Abstract
Melanoma displays frequent activation of RAS/RAF/MAPK and PI3K/AKT signaling pathways as well as inactivation of CDKN2A (INK4a/ARF) and PTEN tumor suppressors via genetic and epigenetic alterations. Pathogenetic roles of these melanoma-prone mutations and their genetic interactions have been established in genetically engineered mouse models. Here, we catalog frequent genetic alterations observed in human melanomas and describe mouse models of melanoma initiation and progression, including our recent study that investigated the genetic interactions of RAS activation and PTEN loss in a CDKN2A (INK4a/ARF) null melanoma prone genetic background. We showed that loss of PTEN cooperates with HRAS activation, leading to increased development of melanoma and emergence of metastasis. Moreover, we observed that RNA i-mediated PTEN inactivation in RAS-driven melanomas enhanced migration and invasion with concomitant downregulation of E-cadherin, the major regulator of epithelial and mesenchymal transition, and enhanced AKT2 phosphorylation, which has been previously linked to invasion and metastasis of several cancer types, including breast and ovary. These data show that activated RAS cooperates with PTEN loss in melanoma genesis and progression.
Key words: melanoma, PTEN, RAS, E-cadherin, mouse model, progression, AKT2
Melanoma is the most aggressive form of skin cancer. Patients with localized melanomas can be cured by surgical excision, but only 14% of patients with metastatic melanoma survive five years due to lack of effective therapies,1 thus underlying the importance of identifying genetic events that drive melanoma initiation and progression.
The small G-protein RAS family is frequently mutated in human solid tumors, including melanoma. Various mouse models that express oncogenic H-, K- or N-RAS specifically in melanocytes have supported the pathogenetic role of RAS proteins in melanoma. Most of these RAS-driven mouse models require additional genetic alterations and/or UV irradiation for melanoma pathogenesis, thus supporting the importance of delineating genetic interactions and synergy between RAS signaling and these lesions on melanomagenesis.
Genetic Alterations Observed in Melanoma
Genetic analyses of melanoma specimens have identified several aberrantly regulated pathways, including INK4a-CDK4/6-RB, ARF-p53-MDM2, RAS-RAF-MAPK, PTEN-PI3K-AKT and aMSH-MC1R-cAMP-MITF via genetic, genomic or epigenetic mechanisms.2 The top five genes mutated in malignant melanomas, as identified by the Sanger Institute COSMIC (Catalogue Of Somatic Mutations In Cancer)3 website (www.sanger.ac.uk/cosmic), are shown in Table 1.
Table 1.
Top 5 genes mutated in malignant melanoma
| Gene name | Sample number | Positive samples | Percent mutated |
| BRAF | 5492 | 2269 | 41% |
| NRAS | 3461 | 686 | 20% |
| CDKN2A | 1056 | 281 | 27% |
| KIT | 1098 | 89 | 8% |
| PTEN | 544 | 85 | 16% |
The mutation data was obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer Web site, (http://www.sanger.ac.uk/cosmic).30
The high frequency of BRAF mutations4 and the presence of the less common yet reciprocal NRAS mutations5 support the importance of the RAS-RAF-MAPK pathway in melanoma. Among the three closely related RAS proteins, NRAS is the most commonly mutated RAS family member (20%), but mutations on KRAS (2%) and HRAS (1%) have also been observed in melanoma specimens.3 The activating mutations on codon 12, 13 or 61 of H-, N- and K-RAS proteins result in constitutive RAS signaling, which activates RAF kinase family, PI3K, RalGDS and phospholipase Cϵ.6 The RAF kinase family, consisting of A-, B- and C-RAF, is a serine/threonine kinase that activates the MAPK pathway. BRAF is the most frequently mutated gene in human melanocytic neoplasms, with frequencies of 82% in benign nevi and 50% in melanomas.4,7,8
Activation of the PI3K-AKT pathway is often achieved by activation of receptor tyrosine kinases, RAS mutation,9,10 inactivation of its negative regulator PTEN (phosphatase and tensin homologue deleted on chromosome 10) or AKT activation.8 PTEN resides on chromosome 10q23-24, the frequently deleted region in melanoma.11,12 Allelic loss or mutation of PTEN has been observed in 5–15% of uncultured melanoma specimens and metastasis, 17% of short-term melanoma cultures, and 30–40% of established melanoma cell lines.5,13–15 PTEN functions as a dual lipid/protein phosphatase and hydrolyzes 3-phosphate on phosphatidylinositol-(3,4,5)-trisphosphate that promotes survival, growth and proliferation, leading to inhibition of AKT activation. When activated, AKT stimulates cell cycle progression, survival, metabolism and migration through phosphorylation of many physiological substrates. Increased AKT3 expression has been reported to accompany DNA copy gain,16,17 whereas amplification or gain of the AKT1 or AKT2 locus has not been observed in melanoma. A rare activating mutation (E17K) was identified on AKT3 in 2 out of 65 melanoma cell lines and 2 out of 137 melanoma specimens as well as on AKT1 in one melanoma sample in the same study. E17K mutation was not observed on AKT2.18
Mouse Models of Melanoma Initiation and Progression
The pathogenetic roles of the genetic alterations identified in human melanoma specimens have been successfully elucidated in genetically engineered mouse models. Why the NRAS mutation prevails in melanoma is not yet clear; however, expression of all three activated RAS isoforms predisposes to melanoma. The melanocyte-directed HRASV12G transgene, combined with inactivating mutations in Ink4a, Arf or p53, promotes development of non-metastatic melanomas.19–21 In contrast, an activated NRASQ61K transgene expression in melanocytic lineages in Ink4a/Arf-deficient mice drives cutaneous melanomas, as well as metastatic spread to lymph nodes and other distal sites (e.g., lung and liver) in one-third of the cases.22 KRASV12G expression in melanocytes utilizing Cre-recombinase/LoxP system (Tyr::CreERT2) has been shown to lead to melanoma formation with median latency of 4 months without additional genetic manipulation or exposure to UV light.23 However, somatic loss of Ink4a p16 and/or p53 in melanocytes24 showed synergism with KRASV12G expression and enhanced melanoma formation. These genetic studies have clearly established the tumorigenic role of activated RAS signaling in melanoma and revealed similarities and dissimilarities of signaling driven by RAS isoforms.
Consistent with the frequent observation of BRAF V600E mutations in benign nevi,4,7 BRAFV600E expression in melanocytes induced a nevoid hyperpigmentation phenotype in mice25–27 with rare progression to melanoma with25 or without26 accompanying Ink4a p16 loss. Similar to observations made in RAS-driven models, oncogenic BRAF expression in melanocytes synergistically interacts with Ink4a/Arf loss and p53 loss25 or Ink4a p16 loss.26 These compound models showed enhanced melanoma formation with decreased latency and increased penetrance.
Contribution of PTEN inactivation in melanoma tumorigenesis has been shown previously. Although neither Pten+/− nor Ink/Arf−/− mice developed melanomas, Pten heterozygosity in Ink4a/Arf null mice caused melanoma formation in a small number of mice (3/46; 6.5%) at 28–31 weeks,28 supporting the cooperative interaction of dual inactivation of PTEN and INK4a/ARF. Moreover, loss of PTEN expression in mouse melanocytes synergized with activated BRAF, leading to robust melanoma formation and development of metastases,27 supporting genetic interactions of the RAS/RAF/MAPK and PTEN/PI3K/AKT pathways in melanoma progression.
Genetic Interaction of HRAS Activation and PTEN Loss in Melanoma Initiation and Progression
Recently, we addressed the genetic interactions of three melanoma-prone genetic elements, namely RAS activation, PTEN loss and INK4a/ARF deficiency, utilizing genetically engineered mouse models. Our results showed evidence of cooperation among these genetic lesions, manifested as accelerated melanoma development and enhanced migration of melanoma cells. Specifically, inactivation of one copy of Pten in mice with melanocyte-directed oncogenic HRASV12G expression and loss of Ink4a/Arf (hereafter RAS-Ink4a/Arf19) led to an earlier onset of melanoma and decreased overall melanoma-free survival (29.6 versus 18.9 weeks in RAS-Ink/Arf mice with Pten+/+ versus Pten+/− genotypes, respectively). Pten+/− Ink/Arf−/− mice without RAS expression did not develop melanoma and succumbed to non-melanoma-related death (median survival of 19.4 weeks). In the period prior to the appearance of non-melanoma tumors, 75% of the Pten+/− RAS-Ink4a/Arf mice developed melanoma compared with 35.7% of mice with wild-type Pten. Histopathologically, these primarily spindle cell melanomas are similar to those observed in the RAS-Ink4a/Arf model.19,29 Moreover, we also observed 2 cases of melanoma metastasis among the 21 tumor-bearing RAS-Ink4a/Arf mice heterozygous for Pten, which was never observed in RAS-Ink4a/Arf mice. At the molecular level, RNAi-mediated Pten inactivation in RAS-Ink/Arf melanoma cells led to E-cadherin downregulation and AKT2 phosphorylation, accompanying enhanced migration and invasion compared to control cells without Pten knock-down. Both the E-cadherin downregulation and the AKT2 activation have been previously shown to promote progression of various tumors.30–32 These data support a role for PTEN as a suppressor of melanoma progression. An inverse correlation of PTEN level to Breslow depth and melanoma progression has been consistently reported.33 Therefore, HRAS activation, loss of PTEN and loss of INK4a/ARF cooperate to drive the genesis and progression of melanoma. Similarly, cooperation of KRAS activation and PTEN loss have been reported in mouse models of lung34 and pancreatic cancer,35 and HRAS activation and PTEN loss have been reported in squamous cell carcinoma model.36
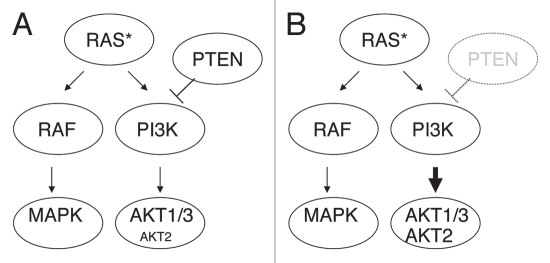
This synergy was unexpected since the activation of RAS and the loss of PTEN have been considered as functionally and genetically redundant events in melanoma, due to the common effects of NRAS and PTEN alterations leading to AKT activation. Moreover, the relative reciprocity of NRAS and PTEN alterations was previously proposed in melanoma specimens5 and cell lines.37 However, in human melanoma, we found that 14% (2/14) of the NRAS-mutated tumor samples harbor PTEN loss or mutation. Conversely, 17% (2/12) of the melanoma samples with PTEN loss or mutation also contain NRAS mutation (Chin L, unpublished data). Thus, concurrent RAS activation and PTEN loss occur in melanoma, and this subtype may have a higher tendency to metastasize considering the phenotype observed in the mouse model. Schematic model for the cooperative interactions of PTEN deficiency and RAS activation in melanoma is shown in Figure 1.
Figure 1.
Schematic model for the cooperative interactions of PTEN deficiency and RAS activation. (A) Activated RAS (RAS*) causes RAF/MAPK activation and PI3K/AKT activation, increasing pAKT1 and 3 preferentially. (B) PTEN inactivation and RAS activation cooperates to increase pAKT2 along with pAKT1 and pAKT3.
Questions that Remain to be Answered
The cooperation of MAPK and AKT pathways in melanoma tumorigenesis and progression is well supported by our study38 employing oncogenic HRAS and by Dankort et al.27 with activated BRAF combined with Pten loss. Considering the prevalence of activating NRAS mutations in melanoma, it is important to confirm whether oncogenic NRAS also synergizes with PTEN loss in melanoma tumorigenesis. Some biological differences exist among highly homologous RAS isoforms, including lipid raft localization of HRAS and NRAS, but not KRAS,39 and more potent activation of PI3K/AKT by NRAS and HRAS than by KRAS in fibroblasts and melanocytes.40,41 However, melanomas developed in similarly designed NRASQ61K and HRASV12G driven mouse models in Ink4a/Arf null background show indistinguishable histological features and similar molecular profiles (Kwong and Chin, unpublished data). In addition, we observed that RNAi-mediated PTEN loss in human melanoma cell line WM1366 harboring NRASQ61L mutation recapitulated the observation made in HRASV12G driven mouse melanoma cells, namely, increased invasion in a Boyden chamber assay accompanied by enhanced AKT2 activation. These data suggest the similarity for the role of HRAS and NRAS activation in melanoma tumorigenesis. Mutation of BRAF, a downstream target of RAS signaling, is frequently observed with concurrent PTEN loss. In one study, 3 out of 7 melanoma specimens with reduced PTEN protein expression showed BRAF mutation, while none of the 7 showed mutation on RAS isoforms.33 About half of the melanoma cell lines with BRAF mutation showed concurrent PTEN loss, whereas only 1 out of 11 melanoma cell lines with NRAS mutation showed PTEN loss.42 Collectively, all of these HRAS, NRAS and BRAF mutations share one feature, MAPK activation, which cooperatively interacts with PTEN loss in melanoma tumorigenesis.
At the molecular level, we reported increased pAKT2 and decreased E-cadherin protein levels upon PTEN inactivation in RAS-activated melanomas and cell lines. RAS proteins positively regulate AKT by directly binding to the p110 catalytic subunit of PI3K9 and by activating autocrine signals involving EGFR family ligands.10 In our study, mouse and human melanoma cells with activating RAS mutations showed AKT activation, and loss of PTEN expression in these cells increased phosphorylation of AKT2 isoform (Fig. 1). It is not clear whether signaling from activated RAS preferentially activates AKT1/3 and represses AKT2, which is activated by PTEN loss. In addition, AKT2 was reported as a downstream target of metabotropic glutamate receptor 1 (GRM1), of which expression in mouse melanocyte leads to melanoma formation, and loss of Akt2 in a Grm1-melanocytic cell line suppressed invasion in vitro.43 However, AKT2 mutation or genomic amplification/gain has not yet been observed in melanomas. On a related note, Stahl et al. showed that RNAi-mediated PTEN loss in melanocytes and WM35 melanoma cells containing BRAF V600E mutation led to increased pAKT3, which inhibited apoptosis.16 Therefore, it will be important to address whether AKT isoforms become differentially activated in specific genetic backgrounds and whether they play a distinct role during melanoma genesis and progression. Similar to that shown for breast cancer,32 low AKT2 activity at early stage may have growth advantage, and AKT2 activation at a later stage may promote progression. Moreover, AKT2 was reported to induce the miR-200 microRNA family, which in turn decreases E-cadherin expression.44 Therefore, it will be of interest to assess whether miR-200 is induced in our model and whether it is responsible for the observed downregulation of E-cadherin. Further systemic and comprehensive work is needed to identify the activity of each AKT isoform in melanoma tumorigenesis.
Acknowledgements
M. Kim is supported by Melanoma Research Foundation, the Bankhead Coley Melanoma Pre-Spore, and American Cancer Society Institutional Research Grant (#93-032-13). We thank Dr. Keiran Smalley for critical reading of this manuscript and Rasa Hamilton for editorial assistance.
Extra View to: Nogueira C, Kim KH, Sung H, Paraiso K, Dannenberg JH, Bosenberg M, et al. Cooperative interactions of PTEN deficiency and RAS activation in melanoma metastasis. Oncogene. 29:6222–6232. doi: 10.1038/onc.2010.349.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/14344
References
- 1.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 3.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Current protocols in human genetics/editorial board. Jonathan L Haines, et al. 2008;10:1. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz ME, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 7.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 8.Chudnovsky Y, Khavari PA, Adams AE. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115:813–824. doi: 10.1172/JCI24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Bardeesy N, Kim M, Xu J, Kim RS, Shen Q, Bosenberg MW, et al. Role of epidermal growth factor receptor signaling in RAS-driven melanoma. Mol Cell Biol. 2005;25:4176–4188. doi: 10.1128/MCB.25.10.4176-4188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163:1765–1770. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 13.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin AF, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 14.Birck A, Ahrenkiel V, Zeuthen J, Hou-Jensen K, Guldberg P. Mutation and allelic loss of the PTEN/MMAC1 gene in primary and metastatic melanoma biopsies. J Invest Dermatol. 2000;114:277–280. doi: 10.1046/j.1523-1747.2000.00877.x. [DOI] [PubMed] [Google Scholar]
- 15.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 17.Madhunapantula SV, Robertson GP. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res. 2009;22:400–419. doi: 10.1111/j.1755-148X.2009.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 22.Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res. 2005;65:4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 23.Milagre C, Dhomen N, Geyer FC, Hayward R, Lambros M, Reis-Filho JS, et al. A mouse model of melanoma driven by oncogenic KRAS. Cancer Res. 2010;70:5549–5557. doi: 10.1158/0008-5472.CAN-09-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monahan KB, Rozenberg GI, Krishnamurthy J, Johnson SM, Liu W, Bradford MK, et al. Somatic p16(INK4a) loss accelerates melanomagenesis. Oncogene. 2010;29:5809–5817. doi: 10.1038/onc.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel VK, Ibrahim N, Jiang G, Singhal M, Fee S, Flotte T, et al. Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice. Oncogene. 2009;28:2289–2298. doi: 10.1038/onc.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You MJ, Castrillon DH, Bastian BC, O'Hagan RC, Bosenberg MW, Parsons R, et al. Genetic analysis of Pten and Ink4a/Arf interactions in the suppression of tumorigenesis in mice. Proc Natl Acad Sci USA. 2002;99:1455–1460. doi: 10.1073/pnas.022632099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 30.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 31.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, et al. Overexpression of AKT2/protein kinase Bbeta leads to upregulation of beta1 integrins, increased invasion and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- 32.Maroulakou IG, Oemler W, Naber SP, Tsichlis PN. Akt1 ablation inhibits, whereas Akt2 ablation accelerates, the development of mammary adenocarcinomas in mouse mammary tumor virus (MMTV)-ErbB2/neu and MMTV-polyoma middle T transgenic mice. Cancer Res. 2007;67:167–177. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]
- 33.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 34.Iwanaga K, Yang Y, Raso MG, Ma L, Hanna AE, Thilaganathan N, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res. 2008;68:1119–1127. doi: 10.1158/0008-5472.CAN-07-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70:7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao D, Alexander CL, Quinn JA, Porter MJ, Wu H, Greenhalgh DA. PTEN loss promotes rasHa-mediated papillomatogenesis via dual upregulation of AKT activity and cell cycle deregulation but malignant conversion proceeds via PTEN-associated pathways. Cancer Res. 2006;66:1302–1312. doi: 10.1158/0008-5472.CAN-05-2341. [DOI] [PubMed] [Google Scholar]
- 37.Tsao H, Zhang X, Fowlkes K, Haluska FG. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 2000;60:1800–1804. [PubMed] [Google Scholar]
- 38.Nogueira C, Kim KH, Sung H, Paraiso KH, Dannenberg JH, Bosenberg M, et al. Cooperative interactions of PTEN deficiency and RAS activation in melanoma metastasis. Oncogene. 2010;29:6222–6232. doi: 10.1038/onc.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prior IA, Hancock JF. Compartmentalization of Ras proteins. Journal of cell science. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- 40.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 41.Whitwam T, Vanbrocklin MW, Russo ME, Haak PT, Bilgili D, Resau JH, et al. Differential oncogenic potential of activated RAS isoforms in melanocytes. Oncogene. 2007;26:4563–4570. doi: 10.1038/sj.onc.1210239. [DOI] [PubMed] [Google Scholar]
- 42.Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12:2301–2307. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- 43.Shin SS, Wall BA, Goydos JS, Chen S. AKT2 is a downstream target of metabotropic glutamate receptor 1 (Grm1) Pigment Cell Melanoma Res. 2010;23:103–111. doi: 10.1111/j.1755-148X.2009.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K, Tsichlis PN. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2:62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]