhibitor concentration, allowing FtsZ assembly there.2
The localization patterns that have been observed thus far in bacterial cells are quite variable. In addition to rapid pole-to-pole oscillation and formation of rings at midcell, bacterial proteins were documented to form dynamic helices along the cell, foci on the cell surface, or clusters at specific intracellular sites. The dynamic internal architecture facilitates behaviors as diverse as symmetric and asymmetric division, motility, morphological differentiation, assembly into multicellular communities, and interactions with animal and plant hosts
Spatial Organization of Bacterial Signaling Systems
The poles of rod-shaped cells are emerging as important sites for the localization of cellular machineries.5,6 The list of proteins that localize to the cell poles is growing rapidly, among them the components of the phosphotransferase (PTS) and the chemotaxis systems that play key roles in governing cell metabolism. The PTS enables hierarchal uptake of carbon sources and appropriate adjustment of cell metabolism via the control of global regulatory systems, such as catabolite repression and inducer exclusion.7 The chemotaxis system enables navigation of bacterial cells along gradients, with carbon source gradients having a major impact.8,9 Together, these systems can be regarded as a microbrain or as the bacterial metabolic nervous system. Obviously, the two systems need to communicate and indeed interaction between the general PTS protein EI and the chemotaxis histidine kinase CheA was suggested to occur.10 How is the information on nutrients availability, which is obtained by receptors of two sensory systems, integrated to generate an optimal metabolic response? Based on recent evidence discussed below, the basis for this crosstalk appears to be the spatial organization of the two signaling systems.
The chemotaxis receptors were shown to cluster at the poles and to form a polar complex with the different Che proteins. Intriguingly, the flagellar motors, whose rotation is controlled by this complex, are distributed around the E. coli cell. The information on environmental signals is communicated by the small CheY protein that shuttles between the polar complex, where it is phosphorylated, and the flagellar motors.11 Why has the cell evolved a polarly localized complex to control organelles that are scattered around the cell periphery? In a study that we recently published in the EMBO Journal,12 we showed that the control center of the PTS system, which consists of the general PTS proteins, EI and HPr, also localizes to the cell poles. The different proteins that are regulated by this polar center via HPr-mediated phosphorylation localize around the cell circumference (sugar permeases) or in the cytoplasm (e.g., IIAglc). The small HPr protein is released from the poles upon sugar stimulation to execute its tasks in the different cellular sub-domains.12 Hence the PTS and the chemotaxis systems utilize a similar strategy; that is, their command centers cluster near the poles, whereas small proteins, which act as messengers, shuttle between the poles and other cellular domains, where they execute their roles. It seems reasonable that localization of the two systems near the poles should facilitate communication between their components and improve the coordination of their actions. Such a strategy seems suitable for the intracellular environment of exponentially growing E. coli cells, which are crowded with macromolecules, mostly proteins, at high concentrations13 that apparently limit diffusion rates and create a need for colocalization of pathways. Hence, I would like to hypothesize that the poles have been selected during the evolution, due to their special features, be it composition, geometry or structure, as hubs for sensory systems. This organization facilitates interactions between proximal clusters to allow for efficient crosstalk between signaling pathways, the aim being to produce an optimal cellular response.
What are the mechanisms that direct assembly of sensory complexes at the poles? For the chemotaxis receptors, clustering and positioning were suggested to rely on stochastic self-assembly.14 In addition, involvement of the general protein translocation machinery (Sec) in their cellular distribution15 and in assembling them into the cytoplasmic membrane16 was suggested. The cues that recruit the PTS components to the poles are utterly unknown. Furthermore, it remains to be studied whether recruitment of the chemotaxis and PTS systems to the poles is interdependent.
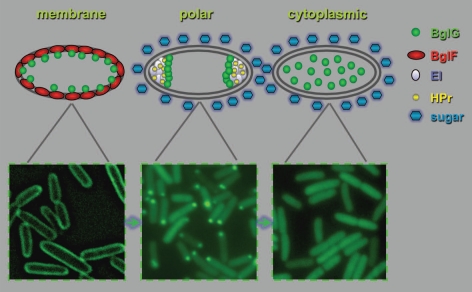
Our recent study also unveiled the spatial basis for the crosstalk between the PTS and other regulatory systems. We showed that localization of the BglG transcription factor, which enables expression of a PTS sugar utilization operon, bgl, is determined by its consecutive interaction with different PTS components, i.e., with the PTS sugar permease BglF, which localizes around the cell circumference, and with the general PTS proteins at the poles.12,17 Hence, upon sugar stimulation, BglG migrates from the cell periphery to the cytoplasm through the poles (Fig. 1). It has been known for quite some time that the activity of BglG homologs depends on the general PTS proteins. In Gram-positive bacteria, activation of BglG-like factors involves their phosphorylation by HPr,7 but in E. coli this activation is phosphorylation-independent.18 Indeed, we have now shown that the interaction of BglG with the different PTS components, which determines its subcellular localization, does not depend on the phosphorylation state of the different players.12 Hence, the PTS components control bgl operon expression by ushering the BglG transcription factor between the different cellular compartments in response to environmental signals. These results clearly indicate that signal transduction in bacteria involves dynamic localization of proteins.
Figure 1.
Spatial and temporal localization of a bacterial regulatory protein in response to environmental stimuli. Following the addition of a stimulating sugar, the BglG transcription factor migrates from the membrane periphery, where is associates with the BglF sensor (left), through the cell poles, where it interacts with the general PTS proteins (EI and HP r , middle), to the cytoplasm, where it activates transcription of the bgl operon (right). Shown are fluorescence microscopy images of cells expressing BglG-GFP. The images were obtained in independent experiments and are shown for illustration only.
How Are Bacterial Proteins Targeted to Their Destination?
There are at least three possible mechanisms to explain deliberate localization of proteins within the bacterial cell. (1) Diffusion and capture. If proteins can diffuse rapidly and freely throughout the cell, then they will eventually encounter other proteins to which they specifically adhere. Whether this encounter is persistent or transient would depend on the protein-protein binding energetics. Similarly, proteins can encounter cues other than proteins, such as geometric cues or certain phospholipids.1 (2) Active protein targeting systems. Bacteria produce homologs of eukaryotic proteins that form filamentous structures, such as actin and tubulin. It is tempting to speculate that these proteins, which form the bacterial cytoskeleton,19 are involved in targeting proteins to their sub-cellular destinations. (3) Targeting of mRNA transcripts to particular positions where their future protein products are required. The advantages of localizing proteins by targeting their mRNA transcripts include (i) cost effectiveness, that is, instead of moving each protein molecule individually, every localized transcript enables many rounds of protein synthesis at the final destination, (ii) restriction of proteins to their site of operation, and prevention of their appearance in sub-compartments where their effects might be harmful, and (iii) facilitation of assembly of protein complexes due to the production of high local concentration of their constituents.
Localization of Bacterial Proteins via mRNA Targeting
Examples of mRNA targeting have been obtained in recent years for different types of eukaryotic cells.20–22 For example, localization of mRNAs in embryos plays an important role in the formation of morphogens gradients and the asymmetric distribution of cell fate determinants.23 However, protein localization via mRNA targeting has been assumed to occur only in eukaryotes, in which transcription and protein synthesis take place in different compartments. Similar mechanisms have not been considered in bacteria due to the alleged coupling between transcription and translation. In line with that, several bacterial mRNAs were shown to localize near the chromosomal loci from which they are expressed.24
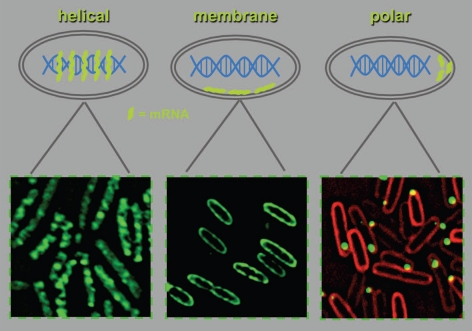
In a recent publication in Science,25 we showed that, contrary to the general thinking in the field, bacterial mRNAs are capable of migrating to particular positions in the cell, where their future protein products are required.25 Our results were based on imaging of live and fixed cells and were validated by a biochemical approach. Thus far, we followed the localization of bacterial mRNAs that code for proteins that localize to the cytoplasm, the inner membrane, or the poles (Fig. 2). In all these cases, localization of the mRNAs correlated with subsequent localization of their cognate protein products. However, we have recently found that proteins can be targeted to the poles, whereas their mRNA demonstrates a typical cytoplasmic distribution (Nevo-Dinur and Amster-Choder, unpublished observations). The implications of this result are: (1) Mechanisms for targeting of proteins, following their translation, operate in bacteria. This is in line with the old dogma of that assumed that protein translocation occurs subsequent to the coupled transcription-translation. (2) mRNA targeting is tied also to processes other than localized translation. One possibility is targeting of mRNA transcripts to degradation, which is in agreement with the documented helical and membrane localization of the E. coli mRNA degradosome components.26,27
Figure 2.
Patterns of mRNA localization in E. coli. Thus far, we have observed three pattern of mRNA localization: helical distribution of cat mRNA (left), membrane localization of bglF mRNA (middle), and polar localization of bglG mRNA (right). Shown are fluorescence microscopy images of cells expressing the coat protein of MS2 bacteriophage fused to GFP (MS2-GFP) and transcripts that contain six binding sites for MS2-GFP. Green, MS2-GFP; red, FM4-64membrane stain.
Similarly unexpected was our finding that, in contrast to the widely held view, localization of bacterial transcripts may occur in a translation-independent manner. This conclusion emerged from a set of experiments in which we uncoupled transcription and translation by different approaches and demonstrated that targeting is a property of the mRNA.25 Therefore, the coupling between transcription and translation, a hallmark of the prokaryotic kingdom, is not an absolute requirement, although de facto the two processes can co-occur in bacterial cells. An important and far-reaching conclusion is that the mechanistic basis for separating transcription and translation has developed in the primitive bacterial cells and did not co-evolve with the nucleus of the eukaryotic cell.
In two cases we mapped the regions that target transcripts to specific cellular domains, i.e., the ‘zip code’-containing sequences. The cis-acting sequence for membrane targeting of bglF transcripts was mapped to the sequence that codes for the first two transmembrane helices of the encoded protein. In the case of bglG, the region that encodes the N′-terminal RNA-binding domain was shown to be sufficient for polar localization of the transcripts. Remarkably, we found a hierarchy in zip codes localization capacity: a membrane-encoding sequence is dominant to a hydrophilic-encoding sequence in determining transcript localization. Intriguingly, the targeting signals seem to be conserved across the eukaryotic-prokaryotic divide. We have shown that transcripts encoding a eukaryotic transmembrane protein, Rhomboid 1 from Drosophila melanogaster, still localized to the membrane when expressed in E. coli.25 It is worth mentioning that Errington and co-workers have shown that a protein targeting signal for division sites in conserved between prokaryotes and eukaryotes.28 Hence, universal mechanisms for targeting both mRNAs and proteins are ancient and have apparently evolved in bacteria.
How do the mRNA transcripts localize to the different sub-cellular domains? In eukaryotes, mRNA localization was suggested to occur by facilitated diffusion in the cytoplasm or by active transport along cytoskeletal filaments.29,30 The average short half-life of bacterial proteins, 3–8 min,31 together with the short generation time of bacterial cells suggest that the involvement of active mechanisms for mRNA targeting should be seriously considered. In our study, we measured the half-life of bglF transcripts encoding an integral membrane protein, which turned out to be less than 2 min,25 reinforcing the need for an active transport mechanism.
Finally, we could show that targeting of transcripts that code for components of a signaling system is in tight correlation with the requirements for complex formation. Specifically, the mRNA that encodes the BglG transcription factor is targeted to the membrane only when coexpressed with the gene that encodes the BglF sugar-sensor, that is, when the two genes that reside in the same operon are transcribed as two cistrons of the same transcript.25 Notably, BglG forms a pre-complex with BglF at the membrane in the absence of the stimulating sugar.17 It is very tempting to suggest that targeting of operon transcripts, which usually code for protein products that operate in the same pathway, is a universal mechanism that facilitates assembly of these products into complexes. Notably, when bglG is transcribed without bglF, its transcripts localize to the poles,25 where the BglG protein forms a complex with the general PTS proteins after it is released from the membrane pre-complex upon sugar stimulation.12 To the best of my knowledge, this is the first demonstration of changes in mRNA localization depending on changes in the site of complex formation in response to environmental stimuli in any organism.
Summarily, our study establishes for the first time the existence of translationindependent mechanisms for targeting bacterial mRNAs to the subcellular regions where their protein products are subsequently detected. Hence, a bacterium like E. coli can now be viewed as a simple model organism for studying spatial regulation of cellular processes and for elucidating the mechanisms that underlie cellular architecture in higher organisms.
Acknowledgments
I thank all past and present members of my lab for fruitful discussions over the years. Work in my lab on protein and RNA localization is supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities.
Addendum to: Lopian L, Elisha Y, Nussbaum-Shochat A, Amster-Choder O. Spatial and temporal organization of the E. coli PTS components. EMBO J. 2010;29:3630–3645. doi: 10.1038/emboj.2010.240. and Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691.
References
- 1.Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Janakiraman A, Goldberg MB. Recent advances on the development of bacterial poles. Trends Microbiol. 2004;12:518–525. doi: 10.1016/j.tim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Ebersbach G, Jacobs-Wagner C. Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol. 2007;15:101–108. doi: 10.1016/j.tim.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 9.Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- 10.Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Lopian L, Elisha Y, Nussbaum-Shochat A, Amster-Choder O. Spatial and temporal organization of the E. coli PTS components. EMBO J. 2010;29:3630–3645. doi: 10.1038/emboj.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 14.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiomi D, Yoshimoto M, Homma M, Kawagishi I. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol Microbiol. 2006;60:894–906. doi: 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert JF, Overhoff B, Manson MD, Boos W. The Tsr chemosensory transducer of Escherichia coli assembles into the cytoplasmic membrane via a SecA-dependent process. J Biol Chem. 1988;263:16652–16660. [PubMed] [Google Scholar]
- 17.Lopian L, Nussbaum-Shochat A, O'Day-Kerstein K, Wright A, Amster-Choder O. The BglF sensor recruits the BglG transcription regulator to the membrane and releases it on stimulation. Proc Natl Acad Sci USA. 2003;100:7099–7104. doi: 10.1073/pnas.1037608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raveh H, Lopian L, Nussbaum-Shochat A, Wright A, Amster-Choder O. Modulation of transcription anti-termination in the bgl operon of Escherichia coli by the PTS. Proc Natl Acad Sci USA. 2009;106:13523–13528. doi: 10.1073/pnas.0902559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 20.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 21.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 22.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du TG, Schmid M, Jansen RP. Why cells move messages: the biological functions of mRNA localization. Semin Cell Dev Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 26.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA. 2007;104:1667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Palacios IM. How does an mRNA find its way? Intracellular localisation of transcripts. Semin Cell Dev Biol. 2007;18:163–170. doi: 10.1016/j.semcdb.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]