Abstract
GTP ases of Immunity-Associated Proteins (GIMAPs) are a family of guanine nucleotide binding (G) proteins which are implicated in the regulation of apoptosis in lymphocytes. GIMAPs are composed of an amino-terminal G domain and carboxy-terminal extensions of varying size. Our recent biochemical and structural analysis of a representative GIMAP family member, GIMAP2, revealed the molecular basis of GTP-dependent oligomerization which involves two interfaces in the G domain. Whereas the amphipathic helix α7 in the C-terminal extension closely folds against the G domain in the GDP-bound state, it might be released in the GTP-bound state to assemble interaction partners. We also showed that the GIMAP2 oligomer functions at the surface of lipid droplets in a Jurkat T cell line. Here, we review our recent work and discuss the GIMAP2 oligomer as a GTP-dependent protein scaffold at the surface of lipid droplets controlling apoptosis.
Key words: GIMAP, GTPase, protein scaffold, lipid droplets, lymphocytes, protein structure, immunity
GTPases of Immunity Associated Proteins (GIMAPs, also known as Immunity-Associated Nucleotide-binding proteins, IANs) comprise a conserved G protein family which appears in plants1 and vertebrates.2 In humans, the seven GIMAP members have molecular masses of 33–75 kDa and are composed of an amino-(N-)terminal guanine nucleotide binding (G) domain containing a unique conserved sequence, the conserved box, followed by carboxy-(C-)terminal extensions of 60–130 amino acids length. GIMAP1, GIMAP2 and GIMAP5 additionally possess one or two predicted helical transmembrane anchors at the very C-terminus, whereas GIMAP8 contains three consecutive GIMAP-specific G domains in a single polypeptide chain.3 Within the TRAnslation FACtor (TRAFAC) class of GTPases, GIMAPs together with the Toc (translocon at the outer envelope membrane of chloroplasts) proteins constitute the paraseptin clade, which is related to the septin family.4
Initial insights into a role of GIMAPs for lymphocyte maintenance originated from the BioBreeding (BB) diabetes-prone rat strain, which shows a severe reduction in the number of peripheral T cells (T lymphopenia),5 and develops spontaneous type 1 diabetes.6 A frameshift mutation in the gimap5 gene was identified as genetic cause of this phenotype.7,8 Subsequently, GIMAP5 was discovered as an anti-apoptotic factor in a human T cell line.9 Knockout studies in mice further corroborated the central function of GIMAPs in the regulation of apoptosis in lymphocytes. Conditional knock out of GIMAP1 in mouse lymphoid tissues results in a severe B and T lymphopenia, and the few remaining lymphocytes exhibit a reduced in vitro survival capacity.10 GIMAP5 knock out mice show a nine-fold decrease in splenic T cells and an increased apoptosis tendency in the residual splenocytes.11 Furthermore, microarray gene expression studies point to an involvement of GIMAPs in human lymphocyte-related diseases; almost the whole GIMAP family is downregulated in CD4+CD25high regulatory T cells of patients suffering from type I diabetes,12 and GIMAP2 and GIMAP7 are within the 15 most downregulated genes in anaplastic large cell lymphoma patient samples, when compared to their progenitor T cells.13
To explore structure and function of GIMAPs, we initiated an expression screen of the human GIMAPs in E. coli. Constructs of GIMAP2 devoid of its two C-terminal hydrophobic segments could be purified to homogeneity.14 The purified proteins still contained GTP from the expression host which could be removed by an additional washing step, attesting to the high nucleotide binding affinity of GIMAP2 (KD = 40 nM for GTP and KD = 600 nM for GDP). We did not observe any GTP hydrolytic activity, but found low affinity GTP-dependent dimerization in solution (KD = 250 µM).
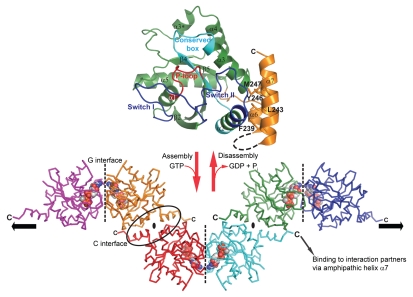
Monomeric structures of GIMAP2 in the nucleotide-free and in the GDP-bound form were solved by X-ray crystallography (Fig. 1 and upper part).15 GIMAP2 exhibits a typical G domain architecture of the TRAFAC class16 composed of a central β-sheet and α-helices at both sides. The C-terminal extension constitutes two amphipathic α-helices, α6 and α7, which bind against switch II of the G domain (Fig. 1 and upper part). We were not able to crystallize this construct in the GTP-bound state; however, our crystallographic analysis of the nucleotide-free protein suggested high flexibility of helix α7. Surprisingly, a construct lacking α7 was already a stable dimer in the presence of GDP and tetramerized with low affinity in the presence of GTP. For this construct, crystals were obtained in the GTP-bound form, in which the G domains oligomerized via two distinct interfaces in the crystal lattice (Fig. 1 and lower part). The ‘G interface’ involves switch I, the conserved box, the guanine nucleotide binding specificity motif (G4) motif and the bound nucleotide itself. Switch I becomes stabilized in the GTP-bound form and mediates nucleotide-dependent interaction with the conserved box of the opposing molecule. Mutations of single amino acid residues in switch I and the conserved box abrogated GTP-dependent dimerization in solution. The second association site, the ‘C interface’, was found both in the GDP- and GTP-bound structures of the shortened GIMAP2 construct and was created by the exposure of hydrophobic residues after removal of α7. It involves residues from α2, α3 and α6, particularly a salt bridge between Arg224 in α6 and Glu131 in α3. Further truncations of α6 including Arg224 abolished stable dimerization via the C-interface.
Figure 1.
GTP-dependent scaffold formation of GIMAP2. (upper part) The structure of the monomeric nucleotide-free GIMAP2 shows a Ras-like G-domain (in green). Additional secondary structure elements compared to the minimal Ras G-domain are helix α3* and the two amphipathic helices α6 and α7 (in orange), which fold against switch II (blue). (lower part) In the absence of α7 and the presence of GTP, GIMAP2 oligomerized via two interfaces in the crystal to form a linear oligomer (the direction of the oligomerization axis is indicated by the black arrows). The conversion between the monomeric and oligomeric states is regulated by GTP binding or hydrolysis (red arrows).
Significant structural changes induced by GTP binding were also observed in the switch II region. In the GDP-bound state, Glu89 from switch II forms a salt bridge to Lys240 in helix α7. Conformational changes in switch II upon GTP-binding lead to a repositioning of Glu89, so that it cannot interact with α7 anymore. Accordingly, our data suggest that association of the G domain with α7 is weakened upon GTP-binding. We propose that in the presence of an adequate hydrophobic acceptor, such as a lipid membrane or an interaction partner exposing a hydrophobic surface, α7 is released from the G domain in a GTP-dependent fashion, allowing oligomerization to proceed via the C-interface.
We also showed that overexpressed GIMAP2 localized to the periphery of lipid droplets (LDs) in the human Jurkat T cell line. The two hydrophobic stretches at the C-terminus of GIMAP2 were necessary and sufficient for lipid droplet targeting. Furthermore, we observed that overexpression of GIMAP2 significantly increased the number of LDs, whereas single amino acid mutations in both the G and C interfaces abrogated this phenotype, indicating a function of the GIMAP2 oligomer during lipid droplet development. The unique architecture of the oligomer, with its C-terminal tails pointing pairwise in opposite directions, is suitable for tethering LDs to each other or for the recruitment of interaction partners to the LD shell (Fig. 1 and lower part).
It has been recognized in recent years that dynamic membrane scaffolds play a major role in organizing the underlying membrane by orchestrating the recruitment of soluble factors in a spatio-temporal fashion. Most importantly, these protein scaffolds allow control and modification of functional outputs of their associated processes. Clathrin-mediated endocytosis is one example where scaffolds of the Eps15 family coordinate the recognition of cargo, membrane remodeling and the recruitment of membrane scission factors.17 Protein scaffolds also assemble components of signal transduction pathways and allow modification of the signal and/or integration of various signalling cascades, e.g., during the assembly of MAP kinases by MAP kinase scaffolds.18 Individual protein-protein interactions within such assemblies are often weak. However, multiple interactions within a two-dimensional matrix create metastable scaffolds by avidity effects, allowing dynamic assembly and disassembly of these scaffolds.17
Oligomers of the GIMAP family are prime examples for such membrane-associated protein scaffolds whose assembly is dynamically regulated by GTP binding and GTP hydrolysis. The high affinity for GTP and the missing GTP hydrolytic activity of GIMAP2,15 and GIMAP5 (our unpublished data) allow stable assembly of these GIMAP scaffolds. Disassembly of these scaffolds might be triggered by accessory proteins which are able to stimulate GTP hydrolysis in a controlled fashion. GIMAP2 is locally concentrated on the LD surface and restricted in its mobility by the two hydrophobic segments, so that the affinity for self-association has to be low to guarantee dynamic assembly and disassembly of the scaffold. Once oligomerized, scaffolds of the GIMAP family might assemble pro- and antiapoptotic B cell lymphoma 2 (Bcl2) family proteins, as shown for GIMAP3, GIMAP4 and GIMAP5,19 via interaction with their amphipathic helix α7. Interestingly, Bcl2 proteins have a similar domain architecture to GIMAPs with a cytoplasmic domain followed by a C-terminal TM region, and they also interact with each other via amphipathic helices which are bound in a hydrophobic cleft of the acceptor protein.20 It is tempting to speculate that the GIMAP2 scaffold employs a similar mechanism to assemble and sequester interaction partners of the Bcl2 family at the surface of lipid droplets, thereby influencing and modifying apoptotic processes. In one scenario, LDs might merely serve as an interaction platform for GIMAP2 to sequester Bcl2 members. Alternatively, GIMAP2 might actively contribute to apoptotic events by controlling the supply of lipids from LDs required for certain aspects of apoptosis. Interestingly, an increase in lipid droplets during apoptosis in the Jurkat T cell line was previously observed,21 but the functional significance remained unclear. Our future studies will address the molecular links between LDs, the GIMAP2 scaffold, the Bcl2 family, and the induction of apoptosis.
Acknowledgements
The idea of GIMAPs as GTP-dependent protein scaffolds on intracellular membranes was intensively discussed during the preparation of a new Collaborative Research Centre “Scaffolding membranes: Molecular Mechanisms and Cellular Functions”, and we would like to thank V. Haucke and L. Aravind for continuous support and ideas. This work was supported by a Career Development Fellowship of The International Human Frontier Science Program Organization to O.D.
Extra View to: Schwefel D, Fröhlich C, Eichhorst J, Wiesner B, Behlke J, Aravind L, Daumke O. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2) Proc Natl Acad Sci USA. 2010;107:20299–20304. doi: 10.1073/pnas.1010322107.
References
- 1.Liu C, Wang T, Zhang W, Li X. Computational identification and analysis of immune-associated nucleotide gene family in Arabidopsis thaliana. J Plant Physiol. 2008;165:777–787. doi: 10.1016/j.jplph.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Krucken J, Schroetel RM, Muller IU, Saidani N, Marinovski P, Benten WP, et al. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 2004;341:291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Krucken J, Epe M, Benten WP, Falkenroth N, Wunderlich F. Malaria-suppressible expression of the anti-apoptotic triple GTPase mGIMAP8. J Cell Biochem. 2005;96:339–348. doi: 10.1002/jcb.20552. [DOI] [PubMed] [Google Scholar]
- 4.Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 5.Naji A, Silvers WK, Kimura H, Bellgrau D, Markmann JF, Barker CF. Analytical and functional studies on the T cells of untreated and immunologically tolerant diabetes-prone BB rats. J Immunol. 1983;130:2168–2172. [PubMed] [Google Scholar]
- 6.Nakhooda AF, Like AA, Chappel CI, Wei CN, Marliss EB. The spontaneously diabetic Wistar rat (the “BB” rat). Studies prior to and during development of the overt syndrome. Diabetologia. 1978;14:199–207. doi: 10.1007/BF00429781. [DOI] [PubMed] [Google Scholar]
- 7.Macmurray AJ, Moralejo DH, Kwitek AE, Rutledge EA, Van YB, Gohlke P, et al. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 2002;12:1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalkiewicz M, Michalkiewicz T, Ettinger RA, Rutledge EA, Fuller JM, Moralejo DH, et al. Transgenic rescue demonstrates involvement of the Ian5 gene in T cell development in the rat. Physiol Genomics. 2004;19:228–232. doi: 10.1152/physiolgenomics.00126.2004. [DOI] [PubMed] [Google Scholar]
- 9.Sandal T, Aumo L, Hedin L, Gjertsen BT, Doskeland SO. Irod/Ian5: an inhibitor of gamma-radiation- and okadaic acid-induced apoptosis. Mol Biol Cell. 2003;14:3292–3304. doi: 10.1091/mbc.E02-10-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders A, Webb LM, Janas ML, Hutchings A, Pascall J, Carter C, et al. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 2010;115:3249–3257. doi: 10.1182/blood-2009-08-237586. [DOI] [PubMed] [Google Scholar]
- 11.Schulteis RD, Chu H, Dai X, Chen Y, Edwards B, Haribhai D, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development and liver failure in mice lacking Gimap5. Blood. 2008;112:4905–4914. doi: 10.1182/blood-2008-03-146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jailwala P, Waukau J, Glisic S, Jana S, Ehlenbach S, Hessner M, et al. Apoptosis of CD4+ CD25(high) T cells in type 1 diabetes may be partially mediated by IL-2 deprivation. PLoS One. 2009;4:6527. doi: 10.1371/journal.pone.0006527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–2138. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 14.Schwefel D, Fröhlich C, Daumke O. Purification, crystallization and preliminary X-ray analysis of human GIMAP2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:725–729. doi: 10.1107/S174430911001537X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwefel D, Fröhlich C, Eichhorst J, Wiesner B, Behlke J, Aravind L, et al. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2) Proc Natl Acad Sci USA. 2010;107:20299–20304. doi: 10.1073/pnas.1010322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 17.Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 18.Dohlman HG. A scaffold makes the switch. Sci Signal. 2008;1:46. doi: 10.1126/scisignal.142pe46. [DOI] [PubMed] [Google Scholar]
- 19.Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006;4:103. doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Al-Saffar NM, Titley JC, Robertson D, Clarke PA, Jackson LE, Leach MO, et al. Apoptosis is associated with triacylglycerol accumulation in Jurkat T-cells. Br J Cancer. 2002;86:963–970. doi: 10.1038/sj.bjc.6600188. [DOI] [PMC free article] [PubMed] [Google Scholar]