Abstract
There is a critical need for biomarkers for early diagnosis, treatment response, and surrogate end point and outcome prediction in organ transplantation, leading to a tailored and individualized treatment. Genomic and proteomic platforms have provided multiple promising new biomarkers during the last few years. However, there is still no routine application of any of these markers in clinical transplantation. This article will discuss the existing gap between biomarker discovery and clinical application in the kidney transplant setting. Approaches to implementing biomarker monitoring into clinical practice will also be discussed.
Keywords: biomarkers, immune monitoring, kidney function, kidney transplantation, long-term outcomes, short-term outcomes
In kidney transplantation, significant improvements in short-term allograft survival have been accomplished [1,2]. However, despite improvement over the years in immunosuppressant strategies, late kidney allograft loss remains the main clinical challenge for long-term graft survival [1,2]. With close to 5000 kidney transplants failing each year in the USA alone, kidney transplant failure is now a leading cause of end-stage renal disease [3].
The Human Genome Project and Genomic Sciences resulted in valuable insights into the mechanisms of the genome and have produced a large quantity of tools [201]. These tools are enabling a detailed analysis of the expression of the complete set of genes encoded in the genome and are collectively referred to as the ‘omics’ technologies. The knowledge base being developed by the genomics revolution, through its associated technologies, is impacting on the practice of all of the biological sciences [4].
There is a critical need for biomarkers for early diagnosis, treatment response, and surrogate end point and outcome prediction in organ transplantation, leading to a tailored and individualized treatment [5–7]. The ‘omics’ technologies have provided a multitude of promising new biomarkers during the last few years. However, there is still no routine application of any of these markers in clinical transplantation. In this article, new potential biomarkers in kidney transplantation are discussed, with particular emphasis on the requirements for implementing their use in clinical kidney transplantation.
Biomarkers: important definitions
Recently, a NIH working group recommended preferred terms and definitions that have broad applications [8]:
Biological marker (biomarker): a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention;
Clinical end point: a characteristic or variable that reflects how a patient feels or functions, or how long a patient survives;
Surrogate end point: a biomarker intended to substitute a clinical end point. A clinical investigator uses epidemiologic, therapeutic, pathophysiologic or other scientific evidence to select a surrogate end point that is expected to predict clinical benefit, harm or lack of benefit or harm.
The introduction of molecular medicine has resulted in an amazing increase in the discovery of new biomarkers. Much of this is related to the advances in new technologies such as genomics, proteomics and imaging. Furthermore, the expansions in technologies and innovative applications towards translational medicine have driven biomarkers into the focus of drug discovery and development in recent years.
Limitations associated with the identification & implementation of biomarkers in transplantation
A major obstacle in the management of transplant recipients is a lack of accurate and specific tests for immune monitoring, as well as predicting long-term graft function. Most accessible methods for evaluating kidney function are either ineffective, inaccurate or highly invasive, such as tissue biopsies.
There are several more important limitations associated with the application of new biomarkers, which are discussed in the following sections.
Lack of a robust ‘gold standard’
In kidney transplantation, despite decades of research, there is still a lack of reliable markers useful for immune monitoring in the clinical routine. This is partly due to the lack of well-defined end points and short follow-up times in validation studies. Furthermore, it reflects the absence of robust metrics and a reliance on ‘subjective’ measures. Measurements of serum creatinine, glomerular filtration rate (GFR), proteinuria (without taking any factors associated with the donor into consideration) and biopsy remain the current gold standards for the evaluation of renal allografts. These tests have significant limitations in predicting which patients are destined for immune tolerance or immune-mediated graft loss, and assisting in the management of long-term immunosuppression. The goal of biomarkers is to allow sensitive and accurate monitoring of graft function, early and specific diagnosis of rejection and the assessment of long-term outcomes, allowing for a tailored immunosuppressive therapy in a noninvasive, cost-effective manner [9].
The ability of serum creatinine measurements at 6 and 12 months to predict 3-year graft survival was analyzed by Fitzsimmons et al. using data from two multicenter clinical studies comparing tacrolimus- and cyclosporin-based immunosuppression [10]. The authors concluded that the levels of serum creatinine evaluated at 6 and 12 months post-transplant in kidney transplant recipients and the change of the levels of serum creatinine between these two time points are important predictors of graft loss over 3 years. However, the inherent limitations of creatinine as an accurate measure of glomerular filtration, such as variability in production or tubular secretion, and the lack of incorporation of donor information in its interpretation makes it an inadequate marker of true kidney function [11,12].
Glomerular filtration rate can be precisely measured by specific filtration markers such as inulin, 125I-iothalamate, 51Cr-EDTA, 99mTc-DTPA and iohexol. However, these measurements are laborious and expensive, and therefore, GFR is commonly estimated in kidney transplantation by using creatinine-based estimation equations [13]. The performance of these equations in kidney transplant recipients is unclear, with conflicting results between studies. Differences in patient populations, baseline GFRs of the study groups, reference standard GFR values used, and creatinine assay calibrations probably account for the heterogeneity in results. These factors need to be considered by investigators and clinicians when interpreting estimates of GFR in kidney transplant recipients [13,14].
After renal transplantation, body composition may undergo some changes, with a high prevalence of obesity and loss of muscle mass owing to steroid use, which could infringe on the assumptions underlying the creatinine-based renal function equations. Thus, for renal transplant recipients, specific validation of renal function equations would be required. Several studies have addressed this issue [15–18], indicating a relatively poor predictive performance of renal function equations in the transplant population. However, these studies included only relatively small populations, and only evaluated a small number of equations.
In a recent publication, Bosma et al. evaluated nine equations with iothalamate GFR at 1 year following transplantation in 798 recipients [19]. Equations were analyzed for precision, bias and accuracy. Sources of bias were analyzed by univariate and multivariate analysis, with BMI, age and sex as independent variables and bias as the dependent variable. The predictive performance of renal function equations is modest in renal transplants, which hampers their use for accurate assessment of renal function in the individual. The role of patient factors in the systematic error suggests that development of better equations should be feasible by better incorporation of these factors.
Kidney biopsies are considered to be the gold standard for evaluating allograft dysfunction and have been shown to change the presumptive diagnosis in 40% of cases [20]. The use of protocol biopsies has provided insights into the pathogenesis of many renal allograft diseases [21]. However, the histological evaluation of biopsies is subjective and there is an associated error among different pathologists evaluating the same tissues [22].
Furthermore, obtaining a renal allograft biopsy is an invasive procedure with some associated morbidity that yields a randomly sampled tiny piece of tissue with an intrinsic sampling error. There are no data to support the suggestion that samples removed from one segment of the transplanted kidney are representative of the whole graft. In a recent report, Piovesan et al. evaluated 200 percutaneous biopsies that were performed on kidney allografts and samples were collected from the upper and lower poles (100 kidneys) [23]. For the diagnosis of AR, the discordance rate between the upper and lower poles was 82.3%, higher than the intrapathologist variation. For the diagnosis of nephrotoxicity, the discordance rate between the upper and lower poles was 28.6%, with no difference compared with the intrapathologist variation. These observations demonstrated that that the histopathological changes in the kidney allograft are not always homogeneous and, furthermore, that this heterogeneity may affect the therapeutic recommendations.
Deficient evaluation of new biomarkers
During multiple recent attempts to identify biomarkers in transplantation, critical characteristics of the new markers were often not included in the analysis. They include, among others, half-lives of the biomarkers, stability, response to treatment patterns and kinetics. A number of developmental objectives must be accomplished before any biomarker can be considered useful for clinical diagnosis. Specifically the biomarker(s) need(s) to be [24,25]:
Evaluated in an independent set of samples/patients
Appraised in group of patients with multiple pathologies to allow determination of marker(s) sensitivity, specificity, positive- and negative-predictive values
Analyzed for intra- and interdetermination variability to establish the coefficient of variation of the measurement
Reproduced by an independent laboratory to demonstrate the robustness/reproducibility of the assay among different groups
Prospectively assessed to determine the ability of a given marker to predict disease progression
Evaluated for the stability of the marker in the sample, influence of sample collection and storage
Appraised of interpatient variability
Unfortunately, most of the potential markers proposed in kidney transplantation to date do not meet the full criteria for biomarker establishment [12].
Lack of integration of clinical factors
The convergence and integration of approaches and technologies are important for developing successful strategies for the discovery, characterization, validation and application of biomarkers. Furthermore, the discovery of human biomarkers requires the association of biological measurements with clinical outcomes. To discover new biomarkers that predict treatment effects and disease progression, it is necessary to analyze a broad spectrum of data from human studies, but there are major challenges. Genomic and proteomic studies generate amazing amounts of data. These studies struggle with patient, disease, platform and study heterogeneity, necessitating detailed annotation. Clinical trials capture a variety of biological samples and hundreds of clinical and laboratory parameters, including multiple potential end points to determine drug efficacy and underappreciated confounders, such as environmental exposures and personal behaviors.
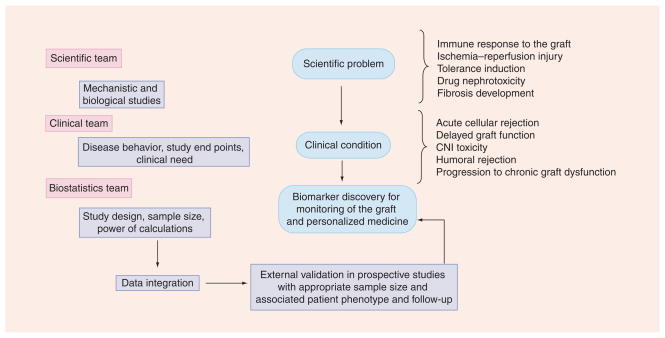
The successful translation of these datasets into new biomarkers will require novel and broad data integration approaches. Data integration can provide value by providing information together with metadata and supplementary information, increasing statistical power, improving the ability to make results comparable across populations, replicating or rejecting findings across studies, and validating results using independent sets of patients [26,27]. Overall, this critical step in biomarker discovery will require the effort of a multidisciplinary team working together in the biological mechanisms associated with the disease, study design, data analysis and validation, and overall data integration (Figure 1).
Figure 1. Possible pathways for biomarker discovery in kidney transplantation.
Both a multidisciplinary team and multiple steps are needed to achieve the goal of moving biomarkers ‘from the bench to the bedside’. The process of discovery and validation of biomarkers that can be used for the routine evaluation of graft function requires a multidisciplinary team able to evaluate the clinical problem, understand the biology behind the clinical condition, and perform data analysis and integration. Moreover, external validation is required in prospective powered studies.
CNI: Calcineurin inhibitor.
The data integration field is currently growing and no clear standards for cross-study and cross-type data storage, integration and analysis methods and systems have emerged. Integration of clinical data with high-throughput expression data is currently an intense area of research, and there are no standard solutions [28].
An approach that combines data from individual research groups conducted on similar disease conditions and then applies this approach for robust cross-validation in a large dataset may be one way to address this issue. In a recent report from our group [29], we applied a nonparametric meta-analytic approach for combining independent microarray datasets pertaining to chronic allograft nephropathy (CAN). The work used a nonparametric meta-analysis approach based on two CAN studies and identified 309 distinct genes that are expressed differently in CAN. With the help of Fisher’s exact test, the study found six Kyoto Encylopedia of Genes and Genomes (KEGG) pathways to be over-represented among the identified genes.
In a recent publication, the authors combined the data from five separate studies to compare AR versus stability after solid organ transplantation, and used this data to examine approaches to multiplex meta-analysis [30]. In their study, the authors demonstrated that a commonly used parametric effect size estimate approach and a commonly used nonparametric method produced very different results in prioritizing genes.
Interlaboratory & intraplatform variability
As this article concentrates on genomics as a tool for biomarker discovery, this section discusses microarray limitations in relation to genomics. However, many of the limitations described in this section are also applicable to proteomics, metabolomics and genome-association studies, especially the ones related to interplatform variation, requested sample size and cost.
There is currently a variety of microarray platforms available to researchers. A number of critical differences exist between the various forms, including length of targeted sequences, global or specific coverage of genes, and glass or membrane support. Furthermore, each platform comes with its specific methodology for sample labeling, hybridization and data analysis. In addition to these issues, there is also the variation associated with sample preparation across laboratories, such as RNA-isolation techniques.
A significant number of studies have investigated the comparability of the data generated from different microarray platforms [31–35]. These studies have demonstrated that although variability is present in interplatform and interlaboratory data, the agreement can be increased using standardized protocols and data analysis. More recently, results were reported from the Microarray Quality Control (MAQC) project [36], evaluating intra- and inter-platform reproducibility of four different microarray platforms. Overall, these studies reported good concordance among platforms at the level of gene-expression changes and gene ontology terms. These results support microarray technology as a reliable tool for biomarker discovery. These studies are limited to transcriptional response to toxin exposure but hopefully are applicable to the wider science fields.
Lack of powered studies
One of the strengths of the ‘omic’ tools is that they have marvelous power to effectively generate a large volume of data dealing with multiple samples at the same time. Unfortunately, many of these studies are measuring thousands of variables/markers in a limited set of patients. In addition to the problem that many of the studies have been performed using small transplant patient groups, there is the added component associated with the high variability of these patients (i.e., donor/recipient characteristics, immunosuppressive treatment, utilization of protocol biopsies versus biopsies for cause, and different time points for analysis, among others). These issues have resulted in a lack of or small degree of overlap between identified markers of disease among different studies. To overcome this problem, many groups decided to work together creating multicenter consortiums. Examples of this approach in the transplant field are the DeKAF group, the A2ALL study and the Immune Tolerance Network ST507 Study Group, among others.
The use of genomics, transcriptomics, proteomics, metabolomics and new imaging techniques will have a critical impact on the transplant field, allowing for an improved understanding of the mechanisms of the diseases affecting the kidney graft. Furthermore, these technologies will lead to the identification of diagnostic, prognostic and therapeutic markers. However, there are also limitations, including the cost of many of these technology platforms and the complexity of the data, which will make the translation of these platforms and results as monitoring tools in the clinic difficult [37].
Biomarkers in renal transplantation ischemia–reperfusion injury & prediction of short-term outcomes
Ischemia–reperfusion injury (IRI) is a composite process leading to delayed graft function (DGF) and reduced long-term graft function. Ischemic injury of the renal allograft is a critical early perioperative insult that augments the risk of acute tubular necrosis and DGF. Early identification of graft recipients at risk would allow modification of post-transplant management, and thereby potentially improve short- and long-term outcomes.
In a recent study, Irish et al. studied a cohort of deceased donor renal transplant recipients (2003–2006) to create a nomogram utilizing both donor and recipient characteristics to aid clinicians in the prediction of DGF prior to transplantation [38]. The authors identified four donor-related characteristics that have a strong impact on the risk of DGF: cold ischemia time, donor creatinine, donation after cardiac death and donor age. A fifth characteristic, recipient BMI, was also identified as predictive of DGF. However, some of the limitations of this study include relying on the use of the United Network for Organ Sharing (UNOS) registry data, which is incomplete and relies on the submission of data that is not independently validated [39].
The recently emerged ‘omics’ technologies together with bioinformatics tools have allowed the integration and analysis of IRI-associated molecular profiles in the context of DGF [40,41].
Hauser et al. reported the genome-wide gene-expression profiles of 32 donor kidney biopsies [42]. In these kidney biopsies, a set of 132 genes that clearly separated living donor from cadaveric donor organs were observed. Many of these genes play a major role in cell communication, growth/survival, inflammation and metabolic pathways. Furthermore, in the same dataset, 48 genes were identified in cadaveric donor kidney biopsies that separated organs with primary function from those with subsequent acute renal failure. The majority of these genes were classified as being involved in signal transduction pathways, cell cycle regulation, cell growth/metabolism and other functions.
In a second report from the same group of investigators, the gene-expression profiles of donor kidney biopsies with good medium-term function and those showing decreased values of GFR at 1 year following transplantation were reported. A set of 52 genes differentially regulated between the two groups was identified. Many of the genes upregulated in the group with poor outcome were involved in immune response, signal transduction cascades and response to stress. These findings concur with those reported previously by Hauser et al. on the genome-expression profiles in cadaveric and living donor organs [43].
Mueller et al. compared 87 consecutive implantation biopsies taken after reperfusion in 42 deceased and 45 living donor kidneys using clinical and histopathology-based scores [44]. Unsupervised analysis separated the kidneys into a living donor group and two deceased donor kidney subgroups. One of the subgroups of kidneys showed a greater incidence of DGF (p < 0.05). However, clinical and histopathological risk scores were not able to discriminate between these two deceased donor kidney subgroups. A total of 1051 transcripts were differentially expressed between the deceased donor kidney subgroups, but no transcripts separated DGF from immediate graft function. The authors concluded that the transcriptome of implantation biopsies (post-reperfusion) reflects kidney quality and susceptibility to DGF better than available clinical and histopathological scoring systems.
In a recent publication, gene-expression profiling was performed on donor kidney tissues collected before implantation from 33 deceased donor kidneys [45]. Samples were classified as grafts with immediate function (non-DGF: n = 21) and grafts with DGF (n = 12). Logistic regression was used to identify genes significantly associated with DGF development. A total of 38 probe sets (n = 36 genes) were univariably differentially expressed in deceased donor kidneys with DGF compared with those kidneys without DGF. A total of 69 probe sets were differentially expressed between these two groups after adjusting for cold ischemia time (α = 0.001). Gene ontology terms classified the overexpressed genes in deceased donor kidneys with DGF as principally related to cell cycle/growth, signal transduction, immune response and metabolism.
Differences in cold-ischemia time between groups may contribute to the differences in gene-expression patterns – for that reason our previously described gene-expression data were adjusted for cold-ischemia time [45]. Future studies demonstrating differential gene-expression patterns between groups of deceased donor recipients with similar cold-ischemia times will strenghten profiles and improve their accuracy in predicting clinical outcome.
Delayed graft function is consequently an important clinical and pathophysiological entity, and better understanding of its trigger mechanisms, in order to find ways to diminish its occurrence is very important. Given the current organ shortage, many of the underlying risk factors for DGF are difficult to control in all circumstances. Strategies to prevent IRI have focused on improving preservation solutions, the use of pulsatile perfusion machines instead of cold storage, additional efforts to avoid hypovolemia of the recipient, and local or systemic administration of vasodilator agents. Understanding and harnessing molecular targets of early DGF injury could potentially modulate more specific and ubiquitously functional pathways, ideally in the form of pretreatment of the donor organ after recovery but before transplantation [46,47].
There are clinical reports supporting the utility of the evaluation of preimplantation biopsies to identify predictors of short-term outcomes. Kainz et al. recently demonstrated in a randomized control trial that steroid pretreatment of the deceased organ donor suppressed inflammation in the transplant organ but did not reduce the rate or duration of DGF [48]. A further study sought to elucidate those factors that caused DGF in the steroid-treated subjects. Genome-wide gene-expression profiles were used from 20 steroid-pretreated donor organs and were analyzed on the level of regulatory protein–protein interaction networks [49]. A total of 63 significantly downregulated sequences associated with DGF were identified that could be functionally categorized according to Protein Analysis Through Evolutionary Relationships ontologies into two main biologic processes (transport and metabolism). The recognized genes suggested hypoxia as the cause of DGF, which cannot be compensated by steroid treatment. The data showed that molecular pathways affected by ischemia, such as transport and metabolism, are associated with DGF. Potential interventional targeted therapy based on these findings includes peroxisome proliferator-activated receptor agonists or caspase inhibitors.
Some of these studies are associated with the limitations of small sample size, lack of independent validation sets and/or insufficient patient follow-up. However, these studies provide an early and critical step in the understanding of possible pathways involved in early renal allograft injury and functionality [42–47]. Nevertheless, a number of potential clinically very useful biomarkers have emerged as a result of these predominantly transcriptome-based studies. In particular, a new set of kidney injury markers such as kidney injury molecule-1, neutrophil gelatinase-associated lipocalin or IL-18 are undergoing validation studies [50–54].
Markers to diagnose acute kidney rejection
Tissue studies
Acute rejection (AR) is an allograft-destructive immune response that may occur at any time during the lifespan of an organ transplant [55]. Histological features of the allograft biopsy are currently used for the differential diagnosis of allograft dysfunction [56]. Needle biopsy is currently used as the gold standard to diagnose AR of renal allografts. As previously mentioned, it is an invasive procedure with an important associated sampling error and interpathologist variation [57,58].
In addition to nucleic acid-based assays (principally micro-array assays and quantitative PCR [QPCR]), proteomics and metabolomics-based approaches represent additional avenues for the development of biomarkers of allograft status [55].
Sarwal et al. provided evidence that molecular heterogeneity in acute renal allograft rejection underlies the variability in the response to antirejection therapy and in graft survival [59]. The authors defined the mRNA-expression profile of RNA isolated from 67 biopsy specimens [59]. The authors argued that the gene-expression profiles could distinguish among samples from patients with acute cellular rejection, nephrotoxic effects of drugs, infection, chronic allograft nephropathy and normal kidneys. A limitation of the study is that a majority of the subjects were pediatric patients.
Reeve et al. distinguished rejection from nonrejection using predictive analysis of microarrays [60]. The authors evaluated 186 consecutive kidney transplant biopsies for cause using micro-arrays. Moreover, a classifier to distinguish rejection from nonrejection using predictive analysis of microarrays (PAM) was built. Most genes selected by PAM were IFN-γ-inducible or cytotoxic T-cell-associated. PAM diagnosis was compared with histopathology (based on the Banff diagnostic criteria). Disagreement occurred in approximately 20% of diagnoses. The problematic diagnosis of ‘borderline rejection’ was resolved by PAM and cases were divided into two distinct classes: rejection and nonrejection.
In a report by Desvaux et al., 43 human renal-allograft biopsies were quantified for mRNA expression of granzyme B, Fas ligand (FasL), IFN-γ, IL-4 and IL-6 with a reverse-transcriptase real-time (RT)-QPCR method [61]. Expression levels were correlated to the histopathologic rejection type according to the Banff 1997 classification criteria, and with sensitivity to antirejection immunosuppressive therapy, by means of receiver operating-characteristic (ROC) curves.
The authors identified that granzyme B and FasL mRNA expression upregulation correlated with all allograft rejection types (p < 0.01). Furthermore, granzyme B showed the highest sensitivity (90%) and specificity (78%) for the potential detection of histologic borderline changes that will require immunosuppressive therapy (AUC = 0.856; p < 0.01).
Studies performed using peripheral blood and/or urine samples
The opportunity to measure gene-expression profiles in kidney transplantation allows the testing of several hypotheses that will directly impact on clinical practice [62–64]. Currently, there is no objective measure for determining the adequacy of immunosuppression, and no objective method of predicting an individual patient’s response to therapy. Therefore, if gene-expression profiling identifies a signature for AR, then a patient on any given immunosuppressive regime could be monitored for that signature as a measure of the adequacy of immunosuppression. Flechner et al. reported an AR signature for both peripheral blood lymphocytes and transplant biopsies, supporting the hypothesis that a prospective approach to monitoring molecular changes in transplant patients could also be used to predict AR [65].
Furthermore, recent attempts to identify noninvasive biomarkers of acute kidney rejection have been performed using genomics and proteomics technologies. Li et al. evaluated the measurement of urinary cell levels of mRNA for perforin and granzyme B as a noninvasive method of diagnosing AR of renal allografts and reported that rejection is predicted with a high degree of accuracy by urinary cell levels of perforin and granzyme B mRNA [66].
However, Yannaraki et al. investigated the predictive and diagnostic interest of target mRNA measurements with a QPCR assay in AR, as well as in other clinical complications occurring (recurrent) in kidney transplantation: urinary tract infection (UTI), cytomegalovirus infection or disease, CAN, DGF and stable graft course (controls) [67]. A total of 162 urine specimens from 37 allograft recipients were investigated. In the case of AR, mRNA levels of all three molecules were significantly higher than in recipients not showing any clinically evident signs of complications. Kinetic studies in three patients with AR revealed that increased perforin, granzyme B and FasL mRNA levels could precede or were concomitant with increased serum creatinine levels. However, perforin, granzyme B and FasL mRNA gene expression in urine specimens were upregulated in AR episodes but also in UTI, cytomegalovirus infection and DGF. Therefore, the authors concluded that this technique would appear to be of limited clinical value as a noninvasive method of diagnosing AR.
Vasconcellos et al. investigated expression patterns of mRNA for perforin, granzyme B and FasL in peripheral blood mononuclear cells (PBMCs) collected from renal transplant recipients and reported that perforin mRNA predicted AR with a sensitivity of 82% and a specificity of 85%; granzyme B mRNA predicted with a sensitivity of 55% and a specificity of 85%; and FasL with a sensitivity of 100% and a specificity of 75% [68]. In this study, the upregulated expression of two or more genes was diagnostic of AR with a positive-predictive value of 100% and a lack of upregulation of any gene ruled out rejection with a negative-predictive value of 95%.
Testing the hypothesis that urine is a potentially rich source of biomarkers for monitoring kidney dysfunction, Ting et al., in a recent publication, monitored urinary soluble human leukocyte antigen (sHLA)-DR levels by sandwich ELIZA in 103 patients with renal diseases or after renal transplantation [69]. sHLA-DR in urine was characterized by Western blotting and mass spectrometry. Acute graft rejection was associated with a significantly elevated level of urinary sHLA-DR (p < 0.0001), compared with recipients with stable graft function or healthy individuals. A ROC curve analysis showed the area under the curve to be 0.88 (p < 0.001). At a selected threshold, the sensitivity was 80% and specificity was 98% for detection of acute renal transplant rejection. The authors concluded that sHLA-DR excreted into urine is a promising indicator of renal transplant rejection.
Although proteomics has the potential to provide insights into complex pathophysiological processes and expose novel diagnostic biomarkers, as well as therapeutic drug targets, the actual status of urine proteomic activities in renal transplantation is still far from reaching these ambitious goals [70,71]. In a study performed by Schaub et al., four patient groups were rigidly defined on the basis of allograft function, clinical course and allograft biopsy result as acute clinical rejection group (n = 18), stable transplant group (n = 22), acute tubular necrosis group (n = 5), and recurrent (or de novo) glomerulopathy group (n = 5) [72]. Urine samples collected on the day of the allograft biopsy were analyzed by mass spectrometry. As a normal control group, 28 urine samples from healthy individuals were analyzed in an identical manner, as well as five urine samples from nontransplanted patients with lower UTI. Furthermore, sequential urine analysis was performed in patients in the acute clinical rejection and the stable transplant group. Three prominent peak clusters were found in 17 out of 18 patients (94%) with AR episodes, but only in four out of 22 patients (18%) without clinical and histological evidence for rejection, and in zero out of 28 normal controls (p < 0.001). The author concluded that the use of proteomic technology, together with a rigorous definition of patient groups, can detect urine proteins associated with acute renal allograft rejection. Furthermore, the results supported the hypothesis that proteins in urine may prove useful as noninvasive diagnostic markers for rejection and for the development of novel therapeutic agents.
In a recent publication, Sigdel et al. used shotgun proteomics applying liquid chromatography (LC)-MS/MS and ELISA to analyze a set of 92 urine samples from patients with AR, stable grafts, proteinuria and healthy controls [73]. The authors identified alterations in a number of specific urinary proteins in AR, primarily relating to major histocompatibility complex antigens, the complement cascade and extracellular matrix proteins. A subset of proteins (UMOD, SERPINF1 and CD44) have been further cross-validated by ELISA in an independent set of urine samples for significant differences in the abundance of these urinary proteins in AR. This label-free, semiquantitative approach for sampling the urinary proteome in normal and disease states provides a robust and sensitive method for detection of urinary proteins for serial, noninvasive clinical monitoring for graft rejection following kidney transplantation.
At the peripheral blood level, in a recent study peripheral blood samples were obtained before transplantation and at days 3 and 7, weeks 2 and 3 and months 1, 2, 3 and 6 post-transplantation [74]. The level of HLA class I (ABC) on peripheral blood CD3+/CD8+ T lymphocytes was measured by flow cytometry. For the 79 transplant recipients, the level of HLA class I (ABC) on peripheral-blood CD3+/CD8+ T lymphocytes was consistently elevated during the first 3 weeks following transplantation, declined gradually to pretransplantation levels, then tapered off and remained stable. Patients experiencing AR or not after transplantation did not differ in level of HLA class I (ABC) up to 6-months follow-up, except at days 14 and 21 after transplantation, when the level was higher for patients experiencing AR (p < 0.01). The authors concluded that the upregulation of HLA class I (ABC) on peripheral-blood CD3+/CD8+ T lymphocytes could be used as an accurate and reliable predictor of AR after renal transplantation.
MiRNAs: new potential biomarkers for AR
miRNAs are small noncoding RNAs approximately 22 nucleotides long that regulate gene expression by inducing translational repression, mRNA degradation, transcriptional inhibition, or all of the above [75]. A single miRNA has the ability to regulate the expression of hundreds of mRNAs. miRNAs have been shown to control processes such as cellular survival, development, differentiation and proliferation [76], as well as modulate both innate and adaptive immunity [77]. Sui et al. described the first comparison between the miRNA expression profiles of AR and control subjects [78]. Through microarray analysis and quantitative real-time (QRT) RT-PCR confirmation, the authors identified 20 miRNAs differently expressed in AR after renal transplantation. Their data indicated that miRNAs were potentially involved in the pathogenesis of AR. Anglicheau et al. recently identified several miRNAs predictive of AR of human renal allografts [79]. The hypothesis that urinary cell and/or peripheral blood cell miRNA-expression profiles are predictive, diagnostic and/or prognostic biomarkers of allografts deserves further investigation [80].
More than 90% of renal allografts transplanted today will still be functioning 1 year from now. This rate represents a remarkable improvement over a 1-year graft survival rate of 70% in 1990 [81]. Despite this improvement in short-term success, which reflects the use of better immunosuppressive medications and the provision of specialized management from teams that care for patients with renal transplants, the long-term deficiencies of current approaches are increasingly apparent: undesirable side effects of immunosuppressive medications and the inexorable loss of grafts due to chronic rejection. Surrogate measures are needed for the prediction of graft survival at the earliest stages of graft deterioration in order to guide effective therapeutic interventions; such surrogates are a first step in unraveling the causes of long-term failure [82].
Markers of chronic allograft dysfunction
Poor donor organ quality is one key cause of inadequate/unsatisfactory long-term outcomes [83,84]. The original nephron mass present in the donor kidney at the time of transplantation is largely dependent on donor quality. After kidney transplantation and as a consequence of surgery itself, preservation methods, ischemia, AR and other stressors, the initial nephron number in the donor kidney will be reduced. Generally, after 6 months post-transplantation, the number of nephrons is stable, resulting in an allograft with normal function. However, for an important number of grafts, the loss of nephrons continues, reflecting continued graft injury. Damaged nephrons cannot be replaced, leading to an accumulation of injury in a progressive and cumulative manner, as confirmed in prospective protocol biopsy studies [85–87]. The damage occurring to the allograft might be the result of a combination of pre-existing donor disease and subsequent insults to the transplant, leading to progressive injury. This condition will end in fibrosis and atrophy, with the final event of loss of graft function [87]. It is unclear why a number of kidneys maintain stable function and others develop injury and inescapable progression to fibrosis development.
The term chronic allograft nephropathy (CAN) was originally coined in 1991 to replace chronic rejection, which was found to be too generalized [88,89]. However, the revised Banff classification, published in 2007 [88], eliminated the term CAN again because it was felt that the term was used too broadly and prevented the search for an underlying cause [85]. Interstitial fibrosis (IF) and tubular atrophy (TA) are integral parts of chronic allograft dysfunction and represent, in the new classification, a separate entity with or without the identification of a specific etiology.
The complex, multistage processes that result in IF/TA are poorly understood. While the determination of organ failure often relies on measurable physiological parameters, the early stages of chronic IF/TA are difficult to diagnose. Detection requires invasive procedures in order to obtain graft biopsies for histological evaluation. Furthermore, current methods for diagnosing allograft dysfunction are inadequate for detecting the significant organ damage that occurs prior to the establishment of clinical manifestations. The development of assays or novel technologies that will be able to detect allograft dysfunction/rejection and predict long-term outcomes is vital for the long-term success of transplantation [83–89]. Histologic abnormalities leading to chronic allograft dysfunction may be observed from the third post-transplant month and are characterized mainly by IF/TA. A protocol biopsy study observed that deterioration in renal function typically appears after the histologic abnormalities were already present [90].
De Matos et al. investigated the histologic pattern associated with proximal tubular dysfunction and its correlation with graft outcome [91]. A total of 49 transplant patients with stable graft function were submitted to biopsy. Simultaneously, urinary retinol-binding protein (uRBP) was measured and creatinine clearance was also determined. Banff’s score and semiquantitative histologic analyses were performed to assess tubulointerstitial alterations. In their study, at biopsy time, mean serum creatinine was 1.43 ± 0.33 mg/dl. A total of 12 patients (24.5%) had uRBP ≥1 mg/l, indicating proximal tubular dysfunction, and 67% of biopsies had some degree of tubulointerstitial injury. At the end of the study period, 18 (36.7%) patients had lost renal function. uRBP levels were not associated with morphologic findings of IF/TA. However, in multivariate analysis, the only variable associated with loss of renal function was uRBP level ≥1 mg/l, determining a risk of 5.290 of loss of renal function (p = 0.003). The authors concluded that kidney transplant patients who present proximal tubular dysfunction have functional alteration, which is not associated with morphologic alteration. This reflects the increasing awareness of dissociation between morphology and function in acute, as well as chronic, kidney diseases [92,93].
Important research progress has been made in understanding the facts about prevalence, clinical significance and pathogenesis of chronic kidney allograft injury. However, minimal improvement in the development of new strategies and/or protocols to improve long-term outcomes has been implemented during the last few years [94]. As a consequence of the complexity of the disease pathogenesis and multifactorial cause, it is highly probable that individualized therapy decisions may be associated with future treatment options. To be able to individualize medicine in these patients, accurate tools are required that can discriminate patients at risk of progression to chronic allograft dysfunction from patients that will keep stable kidney function. A major challenge in the future of kidney transplantation includes the study of the pathogenesis of chronic allograft dysfunction (pathogenesis) to identify early markers of disease progression, as well as potential therapeutic pathways [94].
In recent publications, we and others established the gene-expression signatures that characterize IF/TA [95–101]. However, early diagnosis of IF/TA is a requirement for a timely therapeutic intervention in patients at risk. To evaluate events occurring before evident IF/TA is present in the graft, gene-expression profiling of 3-month protocol biopsies from patients with IF/TA was performed in a patient group who developed mild IF/TA CAN grade I by the Banff scoring system in the subsequent 6-month protocol biopsy (‘progressors’; n = 8), and in 12 patients without IF/TA at 6 months (‘nonprogressors’). Compared with the nonprogressors, the 3-month biopsies of the progressor group demonstrated overexpression of several genes that are important in T- and B-cell activation and immune response [96]. Genes involved in profibrotic processes were identified in the biopsies of the progressors that preceded the observed IF/TA at 6 months. Furthermore, several genes with transporter and metabolic functions were under-represented in the progressors in the 3-month biopsies. The authors described no statistical difference in cyclosporine levels between patient groups [96].
In a previous study, the same group reported on a predictive set of ten probe sets as markers for CAN (Banff 2003) in biopsies 6 months before the diagnosis of CAN [97]. However, in the last study, the authors showed the potential of identifying markers of graft injury leading to IF/TA in early protocol biopsies, demonstrating that these processes already occur before they are detectable with conventional histopathological examination of renal allografts. However, a limitation of the study is the small sample size and these results need to be validated in prospective studies with a higher number of patients.
Mengel et al. described that the atrophy-scarring associated with atrophy-fibrosis correlated with transcripts associated with B cells, plasma cells and mast cells, as well as other transcripts of unknown significance [101]. Furthermore, in a recent publication, Einecke et al. used gene-expression profiles from kidney allograft biopsies for cause to establish a molecular score to predict graft dysfunction [102]. A total of 105 for cause biopsies taken between 1 and 31 years following transplantation were evaluated. Using supervized principal components analysis, the authors derived a molecular classifier to predict graft loss. The genes associated with graft failure were related to tissue injury, epithelial dedifferentiation, matrix remodeling and TGF-β effects, and showed little overlap with rejection-associated genes. The molecular risk score was correlated with IF, TA, tubulitis, interstitial inflammation, proteinuria and GFR. In an independent validation set, the molecular-risk score was the only predictor of graft loss. Based on the previously described findings, the authors proposed that the molecular-risk score reflects active injury and is a good predictor of graft loss.
Park et al. tested whether fibrosis with inflammation at 1 year is associated with the decline of renal function in a low-risk cohort and characterized the nature of the inflammation [103]. The authors evaluated 151 living donor, tacrolimus/ mycophenolate-treated recipients without overt risk factors for reduced graft survival. Transplants with normal histology or fibrosis alone at 1-year protocol biopsy had stable renal function between 1 and 5 years, whereas those with both fibrosis and inflammation exhibited a decline in GFR and reduced graft survival. Immunohistochemistry confirmed increased numbers of interstitial T cells and macrophages/dendritic cells in the group with both fibrosis and inflammation, and there was increased expression of transcripts related to innate and cognate immunity. Pathway- and pathologic process-specific analyses of microarray profiles revealed that potentially damaging immunologic activities were enriched among the overexpressed transcripts. Consequently, the combination of fibrosis and inflammation in 1-year protocol biopsies associates with reduced graft function and survival, as well as a rejection-like gene-expression signature, even among recipients with no clinical risk factors for poor outcome. This study only included living-donor recipients. These patients represent the group with better graft survival at 5 years post-transplantation and have a lower incidence of chronic allograft dysfunction. Further evaluation in deceased-donor recipients is needed to validate these results.
There have also been some attempts to identify noninvasive biomarkers of IF/TA. To this end, Kurian et al. aimed to discover biomarkers in the peripheral blood of kidney transplant patients with biopsy-documented IF/TA (unknown cause) [104]. The authors purposely integrated the results of two independently collected sets of patient samples that were significantly different in multiple clinical elements. Thus, the selection of biomarker candidates is not significantly influenced by the time of biopsy (ranging from 1 to 6 years post-transplant), the specific immunosuppressive protocols (use of different calcineurin inhibitors vs sirolimus) or the technology used to purify the mRNA transcripts (density gradient-separated cells vs whole blood). The results from this study design indicated that whole-genome profiling is certainly not necessary as we obtain very reasonable predictive accuracy, sensitivity and specificity with 150, 100 and 50 total genes per signature. There are now several technology platforms perfectly suitable for clinical service implementation that can measure 100 genes or more cost effectively and within hours. As for application to clinical practice, the authors proposed that the model would be serial, prospective measurements of the signature at regular intervals for the life of the kidney transplant.
In order to identify noninvasive markers of allograft function in kidney transplant patients, our group’s mRNA levels of angiotensinogen (AGT), TGF-β1, EGF receptor (EGFR), IFN-γ thrombospondin (TSP)-1 and IL-10 in urine samples were studied using QRT-PCR [105]. A total of 95 kidney transplant recipients and 111 urine samples were evaluated. Patients were divided as follows: within 6 months (n = 31) and with more than 6 months post-kidney transplantation (n = 64). Kidney transplant patients with more than 6 months post-kidney transplantation were classified as transplant patients with stable kidney function (SKF; n = 32), kidney transplant patients with SKF (creatinine <2 mg/dl) and proteinuria >500 mg/24 h (n = 18), and transplant patients with biopsy-proven IF/TA (n = 14). IL-10 mRNA was decreased in urine samples from transplant patients with less than 6 months post-transplantation (p = 0.005). For transplant patient groups with more than 6 months post-transplantation, AGT and EGFR mRNA were statistically different among kidney transplant patients with SKF, kidney transplant patients with SKF and proteinuria, and IF/TA patients (p = 0.003 and p = 0.01), with transplant patients with SKF having a higher mean expression. TSP-1 mRNA levels were also significantly different among these three groups (p = 0.04), with higher expression observed in IF/TA patients. Using the random forest algorithm, AGT, EGFR and TGF-β1 were identified as predictors of IF/TA, SKF, and SKF with proteinuria. We were able to identify a characteristic pattern of mRNA levels in the different kidney transplant patients groups in urine samples that might reflect allograft function.
Biomarkers of graft tolerance
Currently, a number of studies are trying to identify ‘tolerance signatures’ that might allow a biomarker-based reduction or even cessation of immunosuppressive therapy [95,96]. However, the findings are still preliminary and based on a very small number of highly selected patients. Hence, an application of the identified profiles to actually change therapies is still too premature [106].
One of the principal components in the process leading to allograft loss is the immunologically mediated allograft damage. This immune response is mainly driven by HLA differences between the donor and recipient [107]. To control this sustained response to the graft, allograft recipients require long-term immunosuppression, which results in an increased susceptibility to infection, increased risk of malignancy and damage to graft because of nephrotoxicity. These adverse effects contribute substantially to morbidity and mortality among transplant recipients and limit long-term graft survival [108].
Determining which recipients would benefit from withdrawal or minimization of immunosuppression would be greatly facilitated by biomarkers predictive of tolerance. The search for biomarkers that can be used to identify patients that are tolerant has been encouraged by two recent studies. In the first by Newell et al., the gene-expression profile and peripheral blood lymphocyte subsets of 25 renal transplant patients with stable renal function but no immunosuppression for at least 1 year were compared with 33 stable renal allograft patients on immunosuppression, and 42 healthy controls [109]. Renal allograft tolerance was associated with a signature that included elevated levels of peripheral, naive and transitional B cells, and upregulation of a set of three genes associated with B-cell differentiation. These results were confirmed and enlarged in a second independent study of bioassays and biomarkers by Sagoo et al. [110]. In this study, microarray analysis further revealed a bias toward differential expression of B-cell-related genes and their associated molecular pathways in tolerant recipients. Furthermore, the authors were able to identify tolerant recipients in both the training and the test set. These results support the hypothesis that biomarkers, including the B-cell signatures developed by these two reports [109,110], may help to direct drug withdrawal in selected stable patients, limiting toxicity caused by long-term treatment with immunosuppressive agents. However, only appropriately designed, prospective, randomized trials will determine whether these promising markers can be applied to daily clinical patient care.
Genome-wide association studies in kidney transplantation
Differences in DNA range from a single nucleotide base change to large-scale chromosome rearrangements. There have been an important number of publications describing genetic variability in molecules affecting innate and adaptive immunity, pharmacogenetics and other nonimmunological molecules [111,112]. Studies have indicated some associations between polymorphisms in these candidate genes with outcomes in organ transplantation, and underlined a potential role of genetic variability in transplantation.
Specifically, cytokine genes and innate immune response molecules present popular targets for studies of AR and long-term outcome. Pharmacogenomic studies are mostly focused on the genes of drug targets or the corresponding enzymes metabolizing the drug. Most of the studies are based on recipient, not donor, genotype analysis. Some of the reports might be affected by insufficient study design, including small sample size, lack of adjustment for potential confounders, and multiple comparisons [111–113].
The nature of linkage analysis data means that genomic regions of interest are typically large and may contain many genes. Genome-wide association (GWA) may be used as a complementary approach to linkage studies or where robust linkage analysis is not feasible.
Technical innovations have reduced the cost of single nucleotide polymorphism (SNP)-based genotyping and, together with improved bioinformatic and genetic statistical tools, have made large-scale GWA screens possible. Guidelines have been published for Strengthening the Reporting of Genetic Association (STREGA) studies [114].
There is at least one ongoing multicenter GWA study in clinical kidney transplantation (The United Kingdom and Ireland Renal Transplant Consortium [202]). This study aims to identify the genes that contribute to progressive transplant failure and early kidney transplant damage. The GWA studies have important potential in the discovery of biomarkers. However, to be clinically applicable, large prospective studies must be performed to better define the potential benefits of genotyping on these genetic markers and clinical outcomes. Future research should be directed towards better designed studies, larger sample sizes, and evaluating both recipient and donor genotypes.
Expert commentary
As a consequence of the availability of human genome data and the new technology supporting advances in genomics, proteomics and metabolomics techniques, translational research investigators have great opportunities to discover biomarkers for various health issues, including organ transplantation. The need for better diagnostic and prognostic tools to monitor kidney graft status, as well as treatment course and efficacy, remains one of the most critical requirements in the field. An ideal biomarker would be able to predict outcome onset and the severity of specific events, such as progression to chronic allograft dysfunction even before the damage is evident at the histological level, as well as predict the specific injury responses related to the toxicity associated with the use of immunosuppressive treatment. Despite the important advances achieved so far in the identification of several potentially useful biomarkers of tolerance, rejection and progression to graft dysfunction, validation and demonstration of their clinical utility still needs to be performed. Eventually, in order to facilitate the accomplishment of such important goals, all efforts should probably be performed in the context of multidisciplinary and multi-institutional cooperative networks.
Five-year view
Important progress has been made in improving short-term outcomes in kidney transplantation during the last decades, largely as a result of better use of novel immunosuppressive drugs. However, long term outcomes have not changed. One key reason is the lack of objective biomarkers accurately reflecting allograft status and reliably predicting outcome for the single patient. Successful medicine is practiced by measuring biological markers associated with a specific disease and transplant medicine needs these biomarkers. The new technologies of genomics, metabolomics and proteomics are extremely sensitive allowing the detection of changes in tissue homeostasis associated with disease. However, the lack of rigorous clinical end points and robust gold standards might prevent the full exploitation of the diagnostic and predictive potential of the new technology and might hold back the identification of objective biomarkers. The major challenge to achieve the goal of having accurate biomarkers for monitoring graft function will be to establish accurate study end points, identify homogeneous and comparable patient and control powered groups and to be able to integrate the new methods with the existing diagnostic and theraputic tools. This will require the combined, collaborative effort of multi-institutional and multidisciplinary teams. With this, there is a high possibility of identifying those really needed biomarkers in the kidney as well as other solid organ transplant fields in the next few years.
Key issues.
Kidney transplantation is the treatment of choice for end-stage kidney disease.
Progress in short-term outcomes have not translated into better long-term graft survival and long-term outcomes, with chronic allograft dysfunction, remain the most critical problem post-transplantation.
There are many new approaches that have been able to provide information about the molecular mechanisms involved in the response of the graft to injury. However, none of the candidate markers are currently used for clinical monitoring of the kidney transplant recipients.
Existing evidence suggests significant limitations of currently used clinical end points.
The kidney allograft biopsy still is the ‘gold standard’ for monitoring the graft. However, it is an invasive method associated with patient morbidity and furthermore, with sampling and diagnosis error.
There is a critical need for accurate noninvasive biomarkers to replace the renal transplant biopsy as the gold standard.
Prospective studies with adequate sample size and patient follow-up, as well as external appropriate validation, will biomarkers that might be used to monitor patients in the clinical side.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
The research results included in this report were partially supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R01DK080074. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342(9):605–612. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- 2.Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4(3):378–383. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- 3.Solez K, Vincenti F, Filo RS. Histopathologic findings from 2-year protocol biopsies from a U.S. multicenter kidney transplant trial comparing tarolimus versus cyclosporine: a report of the FK506 Kidney Transplant Study Group. Transplantation. 1998;66 (12):1736–1740. doi: 10.1097/00007890-199812270-00029. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham MJ. Genomics and proteomics: the new millennium of drug discovery and development. J Pharmacol Toxicol Methods. 2000;44(1):291–300. doi: 10.1016/s1056-8719(00)00111-8. [DOI] [PubMed] [Google Scholar]
- 5.Ashton-Chess J, Giral M, Soulillou JP, Brouard S. Using biomarkers of tolerance and rejection to identify high- and low-risk patients following kidney transplantation. Transplantation. 2009;87(9 Suppl):S95–S99. doi: 10.1097/TP.0b013e3181a2e295. [DOI] [PubMed] [Google Scholar]
- 6.Keown PA, McMaster WR, McManus BM. Tools to identify organ rejection and immune quiescence for biological understanding and personalized medical care. Biomark Med. 2010;4(1):115–121. doi: 10.2217/bmm.09.73. [DOI] [PubMed] [Google Scholar]
- 7.Sigdel TK, Sarwal MM. The proteogenomic path towards biomarker discovery. Pediatr Transplant. 2008;12(7):737–747. doi: 10.1111/j.1399-3046.2008.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biomarkers Definitions Working Group. Biomarkers and surrogate end points: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie A, Lee IJ. Biomarkers in renal transplantation. Biomark Med. 2008;2(6):603–612. doi: 10.2217/17520363.2.6.603. [DOI] [PubMed] [Google Scholar]
- 10.Fitzsimmons W, Thompson D, Hariharan S, Van Veldhuisen P. Serum creatinine as a surrogate end point for graft loss in kidney transplantation: validation efforts from multicentre trials. Am J Transplant. 2002;2 (Suppl 3):272. [Google Scholar]
- 11.Kaplan B, Schold J, Srinivas T, et al. Effect of sirolimus withdrawal in patients with deteriorating renal function. Am J Transplant. 2004;4(10):1709–1712. doi: 10.1111/j.1600-6143.2004.00569.x. [DOI] [PubMed] [Google Scholar]
- 12.Schold JD, Kaplan B. The elephant in the room: failings of current clinical end points in kidney transplantation. Am J Transplant. 2010;10(5):1163–1166. doi: 10.1111/j.1600-6143.2010.03104.x. [DOI] [PubMed] [Google Scholar]
- 13.Poggio ED, Batty DS, Flechner SM. Evaluation of renal function in transplantation. Transplantation. 2007;84(2):131–136. doi: 10.1097/01.tp.0000269108.59275.dc. [DOI] [PubMed] [Google Scholar]
- 14.Goerdt PJ, Heim-Duthoy KL, Macres M, Swan SK. Predictive performance of renal function estimate equations in renal allografts. Br J Clin Pharmacol. 1997;44:261–265. doi: 10.1046/j.1365-2125.1997.t01-1-00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoves J, Lindley EJ, Barnfield MC, Burniston MT, Newstead CG. MDRD equation estimates of glomerular filtration rate in potential living kidney donors and renal transplant recipients with impaired graft function. Nephrol Dial Transplant. 2002;17:2036–2037. doi: 10.1093/ndt/17.11.2036. [DOI] [PubMed] [Google Scholar]
- 16.Manotham K, Booranalertpaisarn V, Eiam-Ong S, Chusil S, Praditpornsilpa K, Tungsanga K. Accurately simple estimation of glomerular filtration rate in kidney transplant patients. Transplant Proc. 2002;34:1148–1151. doi: 10.1016/s0041-1345(02)02788-4. [DOI] [PubMed] [Google Scholar]
- 17.Mariat C, Alamartine E, Barthelemy JC, et al. Assessing renal graft function in clinical trials: can tests predicting glomerular filtration rate substitute for a reference method? Kidney Int. 2004;65(1):289–297. doi: 10.1111/j.1523-1755.2004.00350.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaspari F, Ferrari S, Stucchi N, et al. Performance of different prediction equations for estimating renal function in kidney transplantation. Am J Transplant. 2004;4(11):1826–1835. doi: 10.1111/j.1600-6143.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- 19.Bosma RJ, Doorenbos CR, Stegeman CA, van der Heide JJ, Navis G. Predictive performance of renal function equations in renal transplant recipients: an analysis of patient factors in bias. Am J Transplant. 2005;5(9):2193–2203. doi: 10.1111/j.1600-6143.2005.00982.x. [DOI] [PubMed] [Google Scholar]
- 20.Pascual M, Vallhonrat H, Cosimi AB, et al. The clinical usefulness of the renal allograft biopsy in the cyclosporine era: a prospective study. Transplantation. 1999;67(5):737–741. doi: 10.1097/00007890-199903150-00016. [DOI] [PubMed] [Google Scholar]
- 21.Furness PN. Predicting allograft survival: abundant data, but insufficient knowledge? Transplantation. 2007;83(6):681. doi: 10.1097/01.tp.0000262005.84789.de. [DOI] [PubMed] [Google Scholar]
- 22•.Rush D. Can protocol biopsy better inform our choices in renal transplantation? Transplant Proc. 2009;41(6 Suppl):S6–S8. doi: 10.1016/j.transproceed.2009.06.092. Interesting article about the impact of protocol biopsies in discerning allograft status and its impact on biomarker discovery. [DOI] [PubMed] [Google Scholar]
- 23.Piovesan AC, Lucon AM, David DS, Nahas WC, Antonopoulos IM, Srougi M. Multifocal renal allograft biopsy: impact on therapeutic decisions. Transplant Proc. 2008;40(10):3397–3400. doi: 10.1016/j.transproceed.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 24.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5(6):463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Gruttola VG, Clax P, DeMets DL, et al. Considerations in the evaluation of surrogate end points in clinical trials. Summary of a National Institutes of Health workshop. Control Clin Trials. 2001;22(5):485–502. doi: 10.1016/s0197-2456(01)00153-2. [DOI] [PubMed] [Google Scholar]
- 26.Daemen A, Gevaert O, Ojeda F, et al. A kernel-based integration of genome-wide data for clinical decision support. Genome Med. 2009;1(4):39. doi: 10.1186/gm39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losko S, Heumann K. Semantic data integration and knowledge management to represent biological network associations. Methods Mol Biol. 2009;563:241–258. doi: 10.1007/978-1-60761-175-2_13. [DOI] [PubMed] [Google Scholar]
- 28.Furness PN. Predicting allograft survival: abundant data, but insufficient knowledge? Transplantation. 2007;83(6):681. doi: 10.1097/01.tp.0000262005.84789.de. [DOI] [PubMed] [Google Scholar]
- 29.Kong X, Mas V, Archer KJ. A non-parametric meta-analysis approach for combining independent microarray datasets: application using two microarray datasets pertaining to chronic allograft nephropathy. BMC Genomics. 2008;26(9):98. doi: 10.1186/1471-2164-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan AA, Khatri P, Jones RH, Sarwal MM, Butte AJ. Comparison of multiplex meta analysis techniques for understanding the acute rejection of solid organ transplants. BMC Bioinformatics. 2010;11(Suppl 9):S6. doi: 10.1186/1471-2105-11-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockley SL, Mathijs K, Staal YC, et al. Interlaboratory and interplatform comparison of microarray gene expression analysis of HepG2 cells exposed to benzo(a) pyrene. OMICS. 2009;13(2):115–125. doi: 10.1089/omi.2008.0060. [DOI] [PubMed] [Google Scholar]
- 32.Pedotti P, ‘t Hoen PA, Vreugdenhil E, et al. Can subtle changes in gene expression be consistently detected with different microarray platforms? BMC Genomics. 2008;10(9):124. doi: 10.1186/1471-2164-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan J, Feng Y, Niu YS. Nonparametric estimation of genewise variance for microarray data. Ann Stat. 2010;38(5):2723–2750. doi: 10.1214/10-AOS802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Campbell G, Jones WD, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato F, Tsuchiya S, Terasawa K, Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One. 2009;4(5):e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao S, Wang C, Dong G. Evaluation of inter-laboratory and cross-platform concordance of DNA microarrays through discriminating genes and classifier transferability. J Bioinform Comput Biol. 2009;7(1):157–173. doi: 10.1142/s0219720009004011. [DOI] [PubMed] [Google Scholar]
- 37.Sarwal MM. Deconvoluting the ‘omics’ for organ transplantation. Curr Opin Organ Transplant. 2009;14(5):544–551. doi: 10.1097/MOT.0b013e32833068fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irish WD, Ilsley JN, Schnitzler MA, Feng S, Brennan DC. A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant. 2010;10(10):2279–2286. doi: 10.1111/j.1600-6143.2010.03179.x. [DOI] [PubMed] [Google Scholar]
- 39.Bartlett ST, Schweitzer EJ, Cooper M. Prediction model for delayed kidney transplant function: no need for new regulation. Am J Transplant. 2010;10(10):2191–2192. doi: 10.1111/j.1600-6143.2010.03253.x. [DOI] [PubMed] [Google Scholar]
- 40.Mühlberger I, Perco P, Fechete R, Mayer B, Oberbauer R. Biomarkers in renal transplantation ischemia reperfusion injury. Transplantation. 2009;88(3 Suppl):S14–S19. doi: 10.1097/TP.0b013e3181af65b5. [DOI] [PubMed] [Google Scholar]
- 41.Basile DP. Novel approaches in the investigation of acute kidney injury. J Am Soc Nephrol. 2007;18(1):7–9. doi: 10.1681/ASN.2006111228. [DOI] [PubMed] [Google Scholar]
- 42.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84(3):353–361. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 43.Kainz A, Mitterbauer C, Hauser P, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4(10):1595–1604. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 44.Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8(1):78–85. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 45.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85(4):626–635. doi: 10.1097/TP.0b013e318165491f. [DOI] [PubMed] [Google Scholar]
- 46.Naesens M, Sarwal M. Looking into the crystal chip: can microarrays predict graft function? Transplantation. 2008;85(4):499–500. doi: 10.1097/TP.0b013e31816548c8. [DOI] [PubMed] [Google Scholar]
- 47.Korbély R, Wilflingseder J, Perco P, et al. Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transpl Int. 2010;24(2):143–149. doi: 10.1111/j.1432-2277.2010.01162.x. [DOI] [PubMed] [Google Scholar]
- 48.Kainz A, Wilflingseder J, Mitterbauer C, et al. Steroid pretreatment of organ donors to prevent postischemic renal allograft failure: a randomized, controlled trial. Ann Intern Med. 2010;153(4):222–230. doi: 10.7326/0003-4819-153-4-201008170-00003. [DOI] [PubMed] [Google Scholar]
- 49.Wilflingseder J, Kainz A, Mühlberger I, et al. Impaired metabolism in donor kidney grafts after steroid pretreatment. Transpl Int. 2010;23(8):796–804. doi: 10.1111/j.1432-2277.2010.01053.x. [DOI] [PubMed] [Google Scholar]
- 50.Hall IE, Yarlagadda SG, Coca SG, et al. IL-18 and urinary NGAL predict dialysis and graft recovery after kidney transplantation. J Am Soc Nephrol. 2010;21(1):189–197. doi: 10.1681/ASN.2009030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra J, Ma Q, Kelly C, et al. Kidney NGAL is a novel early marker of acute injury following transplantation. Pediatr Nephrol. 2006;21(6):856–863. doi: 10.1007/s00467-006-0055-0. [DOI] [PubMed] [Google Scholar]
- 52.Zhang PL, Rothblum LI, Han WK, Blasick TM, Potdar S, Bonventre JV. Kidney injury molecule-1 expression in transplant biopsies is a sensitive measure of cell injury. Kidney Int. 2008;73(5):608–614. doi: 10.1038/sj.ki.5002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nijboer WN, Schuurs TA, Damman J, et al. Kidney injury molecule-1 is an early noninvasive indicator for donor brain death-induced injury prior to kidney transplantation. Am J Transplant. 2009;9(8):1752–1759. doi: 10.1111/j.1600-6143.2009.02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coca SG, Yalavarthy R, Concato J, Parikh CR. Biomarkers for the diagnosis and risk stratification of acute kidney injury: a systematic review. Kidney Int. 2008;73(9):1008–1016. doi: 10.1038/sj.ki.5002729. [DOI] [PubMed] [Google Scholar]
- 55.Hartono C, Muthukumar T, Suthanthiran M. Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant. 2010;15(1):35–41. doi: 10.1097/MOT.0b013e3283342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 57.Sorof JM, Vartanian RK, Olson JL, et al. Histopathological concordance of paired renal allograft biopsy cores. Effect on the diagnosis and management of acute rejection. Transplantation. 1995;60:1215–1219. [PubMed] [Google Scholar]
- 58.Furness PN, Taub N. International variation in the interpretation of renal transplant biopsies: report of the CERTRAP project. Kidney Int. 2001;60:1998–2012. doi: 10.1046/j.1523-1755.2001.00030.x. [DOI] [PubMed] [Google Scholar]
- 59•.Sarwal M, Fernandez-Fresnedo G, Rodrigo E, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349(2):125–138. doi: 10.1056/NEJMoa035588. Pioneer article on the use of microarrays for detecting gene-expression profiles in allograft biopsies of patients with acute rejection. [DOI] [PubMed] [Google Scholar]
- 60.Reeve J, Einecke G, Mengel M, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am J Transplant. 2009;9(8):1802–1810. doi: 10.1111/j.1600-6143.2009.02694.x. [DOI] [PubMed] [Google Scholar]
- 61.Desvaux D, Schwarzinger M, Pastural M, et al. Molecular diagnosis of renal-allograft rejection: correlation with histopathologic evaluation and antirejection-therapy resistance. Transplantation. 2004;78(5):647–653. doi: 10.1097/01.tp.0000133530.26680.dc. [DOI] [PubMed] [Google Scholar]
- 62.Chua MS, Mansfield E, Sarwal M. Applications of microarrays to renal transplantation: progress and possibilities. Front Biosci. 2003;8:S913–S923. doi: 10.2741/1175. [DOI] [PubMed] [Google Scholar]
- 63.Jurcevic S, Sacks S. Gene microarrays in transplantation. Curr Opin Nephrol Hypertens. 2003;12(6):577–579. doi: 10.1097/00041552-200311000-00001. [DOI] [PubMed] [Google Scholar]
- 64.Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 2009;24(9):1643–1659. doi: 10.1007/s00467-008-0808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4(9):1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 67.Yannaraki M, Rebibou JM, Ducloux D, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006;19(9):759–768. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 68.Vasconcellos LM, Schachter AD, Zheng XX, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66(5):562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 69.Ting YT, Coates PT, Marti HP, et al. Urinary soluble HLA-DR is a potential biomarker for acute renal transplant rejection. Transplantation. 2010;89(9):1071–1078. doi: 10.1097/TP.0b013e3181d15492. [DOI] [PubMed] [Google Scholar]
- 70.Nickerson P. Post-transplant monitoring of renal allografts: are we there yet? Curr Opin Immunol. 2009;21(5):563–568. doi: 10.1016/j.coi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 71.Schaub S, Wilkins JA, Nickerson P. Proteomics and renal transplantation: searching for novel biomarkers and therapeutic targets. Contrib Nephrol. 2008;160:65–75. doi: 10.1159/000125934. [DOI] [PubMed] [Google Scholar]
- 72.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol. 2004;15(1):219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 73.Sigdel TK, Kaushal A, Gritsenko M, et al. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl. 2010;4(1):32–47. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tian J, Shi WF, Zhang LW, et al. HLA class I (ABC) upregulation on peripheral blood CD3+/CD8+ T lymphocyte surface is a potential predictor of acute rejection in renal transplantation. Transplantation. 2009;88(12):1393–1397. doi: 10.1097/TP.0b013e3181bc5c94. [DOI] [PubMed] [Google Scholar]
- 75.Hoefig KP, Heissmeyer V. MicroRNAs grow up in the immune system. Curr Opin Immunol. 2008;20:281–287. doi: 10.1016/j.coi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Lodish HF, Zhou B, Liu G, et al. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 77.Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translational and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 78.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008;19(1):81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 79•.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. Pioneer article in the elucidation of miRNA profiles associated with acute renal allograft rejection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anglicheau D, Muthukumar T, Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation. 2010;90(2):105–112. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serón D, Moreso F, Ramón JM, et al. Protocol renal allograft biopsies and the design of clinical trials aimed to prevent or treat chronic allograft nephropathy. Transplantation. 2000;69(9):1849–1855. doi: 10.1097/00007890-200005150-00019. [DOI] [PubMed] [Google Scholar]
- 82.de Matos AC, Câmara NO, de Oliveira AF, et al. Functional and morphologic evaluation of kidney proximal tubuli and correlation with renal allograft prognosis. Transpl Int. 2010;23(5):493–499. doi: 10.1111/j.1432-2277.2009.01005.x. [DOI] [PubMed] [Google Scholar]
- 83.Vazquez MA, Jeyarajah DR, Kielar ML, Lu CY. Long-term outcomes of renal transplantation: a result of the original endowment of the donor kidney and the inflammatory response to both alloantigens and injury. Curr Opin Nephrol Hypertens. 2000;9(6):643–648. doi: 10.1097/00041552-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 84.Giral M, Nguyen JM, Karam G, et al. Impact of graft mass on the clinical outcome of kidney transplants. J Am Soc Nephrol. 2005;16(1):261–268. doi: 10.1681/ASN.2004030209. [DOI] [PubMed] [Google Scholar]
- 85.Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349(24):2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 86.2008 Annual Report of the US Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients: transplant data 1998–2007. US Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau, Division of Transplantation; Rockville, MD, USA: 2008. [Google Scholar]
- 87.Halloran PF, Langone AJ, Helderman JH, Kaplan B. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am J Transplant. 2004;4(11):1729–1730. doi: 10.1111/j.1600-6143.2004.00662.x. [DOI] [PubMed] [Google Scholar]
- 88•.Mengel M. The kidney transplant: new horizons. Curr Opin Nephrol Hypertens. 2010;19(3):260–265. doi: 10.1097/MNH.0b013e3283381ea5. Very interesting article on current knowledge and future applications. [DOI] [PubMed] [Google Scholar]
- 89••.Bestard O, Cruzado JM, la Franquesa M, Grinyó JM. Biomarkers in renal transplantation. Curr Opin Organ Transplant. 2010;15(4):467–473. doi: 10.1097/MOT.0b013e32833b9ccb. Interesting article about biomarkers in transplantation and their associated advantages and limitations. [DOI] [PubMed]
- 90.Serón D, Moreso F, Ramón JM, et al. Protocol renal allograft biopsies and the design of clinical trials aimed to prevent or treat chronic allograft nephropathy. Transplantation. 2000;69(9):1849–1855. doi: 10.1097/00007890-200005150-00019. [DOI] [PubMed] [Google Scholar]
- 91.de Matos AC, Câmara NO, de Oliveira AF, et al. Functional and morphologic evaluation of kidney proximal tubuli and correlation with renal allograft prognosis. Transpl Int. 2010;23(5):493–499. doi: 10.1111/j.1432-2277.2009.01005.x. [DOI] [PubMed] [Google Scholar]
- 92.Rosen S, Stillman IE. Acute tubular necrosis is a syndrome of physiologic and pathologic dissociation. J Am Soc Nephrol. 2008;19(5):871–875. doi: 10.1681/ASN.2007080913. [DOI] [PubMed] [Google Scholar]
- 93.Schlondorff DO. Overview of factors contributing to the pathophysiology of progressive renal disease. Kidney Int. 2008;74(7):860–866. doi: 10.1038/ki.2008.351. [DOI] [PubMed] [Google Scholar]
- 94.Schröppel B, Heeger PS. Gazing into a crystal ball to predict kidney transplant outcome. J Clin Invest. 2010;120(6):1803–1806. doi: 10.1172/JCI43286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mas VR, Archer KJ, Scian M, Maluf DG. Molecular pathways involved in loss of graft function in kidney transplant recipients. Expert Rev Mol Diagn. 2010;10(3):269–284. doi: 10.1586/erm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scherer A, Gwinner W, Mengel M, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol Dial Transplant. 2009;24(8):2567–2575. doi: 10.1093/ndt/gfp183. [DOI] [PubMed] [Google Scholar]
- 97.Scherer A, Krause A, Walker JR, Korn A, Niese D, Raulf F. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003;75(8):1323–1330. doi: 10.1097/01.TP.0000068481.98801.10. [DOI] [PubMed] [Google Scholar]
- 98.Hotchkiss H, Chu TT, Hancock WW, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation. 2006;81(3):342–349. doi: 10.1097/01.tp.0000195773.24217.95. [DOI] [PubMed] [Google Scholar]
- 99.Mas V, Maluf D, Archer K, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 2007;83(4):448–457. doi: 10.1097/01.tp.0000251373.17997.9a. [DOI] [PubMed] [Google Scholar]
- 100.Maluf DG, Mas VR, Archer KJ, et al. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med. 2008;14(5–6):276–285. doi: 10.2119/2007-00111.Maluf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mengel M, Reeve J, Bunnag S, Einecke G, et al. Molecular correlates of scarring in kidney transplants: the emergence of mast cell transcripts. Am J Transplant. 2009;9(1):169–178. doi: 10.1111/j.1600-6143.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- 102.Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120(6):1862–1872. doi: 10.1172/JCI41789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010;21(11):1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurian SM, Heilman R, Mondala TS, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4(7):e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mas VR, Mas LA, Archer KJ, et al. Evaluation of gene panel mRNAs in urine samples of kidney transplant recipients as a non-invasive tool of graft function. Mol Med. 2007;13(5–6):315–324. doi: 10.2119/2007-00017.Mas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schröppel B, Heeger PS. Gazing into a crystal ball to predict kidney transplant outcome. J Clin Invest. 2010;120(6):1803–1806. doi: 10.1172/JCI43286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Afzali B, Lombardi G, Lechler RI. Pathways of major histocompatibility complex allorecognition. Curr Opin Organ Transplant. 2008;13:438–444. doi: 10.1097/MOT.0b013e328309ee31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernandez-Fuentes MP, Lechler RI. A ‘biomarker signature’ for tolerance in transplantation. Nat Rev Nephrol. 2010;6(10):606–613. doi: 10.1038/nrneph.2010.112. [DOI] [PubMed] [Google Scholar]
- 109.Newell KA, Asare A, Kirk AD, et al. Immune Tolerance Network ST507 Study Group. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120(6):1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sagoo P, Perucha E, Sawitzki B. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest. 2010;120(6):1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zaza G, Granata S, Sallustio F, Grandaliano G, Schena FP. Pharmacogenomics: a new paradigm to personalize treatments in nephrology patients. Clin Exp Immunol. 2010;159(3):268–280. doi: 10.1111/j.1365-2249.2009.04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goldfarb-Rumyantzev AS, Naiman N. Genetic prediction of renal transplant outcome. Curr Opin Nephrol Hypertens. 2008;17(6):573–579. doi: 10.1097/MNH.0b013e32830f4579. [DOI] [PubMed] [Google Scholar]
- 113.McKnight AJ, Currie D, Maxwell AP. Unravelling the genetic basis of renal diseases; from single gene to multifactorial disorders. J Pathol. 2010;220(2):198–216. doi: 10.1002/path.2639. [DOI] [PubMed] [Google Scholar]
- 114.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the Reporting of Genetic Association Studies (STREGA) – an extension of the STROBE statement. Genet Epidemiol. 2009;33:581–598. doi: 10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
Websites
- 201.Human Genome Project Information. www.ornl.gov/sci/techresources/Human_Genome/project/info.shtml.
- 202.WTCCC3 – Wellcome Trust Case–Control Consortium 3: defining the genetic basis of interactions between donor and recipient DNA that determine early and late renal transplant dysfunction. 2010 www.ukirtc.org/site.