Abstract
During the torpor phase of mammalian hibernation when core body temperature is near 4°C, the autonomic system continues to maintain respiration, blood pressure and heartbeat despite drastic reductions in brain activity. Additionally, the hibernator’s neuronal tissues enter into a protected state in which the potential for ischemia-reperfusion injury is markedly minimized. Evolutionary adaptations for continued function and neuroprotection throughout cycles of torpor and euthermia in winter are predicted to manifest themselves partly in changes in the brainstem proteome. Here we compare the soluble brainstem protein complement from six summer active ground squirrels and six in the early torpor phase of hibernation. Thirteen percent of the ~1500 quantifiable 2D gel spots alter significantly from summer to early torpor; the proteins identified in these differing spots are known to play roles in energy homeostasis via the tricarboxylic acid cycle (eight proteins), cytoarchitecture and cell motility (14 proteins), anabolic protein processes (13 proteins), redox control (11 proteins) and numerous other categories including protein catabolism, oxidative phosphorylation, signal transduction, glycolysis, intracellular protein trafficking and antiapoptotic function. These protein changes represent, at least in part, the molecular bases for restructuring of cells in the brainstem, a shift away from glucose as the primary fuel source for brain in the winter, and the generation of a streamlined mechanism capable of efficient and rapid energy production and utilization during the torpor and arousal cycles of hibernation.
Keywords: Thirteen-lined ground squirrel, Spermophilus tridecemlineatus, Ictidomys tridecemlineatus, DIGE proteomics, hibernation
Introduction
The yearly thermoregulatory cycle of a ground squirrel includes two phases. The summer active phase is similar to that of any non-hibernating homeothermic mammal in that the core body temperature (Tb) is maintained at ~35–37°C. In contrast, the winter phase is characterized by dramatic heterothermy in which Tb drops to ~ −3°C to 5°C (torpor) with the exception of spontaneous arousals to euthermy for ~12–15 hr every 3–16 days (Fig. 1). In addition to low body temperature, the torpid animal demonstrates a tremendously reduced rate of metabolism, respiration and cardiac rhythm (reviewed in Andrews 2007; Carey et al. 2003; reviewed in Lyman et al. 1982).
Fig. 1.
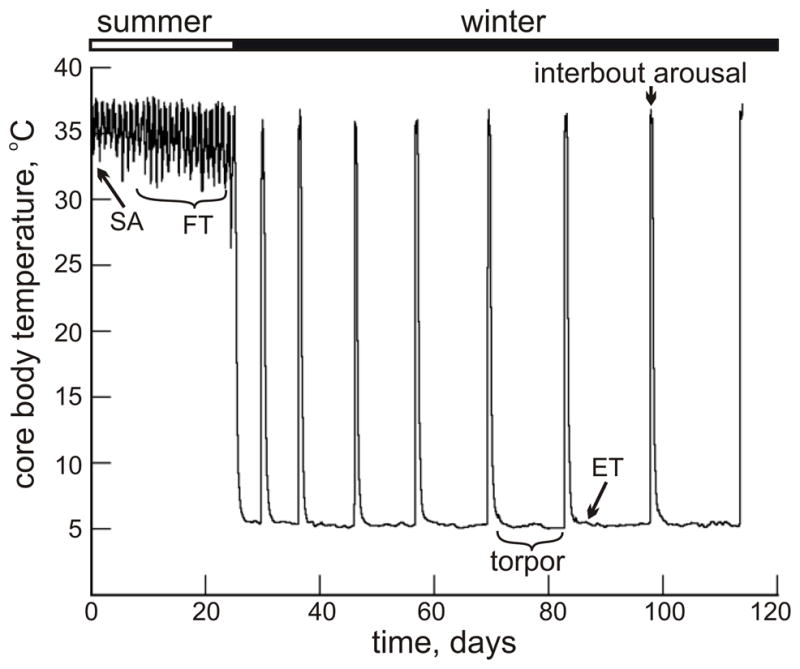
Body temperature (Tb) of a 13-lined ground squirrel for several weeks in late fall and into the first three months of winter heterothermy, i.e. hibernation; SA, summer active; FT, fall transition; ET, early torpor.
Life-sustaining brainstem activity, including regulation of heartbeat, respiration and blood pressure, must meet demands throughout the varied and dynamic phases of hibernation, and indeed distinct patterns of brainstem activity are found in torpor and arousal cycles (Bitting et al. 1994; Bratincsák et al. 2007; Kilduff et al. 1990; Kilduff et al. 1982; Milsom et al. 1999). Additionally, brain tissue assumes a state of protection in hibernation. Hippocampal slices tolerate hypoxia and aglycemia (Frerichs and Hallenbeck 1998), the inflammatory response to traumatic brain injury is minimal (Zhou et al. 2001), neuronal damage does not accrue after cerebral ischemia by cardiac arrest (Dave et al. 2006), and as metabolic rate and Tb increase during arousal from torpor, brain tissues undergo a period of mild hypoxia but remain resistant to stress and damage (Ma and Wu 2008; Ma et al. 2005; Tøien et al. 2001). Shifts in brain activity, cytoarchitecture (Arendt et al. 2003; Ruediger et al. 2007; von der Ohe et al. 2007), energy demand and protection from damage are likely to result from seasonal alterations in gene expression.
Broad-spectrum unbiased screens for changes in gene expression in hibernating sciurids across the annual cycle have been performed at the mRNA level in a variety of tissues including liver, heart, brain (reviewed in Andrews 2007; reviewed in Carey et al. 2003), and five tissues of the arctic ground squirrel (Yan et al. 2008). At the protein level, differences have been detected in hibernators using liver, heart, muscle and intestinal tissues (Eddy et al. 2005; Epperson et al. 2004; Lee et al. 2008; Martin et al. 2008; Russeth et al. 2006). Current data support a two-switch model (Serkova et al. 2007) in which a reprogramming of gene expression is the primary facilitator of the first switch from summer to winter, and the less time- and energy-consuming regulation of small biomolecules controlling protein activity drives the second switch of arousal and torpor.
To better understand the molecular events that underlie the hibernator’s ability to maintain neuronal function and resist damage from the extreme changes associated with winter heterothermy, we used a quantitative 2D gel approach to compare the proteins present in summer active (SA) ground squirrel brainstem to those in early torpor (ET). Many of the changes found are consistent with the known dramatic restructuring of cellular architecture. In addition, the ET animals demonstrated steady-state upregulation of proteins in the pyruvate dehydrogenase complex, tricarboxylic acid cycle, and oxidative phosphorylation pathways and downregulation of proteins involved in glucose metabolism. Based on these protein alterations, a model is proposed to explain the remarkable ability of hibernators to meet brain ATP requirements during arousal.
Methods
Animals and acquisition of tissue
Thirteen-lined ground squirrels (Ictidomys tridecemlineatus, recently revised from genus Spermophilus (Helgen et al. 2009)) were trapped in the summer in the vicinity of Madison, Wisconsin, and were housed, abdominally implanted with telemeters, and euthanized as described previously, (Fleck and Carey 2005). SA animals were anesthetized with isoflurane and decapitated in late June-August when Tb≈37°C, ET animals after at least 4 torpor-arousal cycles and within 24h of having cooled to a Tb of 5–7°C. Brainstems (pons and medulla only) were removed and snap frozen in liquid nitrogen. The University of Wisconsin Institutional Animal Care and Use Committee approved all animal procedures. The tissues were shipped on dry ice to the University of Colorado and stored at −80°C until use.
Sample preparation
Frozen brainstem was pulverized in liquid nitrogen. Approx. 100mg of frozen powdered tissue was taken from the total, placed in homogenization buffer and processed as previously described for liver in a Polytron (Brinkmann) (Epperson et al. 2004). The homogenate was transferred to a microfuge tube and nuclei were pelleted at 500Xg, 4°C for ten min. The post-nuclear supernatant was gently transferred to another tube and divided into 15μl aliquots. These were snap frozen in liquid nitrogen and stored at -80°C. Each aliquot was used only once to avoid freeze-thaw cycles. One aliquot from each animal was used for protein quantitation by BCA protein assay (Pierce). To prepare the reference standard, 230 μg of each sample (12 total: six SA, six ET) were combined into a single tube and mixed thoroughly. This pooled sample was divided into five aliquots, snap frozen and placed at −80°C until use.
90μg of each sample were denatured overnight at room temperature in denaturing buffer: 8M urea, 2M thiourea, 4% CHAPS, 25mM Tris pH 8.8, and subsequently labeled with Cy2, Cy3 or Cy5 (GE Healthcare). Cy2 was always used to label the reference standard and Cy3 and Cy5 were alternated between SA and ET samples to control for dye bias (“dye swap”). Labeling was done according to the manufacturer’s protocol except that 80nmol rather than 400nmol was used to label 50μg of protein. For each gel, the Cy3 and Cy5-labeled samples were combined with the reference standard (Cy2) and this combined sample was methanol/chloroform precipitated as described (Epperson et al. 2004). The pellet was resuspended in 150μl iso and 150μl 3; iso: 9M urea, 4% CHAPS, 65mM DTT, 35mM Tris base, 0.0025% bromophenol blue. 3: 7M urea, 4% CHAPS, 100mM DTT, 0.0025% bromophenol blue, 2M thiourea, 0.8% 3.5–10 ampholytes (“Resolyte”, Gallard-Schlesinger, Plainview, NY), pipetted 10X with a gel loading tip and shaken at 900rpm, 25°C in a Thermomixer (Eppendorf) for at least two hr to solubilize.
Two-dimensional gel electrophoresis
The solubilized proteins were used to swell Immobiline DryStrips pH 3–10 NL, 18cm (GE Healthcare) for 21–24h at RT, after which isoelectric focusing (IEF) and 2D separation were conducted as previously detailed (Bernard et al. 2004) with 20mg/ml fresh DTT in water added to the basic end filter paper. The IEF program was as follows with 2mA and 5W for all steps: 1) ramp to 500V, 30 min, 2) ramp to 3500V, 4hr, 3) hold at 3500V, 14–17hr total. After focusing, the strips were incubated 15 min each in reducing (50mM Tris pH 6.8, 2% SDS, 15% glycerol, 6M urea, 1% DTT) and alkylating (50mM Tris pH 6.8, 2% SDS, 15% glycerol, 6M urea, 1.25% iodoacetamide, 0.05% bromophenol blue) buffers to covalently modify sulfhydryl groups and prevent subsequent disulfide bridge formation. For the second dimension, proteins were separated by SDS-PAGE on a 9–16% acrylamide gradient, and gels were scanned in the glass plates within four hours of completion of electrophoresis.
Three channels were used on a Typhoon 9400 (GE Healthcare) to collect the Cy2, Cy3, and Cy5 images. For each gel image, a prescan was performed and the photomultiplier voltage adjusted until the spot of maximum volume on each image was reduced to approximately one-third of saturation. Images used for analysis were collected at a pixel size of 100μm. Two of these six gels were “pick” gels that were affixed to glass plates previously marked with reference markers (GE Healthcare, product #18-1143-34) and treated with bind-silane (PlusOne, GE Healthcare) for the purpose of accurate robotic picking and digestion of the protein spots. These two gels were fixed and stained with Sypro Ruby (BioRad) according to Epperson, et al. after scanning for Cy2, 3 and 5 (Epperson et al. 2004).
All 20 images (six gels with three Cy images each: Cy2, Cy3 and Cy5, and two of these same gels poststained with Sypro Ruby to make pick gel images) were imported into DeCyder 2D 6.5 software (GE) for spot matching. T-tests were run using the Cy images in which the Cy3 and Cy5 spot values (pixel volumes) were normalized to their corresponding Cy2 spot value. The BVA module also includes a statistical post hoc algorithm “False Discovery Rate” (FDR) (Benjamini and Hochberg 1995) which is a stringent modifier of p-values and greatly reduces false positives in a large set of comparisons with a relatively small sample size, the resulting probability is called a “q value”. Only spots whose t-tests retained q≤0.05 that were found to be reproducible and robust by individual inspection on all Cy2 images were excised for identification.
Spot picking and mass spectral analysis
Picks and digests were done in the UCDHSC Proteomics Shared Facility (hsc-proteomics.uchsc.edu/mscore) using an Ettan spot picker and an Ettan spot digester (GE Healthcare). Tryptic fragments were separated in a 3–45% hydrophobicity gradient of buffer B (90% ACN, 0.1% formic acid) over 45 min, followed by a 5 min wash in 90% ACN for analysis of full and tandem mass spectra by LC-MS/MS in an ESI ion trap (LC/MSD XCT Plus, Agilent Technologies) in positive ion mode in the Ultra Scan setting. A full MS scan was followed by tandem mass spectral (MS/MS) scans of the two highest peaks with a dynamic exclusion of 1 min. Raw mass spectra were analyzed using Spectrum Mill MS Proteomics Workbench Rev A.03.02.060a ETD-65. The range of mass limits for the precursor ions was 600–4000 Da, parent and fragment masses were both set to monoisotopic, precursor peptide mass tolerance was 2.5 Da, fragment ion tolerance was 0.7, the enzyme specified was trypsin, maximum number of internal missed cleavage sites was two, and cysteines were given a fixed modification of +57 for carbamidomethylation. The database used was an in-house compilation of all mammal sequences in the NCBI nr database in August 2008 and contained 704,905 entries. Spectral data from the same spot on the two pick gels were combined to increase confidence in the identification.
Protein identifications and functional assignments
Protein IDs were acquired for each spot. A program called ExtracTags was written in-house to collate necessary information from Spectrum Mill results about the peptides on which protein IDs were based. For each peptide, the NCBI GI#, species of the organism to which that peptide was matched, predicted peptide sequence, Spectrum Mill score, SPI (scored peak intensity), parent mass and charge, and delta mass were all recorded. For each protein ID obtained, a combined score from all peptides and the total amino acid coverage by those peptides were also calculated. In all tables, protein IDs relying on a single peptide match were eliminated along with any IDs that were > ±15% of their predicted molecular weight based on the second dimension size. Additionally, for spots with multiple credible identifications where one protein’s peptides demonstrated an average spectral intensity that exceeded those of another protein in the same spot by four fold or more, the protein with lower average spectral intensity was discarded (Martin et al. 2008). If the difference in both score and coverage was four-fold or greater, the lower quality identification was discarded. Trypsin and keratin were eliminated from the lists. Identifications that passed these criteria were considered valid, and spots with single valid protein IDs after these eliminations were considered unique IDs. In cases where a protein was “hypothetical” or “unnamed protein product”, the name of the human homolog as found using the NCBI BLink blast tool was used. Potential biological function was determined using NCBI’s Entrez Gene and links therein.
Western blot analysis
20μg of total brainstem protein were separated by SDS-PAGE. One of the SA samples had been depleted and was therefore omitted from the western blot analysis. The gel was cut at the 43kDa marker. The lower portion was stained using Sypro Ruby (Bio-Rad) and scanned on a Typhoon 9400 (GE) to assess gel loading consistency among lanes. Proteins in the upper gel portion were transferred to Immobilon-FL transfer membrane (Millipore), which was blocked with 3% milk, 1% BSA in TBST and incubated with OGDH goat polyclonal antibody (1:100, SC-49589, Santa Cruz Biotech) at 4°C overnight followed by anti-goat Cy5-conjugated secondary antibody (1:400, Jackson ImmunoResearch). The image was captured using a Typhoon 9400 and analyzed with ImageQuant software.
Results
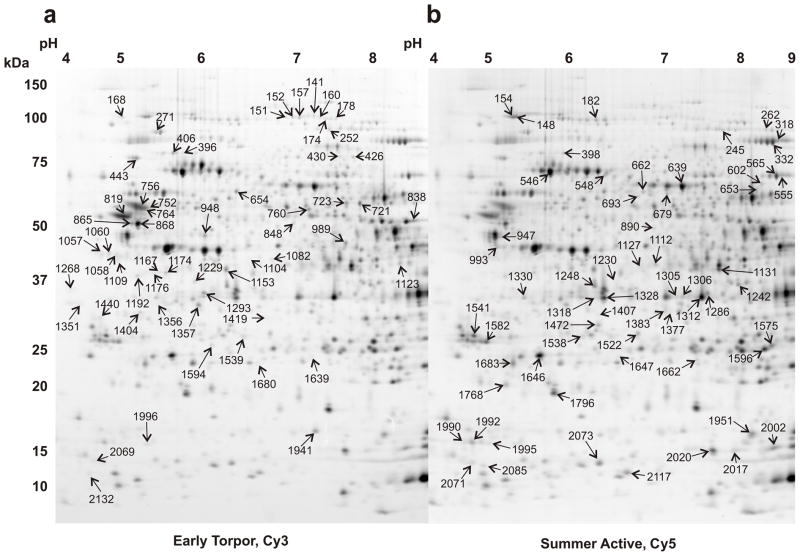
Protective and functional alterations in the brain are likely to manifest themselves in large part at the protein level in a manner that is dependent on hibernation season (Epperson and Martin 2002; Martin and Epperson 2008; von der Ohe et al. 2007; Zhao et al. 2006). Evidence from previous two-dimensional gel electrophoresis (2DE) studies suggests a loss of protein quality during torpor that is corrected during interbout arousal (Epperson et al. 2004; Martin et al. 2004). Therefore, animals in ET were used in the present study to represent the brainstem proteome of the winter season. Soluble brainstem proteins from six SA and six ET animals were resolved first by isoelectric point and then by size using CyDye labels and 2DE to assess changes in the brainstem proteome associated with the summer to winter alterations in hibernation biochemistry (Fig. 2). Biological replicates were labeled and subjected to 2DE separately rather than pooled to increase statistical power (Karp et al. 2005). The DIGE approach includes a pooled reference labeled with Cy2 which acts as a common denominator, improving quantitation and gel matching (reviewed in Marouga et al. 2005). The most populated gel, the “master” was determined to have 2314 Cy2 spots; these spots were matched to the other Cy2 images allowing for quantitative analysis of their respective Cy3 and Cy5 spots by analysis of their ratios to Cy2, i.e. Cy5/Cy2 compared to Cy3/Cy2 for a single spot. T-tests were run on 2196 spots according to the algorithm present in the DeCyder software. A post hoc multiple test correction algorithm (False Discovery Rate, FDR) was applied to reduce the occurrence of false positives, and only spots that passed this stringent filter were considered further. 633 of the 2196 t-tests demonstrated a p-value of less than 0.05; 286 of these remained significant after FDR (q< 0.05).
Fig. 2.
Brainstem protein differences between SA and ET. Two different Cy images from a single gel separation of soluble 13-lined ground squirrel brainstem proteins by 2DE are shown. (a) ET brainstem proteins labeled with Cy3, master numbers are indicated for spots elevated in ET. (b) SA proteins labeled with Cy5, master numbers are indicated for spots elevated in SA over ET. On the left side of the figure are markers of approximate molecular mass in kDa, and across the top are indicated the approximate isoelectric (pI) values in pH.
Of the 286 significantly changing spots, 206 were found to match well on the pick gels and to be reproducible on all gels. These 206 were picked, digested, and their tryptic peptides analyzed by LC-MS/MS mass spectrometry. Protein identifications were obtained for all but seven of the 206 spots analyzed. Information about each uniquely identified protein gene product was found using NCBI’s “Gene” link, and possible functional roles are listed in Tables 1 and 2. 102 (51%) of the 199 identified spots were up in ET relative to SA, 58 of which gave a unique protein ID. 97 (49%) of the 199 identified spots were down in ET, 61 of which gave a unique protein ID. Eighty spots contained multiple valid IDs. Because protein identification by this method depends on the availability of sequence information, the lack of a sequence database for this or a closely related organism lowers the recovery of exact matches. However, high quality spectral information and use of the largest mammalian database available enable positive identification of almost all protein spots with current methods (Epperson et al. 2004; Russeth et al. 2006). One hundred nineteen spots contained a single protein and these unique IDs are reported in Tables 1 and 2. Further information regarding these spots and their corresponding peptides is listed in the Supplementary Table. The fold changes for those spots that were elevated in ET were on average 17% higher than those spots found to be higher in SA (p<3X10−9, Student’s t-test; average fold change for ET, 1.55X; for SA, 1.33X). Eighty spots revealed the presence of multiple robust protein identifications, consistent with the high complexity of the starting sample. Proteins identified as changing in steady-state level encompassed many functions including roles in the citric acid cycle, cellular architecture, intracellular transport, protein metabolism and ATP synthesis.
Table 1.
Proteins higher in ET. ET, n=6; SA, n=6; q<0.05. Listed are the 58 spots for which a unique ID was recovered based on criteria summarized in methods. Columns are as follows: master number of spot (spot), average normalized volume ratio of ET to SA (fold), False Discovery Rate-modified p-value for the t-test (q), identification of match (protein ID), official symbol (gene), NCBI GenInfo Identifier (GI), species for this GI (species), molecular mass in Daltons for this GI (Da), number of distinct peptides matched (peps), Spectrum Mill protein score (score), amino acid coverage with ratio of quantity of recovered amino acids over total amino acids in the first column and percent coverage in the second column (AA coverage), possible area of function based on information from NCBI “gene” (functional grouping); Ub, ubiquitin; ER, endoplasmic reticulum; UPR, unfolded protein response; TCA, tricarboxylic acid cycle; ROS, reactive oxygen species; dev, development
| spot | fold | q | protein ID | gene | GI | species | Da | peps | score | AA coverage | functional grouping | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 141 | 1.53 | 0.02 | oxoglutarate dehydrogenase | OGDH | 29145087 | mouse | 116118 | 4 | 58 | 55/1019 | 5 | TCA cycle, lysine degradation, tryptophan metabolism |
| 151 | 1.75 | 0.048 | oxoglutarate dehydrogenase-like | OGDH | 8922716 | human | 114480 | 5 | 84 | 76/1010 | 8 | TCA cycle, lysine degradation, tryptophan metabolism |
| 152 | 1.82 | 0.012 | alpha-ketoglutarate dehydrogenase-like | OGDH | 160419019 | human | 114482 | 13 | 207 | 166/1010 | 16 | TCA cycle, lysine degradation, tryptophan metabolism |
| 157 | 1.54 | 0.033 | alpha-ketoglutarate dehydrogenase-like | OGDH | 160419019 | human | 114482 | 7 | 114 | 100/1010 | 10 | TCA cycle, lysine degradation, tryptophan metabolism |
| 160 | 1.56 | 0.012 | fragment: oxoglutarate dehydrogenase | OGDH | 90076414 | macaque | 40206 | 2 | 31 | 22/358 | 6 | TCA cycle, lysine degradation, tryptophan metabolism |
| 168 | 1.39 | 0.017 | ubiquitin specific peptidase 5 (isopeptidase T) | USP5 | 74150576 | mouse | 93355 | 9 | 136 | 109/835 | 13 | Ub mediated protein catabolism, metal ion binding |
| 174 | 1.61 | 0.012 | ogdh protein | OGDH | 29145087 | mouse | 116118 | 4 | 58 | 55/1019 | 5 | TCA cycle, lysine degradation, tryptophan metabolism |
| 178 | 1.53 | 0.02 | oxoglutarate dehydrogenase (lipoamide) | OGDH | 85861164 | mouse | 116450 | 15 | 220 | 147/1023 | 14 | TCA cycle, lysine degradation, tryptophan metabolism |
| 252 | 1.41 | 0.019 | dynamin 1 | DNM1 | 18093102 | rat | 95928 | 18 | 276 | 201/851 | 24 | receptor-mediated endocytosis |
| 271 | 1.33 | 0.045 | valosin-containing protein | VCP | 30023842 | mouse | 89364 | 23 | 354 | 303/806 | 38 | caspase activation, ER UPR, protein localization |
| 396 | 1.29 | 0.044 | aconitase 2, mitochondrial | ACO2 | 27806769 | cow | 85358 | 28 | 471 | 314/780 | 40 | TCA cycle, precursor metabolites |
| 406 | 1.63 | 0.029 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | NDUFS1 | 67970033 | macaque | 79549 | 15 | 240 | 186/727 | 26 | ETC, ATP metabolism, apoptosis, ROS metabolism |
| 426 | 1.92 | 0.012 | N-ethylmaleimide-sensitive factor | NSF | 10257494 | human | 82653 | 16 | 251 | 187/744 | 25 | protein transport, ATP binding, hydrolase activity |
| 430 | 1.87 | 0.012 | N-ethylmaleimide-sensitive factor | NSF | 10257494 | human | 82653 | 15 | 234 | 170/744 | 23 | protein transport, ATP binding, hydrolase activity |
| 443 | 1.22 | 0.038 | 78 kDa glucose-regulated protein precursor (BiP) | HSPA5 | 115502217 | ground squirrel | 72329 | 19 | 320 | 224/654 | 34 | ER overload and glucose response, anti-apoptosis |
| 654 | 1.16 | 0.024 | chaperonin containing TCP1, subunit 5 (epsilon) | CCT5 | 194676636 | cow | 59615 | 15 | 241 | 163/541 | 30 | ATP-dependent protein folding |
| 721 | 1.38 | 0.012 | dihydrolipoamide dehydrogenase | DLD | 75040928 | orangutan | 54122 | 11 | 187 | 133/509 | 26 | pyruvate, alpha-ketoglutarate dehydrogenase complex |
| 723 | 1.29 | 0.018 | dihydrolipoamide dehydrogenase | DLD | 75040928 | orangutan | 54122 | 7 | 113 | 90/509 | 18 | pyruvate, alpha-ketoglutarate dehydrogenase complex |
| 752 | 1.77 | 0.027 | tubulin, alpha 1B | TUBA1B | 34740335 | mouse | 50152 | 10 | 178 | 142/451 | 31 | microtubule-based movement, protein polymerization |
| 756 | 1.73 | 0.02 | tubulin, alpha 1 | TUBA1B | 6755901 | mouse | 50136 | 15 | 258 | 201/451 | 45 | microtubule-based movement, protein polymerization |
| 760 | 1.26 | 0.019 | chaperonin containing TCP1, subunit 2 | CCT2 | 149632291 | platypus | 60820 | 11 | 177 | 128/563 | 23 | protein folding, specifically actin and tubulin |
| 764 | 1.94 | 0.018 | tubulin alpha 6 | TUBA1C | 14389309 | human | 49896 | 12 | 199 | 180/449 | 40 | microtubule-based movement, polymerization |
| 819 | 1.68 | 0.03 | tubulin, beta 2 | TUBB2C | 4507729 | human | 49907 | 15 | 249 | 198/445 | 44 | microtubule-based movement, polymerization |
| 838 | 1.67 | 0.019 | ATP synthase subunit alpha, mitochondrial precursor | ATP5A1 | 75041057 | orangutan | 59781 | 18 | 283 | 203/553 | 37 | oxidative phosphorylation |
| 848 | 1.45 | 0.014 | dihydrolipoamide dehydrogenase-binding protein | PDHX | 90075986 | macaque | 54055 | 5 | 72 | 57/501 | 11 | pyruvate, alpha-ketoglutarate dehydrogenase complex |
| 865 | 1.97 | 0.014 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta | ATP5B | 32189394 | human | 56560 | 15 | 250 | 198/529 | 37 | oxidative phosphorylation |
| 868 | 1.79 | 0.015 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta | ATP5B | 28940 | human | 57956 | 11 | 194 | 144/539 | 27 | oxidative phosphorylation |
| 948 | 1.59 | 0.023 | ubiquinol-cytochrome c reductase core protein I | UQCRC1 | 194041191 | pig | 52699 | 14 | 225 | 174/480 | 36 | oxidative phosphorylation, electron transport chain |
| 989 | 1.46 | 0.012 | Tu translation elongation factor, mitochondrial | TUFM | 73958590 | dog | 63130 | 14 | 218 | 170/583 | 29 | translation in mitochondria |
| 1057 | 1.61 | 0.019 | N-myc downstream-regulated gene 2 isoform a | NDRG2 | 73977281 | dog | 40718 | 4 | 55 | 61/371 | 16 | neurite outgrowth |
| 1058 | 1.39 | 0.021 | N-myc downstream-regulated gene 2 isoform a | NDRG2 | 73977281 | dog | 40718 | 5 | 75 | 67/371 | 18 | neurite outgrowth |
| 1060 | 1.64 | 0.012 | fragment: 40S ribosomal protein SA (p40) | RPSA | 73986556 | dog | 29778 | 7 | 119 | 89/267 | 33 | cell growth and motility |
| 1082 | 1.33 | 0.047 | purine-rich element binding protein A | PURA | 149269977 | mouse | 33476 | 6 | 88 | 100/303 | 33 | control of DNA replication and transcription |
| 1104 | 1.66 | 0.018 | purine-rich element binding protein A | PURA | 73949258 | dog | 42792 | 4 | 53 | 46/404 | 11 | control of DNA replication and transcription |
| 1109 | 1.56 | 0.012 | arsA arsenite transporter, ATP-binding | ASNA1 | 73986513 | dog | 40091 | 7 | 102 | 70/360 | 19 | anion transport |
| 1123 | 1.62 | 0.021 | creatine kinase, mitochondrial 1B | CKMT1B | 57108147 | dog | 47076 | 9 | 137 | 93/417 | 22 | energy homeostasis |
| 1153 | 1.44 | 0.045 | NAD+-isocitrate dehydrogenase, alpha subunit | IDH3A | 1182011 | macaque | 36801 | 9 | 145 | 100/340 | 29 | tricarboxylic acid cycle, rate-limiting step |
| 1167 | 1.38 | 0.033 | guanine nucleotide binding protein, alpha o isoform B | GNAO1 | 164607137 | mouse | 40037 | 3 | 44 | 39/354 | 11 | signal transduction, G-protein coupled |
| 1174 | 1.58 | 0.044 | guanine nucleotide binding protein, alpha o isoform B | GNAO1 | 164607137 | mouse | 40037 | 9 | 137 | 110/354 | 31 | signal transduction, G-protein coupled |
| 1176 | 1.6 | 0.028 | guanine nucleotide binding protein, alpha o isoform B | GNAO1 | 164607137 | mouse | 40037 | 5 | 80 | 71/354 | 20 | signal transduction, G-protein coupled |
| 1192 | 1.19 | 0.039 | serine/threonine kinase receptor associated protein | STRAP | 57106953 | dog | 38533 | 12 | 196 | 181/350 | 52 | RNA splicing, mRNA processing, protein binding |
| 1229 | 1.99 | 0.014 | gamma-soluble NSF attachment protein | GSNAP | 73962213 | dog | 34588 | 6 | 93 | 69/311 | 22 | intracellular protein transport, Golgi and ER |
| 1268 | 1.62 | 0.024 | clathrin, light chain (Lca) | CLTA | 73971342 | dog | 27119 | 2 | 23 | 17/248 | 7 | intracellular protein transport |
| 1293 | 1.54 | 0.014 | G protein beta 1 subunit | GNB1 | 984553 | rat | 37393 | 9 | 156 | 106/340 | 31 | signal transduction, G-protein coupled |
| 1351 | 1.9 | 0.013 | clathrin, light chain (Lca) | CLTA | 73953331 | dog | 23174 | 3 | 42 | 29/207 | 14 | intracellular protein transport |
| 1356 | 1.61 | 0.013 | N-ethylmaleimide-sensitive factor attachment protein, alpha | NAPA | 148232359 | cow | 33205 | 9 | 138 | 124/295 | 42 | intracellular protein transport, Golgi and ER, brain dev. |
| 1357 | 1.57 | 0.013 | pyruvate dehydrogenase E1 component beta subunit | PDHB | 73985155 | dog | 37218 | 4 | 56 | 42/341 | 12 | glycolysis, TCA cycle |
| 1404 | 1.35 | 0.012 | annexin A5 | ANXA5 | 57100553 | dog | 35944 | 11 | 151 | 143/321 | 45 | anti-coagulation, signal transduction |
| 1419 | 1.5 | 0.012 | voltage-dependent anion channel 2 | VDAC2 | 90076508 | macaque | 31607 | 4 | 56 | 43/294 | 15 | anion transport, Eukaryotic porin |
| 1440 | 1.26 | 0.037 | tropomyosin 3 | TPM3 | 73961103 | dog | 28760 | 14 | 230 | 90/248 | 36 | cytoskeleton, cell motility |
| 1539 | 1.25 | 0.02 | endoplasmic reticulum protein 29 | ERP29 | 115495555 | cow | 28806 | 3 | 49 | 32/258 | 12 | secretory protein processing, ER lumen |
| 1594 | 1.78 | 0.029 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa | NDUFS3 | 73983298 | dog | 30158 | 10 | 182 | 110/263 | 42 | ETC, ATP metabolism, apoptosis, ROS metabolism |
| 1639 | 1.69 | 0.02 | heat shock protein beta-1 | HSPB1 | 85542053 | cow | 22393 | 2 | 35 | 26/201 | 13 | oxidative protection, cytoskeleton, chaperone |
| 1680 | 1.42 | 0.017 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | NDUFV2 | 115502498 | orangutan | 27355 | 3 | 52 | 34/249 | 14 | ETC/ oxidative phosphorylation |
| 1941 | 1.87 | 0.029 | cofilin 1 | CFL1 | 73983054 | dog | 16812 | 7 | 117 | 77/149 | 52 | cytoskeleton; actin polymerization |
| 1996 | 1.59 | 0.021 | eukaryotic translation initiation factor 5A | EIF5A | 4503545 | human | 16832 | 4 | 61 | 54/154 | 35 | protein synthesis and folding (anabolism) |
| 2069 | 1.26 | 0.038 | transcription elongation factor B (SIII), polypeptide 2 | TCEB2 | 57088081 | dog | 12564 | 2 | 32 | 21/113 | 19 | transcription, part of ubiquitin ligase complex |
| 2132 | 1.31 | 0.045 | chain H, Cytochrome Bc1 Complex | UQCRH | 4139399 | cow | 9175 | 3 | 43 | 28/78 | 36 | ETC, mitochondrial respiration |
Table 2.
Proteins lower in ET. ET, n=6; SA, n=6; q<0.05. Listed are the 61 spots for which a unique ID was recovered based on criteria summarized in methods. For explanation of columns, please see legend to Table 1, except average normalized volume is ratio of SA to ET (fold); UPR, unfolded protein response; NA, nucleic acid
| spot | fold | q | protein ID | gene | GI | species | MW | peps | score | AA coverage | functional grouping | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 148 | 1.42 | 0.02 | heat shock 70kDa protein 4 isoform 6 | HSPA4 | 114601656 | chimp | 94370 | 48 | 733 | 306/840 | 36 | UPR, ATP binding, nucleotide binding |
| 154 | 1.42 | 0.035 | heat shock 70 kDa protein 4 | HSPA4 | 75061973 | orangutan | 94299 | 15 | 240 | 152/840 | 18 | UPR, ATP binding, nucleotide binding |
| 182 | 1.3 | 0.026 | heat shock 70 kDa protein 4 | HSPA4 | 194208465 | horse | 94749 | 16 | 265 | 223/841 | 27 | UPR, ATP binding, nucleotide binding |
| 245 | 1.28 | 0.019 | brain glycogen phosphorylase | PYGB | 21361370 | human | 96697 | 14 | 206 | 146/843 | 17 | glycogen catabolism |
| 262 | 1.26 | 0.028 | aconitase 1 | ACO1 | 126030781 | rabbit | 98344 | 10 | 154 | 142/888 | 16 | iron homeostasis |
| 318 | 1.22 | 0.029 | aconitase 2, mitochondrial | ACO2 | 40538860 | rat | 85434 | 21 | 344 | 230/780 | 29 | TCA cycle, precursor metabolites |
| 332 | 1.21 | 0.021 | aconitase 2, mitochondrial | ACO2 | 40538860 | rat | 85434 | 24 | 388 | 265/780 | 34 | TCA cycle, precursor metabolites |
| 398 | 1.26 | 0.025 | N-acylaminoacyl-peptide hydrolase | APEH | 149728649 | horse | 81243 | 7 | 102 | 79/732 | 11 | proteolysis, serine-type endopeptidase |
| 546 | 1.13 | 0.046 | heat shock 70 kDa protein 8 | HSPA8 | 123647 | hamster | 70805 | 25 | 424 | 283/646 | 44 | protein folding, UPR |
| 548 | 1.27 | 0.049 | collapsin response mediator protein-2A isoform 2 | DPYSL2 | 149746221 | horse | 73531 | 15 | 241 | 223/677 | 33 | axon guidance, |
| 555 | 1.26 | 0.02 | transketolase | TKT | 67971086 | macaque | 67848 | 8 | 138 | 109/623 | 18 | anabolic precursors, regulation of growth |
| 565 | 1.27 | 0.021 | transketolase | TKT | 75054695 | orangutan | 67897 | 15 | 236 | 216/623 | 35 | anabolic precursors, regulation of growth |
| 602 | 1.25 | 0.029 | aldehyde dehydrogenase 4 family, member A1 | ALDH4A1 | 194207963 | horse | 62073 | 10 | 159 | 128/562 | 23 | arginine, proline, glutamate metabolism |
| 639 | 1.27 | 0.035 | collapsin response mediator protein-2A isoform 2 | DPYSL2 | 149746221 | horse | 73531 | 20 | 348 | 274/677 | 40 | axon guidance |
| 653 | 1.37 | 0.012 | dihydropyrimidinase-like 5 | DPYSL5 | 62822083 | human | 52094 | 9 | 140 | 112/477 | 23 | nervous system dev., cone guidance |
| 662 | 1.37 | 0.017 | dihydropyrimidinase-like 3 | DPYSL3 | 4503379 | human | 61964 | 10 | 139 | 143/570 | 25 | nervous system dev., NA metabolism |
| 679 | 1.24 | 0.012 | dihydropyrimidinase related protein-3 | DPYSL3 | 73949646 | dog | 94582 | 7 | 97 | 84/881 | 10 | nervous system dev., NA metabolism |
| 693 | 1.34 | 0.016 | dihydropyrimidinase related protein-2 | DPYSL2 | 73993705 | dog | 62262 | 13 | 200 | 180/572 | 31 | axon guidance, |
| 890 | 1.47 | 0.048 | galactokinase 2 | GALK2 | 11041513 | macaque | 40195 | 2 | 29 | 25/363 | 7 | galactose metabolism |
| 947 | 1.26 | 0.038 | enolase 2 | ENO2 | 5803011 | human | 47269 | 11 | 178 | 175/434 | 40 | glycolysis |
| 993 | 1.27 | 0.014 | branched chain aminotransferase 1, cytosolic | BCAT1 | 73997380 | dog | 48242 | 3 | 41 | 21/437 | 5 | protein anabolism/ nitrogen metabolism |
| 1112 | 1.29 | 0.029 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 | SERPINB1 | 13489087 | human | 42742 | 2 | 33 | 21/379 | 6 | serine protease inhibitor |
| 1127 | 1.35 | 0.035 | guanine nucleotide binding protein | GNAO1 | 164607137 | mouse | 40037 | 6 | 84 | 74/354 | 21 | signal transduction |
| 1131 | 1.23 | 0.035 | aldolase C, fructose-bisphosphate | ALDOC | 74355075 | human | 43010 | 16 | 262 | 174/398 | 44 | glycolysis |
| 1230 | 1.48 | 0.012 | aldolase C | ALDOC | 229506 | rabbit | 39019 | 2 | 30 | 36/361 | 10 | glycolysis |
| 1242 | 1.26 | 0.014 | aldo-keto reductase family 1, member A1 isoform 2 | AKR1A1 | 109020298 | macaque | 36631 | 6 | 101 | 64/325 | 20 | aldehyde catabolism |
| 1248 | 1.22 | 0.018 | dimethylarginine dimethylaminohydrolase 1 | DDAH1 | 57088769 | dog | 31307 | 7 | 112 | 72/285 | 25 | nitric oxide generation |
| 1286 | 1.38 | 0.026 | aldo-keto reductase family 1, member B3 | AKR1B3 | 160707894 | mouse | 35732 | 7 | 103 | 57/316 | 18 | aldose reductase |
| 1305 | 1.41 | 0.027 | malate dehydrogenase 1, NAD (soluble) | MDH1 | 31982178 | mouse | 36468 | 3 | 46 | 31/334 | 9 | upstream of TCA cycle, cytosolic |
| 1306 | 1.35 | 0.043 | malate dehydrogenase 1, NAD (soluble) | MDH1 | 31982178 | mouse | 36468 | 5 | 77 | 67/334 | 20 | upstream of TCA cycle, cytosolic |
| 1312 | 1.38 | 0.024 | malate dehydrogenase 1, NAD (soluble) | MDH1 | 92087001 | mouse | 36511 | 7 | 98 | 75/334 | 22 | upstream of TCA cycle, cytosolic |
| 1318 | 1.37 | 0.012 | lactate dehydrogenase B | LDHB | 4557032 | human | 36639 | 7 | 109 | 86/334 | 26 | glycolysis, pyruvate to lactate |
| 1328 | 1.45 | 0.015 | lactate dehydrogenase B | LDHB | 4557032 | human | 36639 | 16 | 268 | 157/334 | 47 | glycolysis, pyruvate to lactate |
| 1330 | 1.42 | 0.019 | pyridoxal (pyridoxine, vitamin B6) phosphatase | PDXP | 109127253 | macaque | 47912 | 6 | 91 | 69/453 | 15 | glycolysis, catabolism of vitamin B6 |
| 1377 | 1.37 | 0.013 | pirin | PIR | 57111643 | dog | 32150 | 4 | 57 | 43/290 | 15 | transcription |
| 1383 | 1.21 | 0.019 | proteasome 26S subunit, non-ATPase, 14 | PSMD14 | 74004396 | dog | 33384 | 2 | 29 | 23/299 | 8 | protein turnover |
| 1407 | 1.17 | 0.037 | pyridoxal (pyridoxine, vitamin B6) phosphatase | PDXP | 10092677 | human | 31698 | 6 | 89 | 62/296 | 21 | glycolysis, catabolism of vitamin B6 |
| 1472 | 1.27 | 0.012 | 3-hydroxyisobutyrate dehydrogenase | HIBADH | 114612561 | chimp | 30295 | 7 | 120 | 71/287 | 25 | valine catabolism, mitochondrial |
| 1522 | 1.31 | 0.037 | haloacid dehalogenase-like hydrolase domain containing 2 | HDHD2 | 57089151 | dog | 28942 | 3 | 50 | 40/263 | 15 | redox |
| 1538 | 1.24 | 0.022 | chloride intracellular channel 4 | CLIC4 | 7330335 | human | 28772 | 6 | 85 | 70/253 | 28 | membrane potential |
| 1541 | 1.14 | 0.018 | 14-3-3 protein gamma | YWHAG | 71153781 | cow | 28253 | 9 | 144 | 102/247 | 41 | signal transduction |
| 1575 | 2.2 | 0.044 | glutathione S-transferase mu 2 | GSTM2 | 13936373 | guinea pig | 25695 | 5 | 75 | 40/218 | 18 | glutathione-mediated detoxification |
| 1582 | 1.63 | 0.012 | calbindin 2 | CALB2 | 34098931 | mouse | 31373 | 9 | 139 | 93/271 | 34 | calcium ion modulation |
| 1596 | 1.31 | 0.018 | triosephosphate isomerase 1 variant | TPI1 | 62896835 | human | 26713 | 11 | 204 | 158/249 | 63 | glycolysis, pentose phosphate |
| 1646 | 1.35 | 0.017 | ubiquitin carboxyl-terminal esterase L1 | UCHL1 | 114051423 | cow | 28336 | 11 | 182 | 145/252 | 58 | stress response, walking and eating behaviors |
| 1647 | 1.33 | 0.029 | hypoxanthine phosphoribosyltransferase 1 | HPRT1 | 4504483 | human | 24580 | 2 | 27 | 26/218 | 12 | purine metabolism |
| 1662 | 1.35 | 0.018 | protein-L-isoaspartate (D-aspartate) O-methyltransferase | PCMT1 | 67970625 | macaque | 30318 | 3 | 42 | 31/285 | 11 | anabolism, substrate-specific methylation |
| 1683 | 1.26 | 0.037 | Rho GDP dissociation inhibitor (GDI) alpha | ARHGDIA | 31982030 | mouse | 23408 | 5 | 80 | 55/204 | 27 | signal transduction |
| 1768 | 1.17 | 0.037 | glyoxalase I | GLO1 | 134085635 | cow | 20766 | 2 | 21 | 14/184 | 8 | apoptosis/anti-apoptosis |
| 1796 | 1.24 | 0.024 | peroxiredoxin 2 isoform 1 | PRDX2 | 109123569 | macaque | 19470 | 8 | 138 | 80/176 | 45 | redox |
| 1951 | 1.29 | 0.012 | cofilin 2 | CFL2 | 14719392 | human | 18737 | 6 | 98 | 68/166 | 41 | cytoskeleton; actin polymerization |
| 1990 | 1.37 | 0.033 | synuclein, gamma | SNCG | 90110074 | human | 13331 | 3 | 46 | 37/127 | 29 | cell motility, metastasis |
| 1992 | 1.51 | 0.024 | synuclein, gamma | SNCG | 149034108 | rat | 13046 | 6 | 97 | 53/123 | 43 | cell motility, metastasis |
| 1995 | 1.41 | 0.018 | synuclein, gamma | SNCG | 90110074 | human | 13331 | 2 | 30 | 37/127 | 29 | cell motility, metastasis |
| 2002 | 1.23 | 0.024 | peptidylprolyl isomerase A | PPIA | 6679439 | mouse | 17971 | 5 | 75 | 47/164 | 29 | chaperone-dependent protein folding |
| 2017 | 1.28 | 0.025 | peptidylprolyl isomerase A | PPIA | 74146841 | mouse | 17944 | 4 | 59 | 31/164 | 19 | chaperone-dependent protein folding |
| 2020 | 1.19 | 0.045 | peptidylprolyl isomerase A | PPIA | 73962252 | dog | 17969 | 2 | 26 | 18/164 | 11 | chaperone-dependent protein folding |
| 2071 | 1.56 | 0.012 | phosphoprotein enriched in astrocytes 15 | PEA15 | 74006311 | dog | 15781 | 3 | 51 | 27/137 | 20 | apoptosis/anti-apoptosis |
| 2073 | 1.27 | 0.05 | programmed cell death 5 | PDCD5 | 73948546 | dog | 15465 | 2 | 27 | 25/138 | 18 | apoptosis/anti-apoptosis |
| 2085 | 1.6 | 0.024 | phosphoprotein enriched in astrocytes 15 | PEA15 | 74006311 | dog | 15781 | 3 | 46 | 36/137 | 26 | apoptosis/anti-apoptosis |
| 2117 | 1.25 | 0.013 | profilin 2 | PFN2 | 16753215 | human | 15046 | 4 | 59 | 37/140 | 26 | cytoskeleton; actin polymerization |
In all cases except two, when more than one spot contained the same protein, those spots were either elevated or reduced coordinately from SA to ET. Only guanine nucleotide binding protein (GNAO1) and aconitase 2 (ACO2), showed a disparate expression: for GNAO1, spots 1167, 1174, and 1176 were elevated in ET (Table 1), and spot 1127 was reduced in ET (Table 2); in the case of ACO2, spot 396 was elevated in ET (Table 1), and spots 318 and 332 were reduced in ET (Table 2). These isoforms likely represent alternative post-translational modifications or a variation earlier in the gene expression process such as alternative splicing.
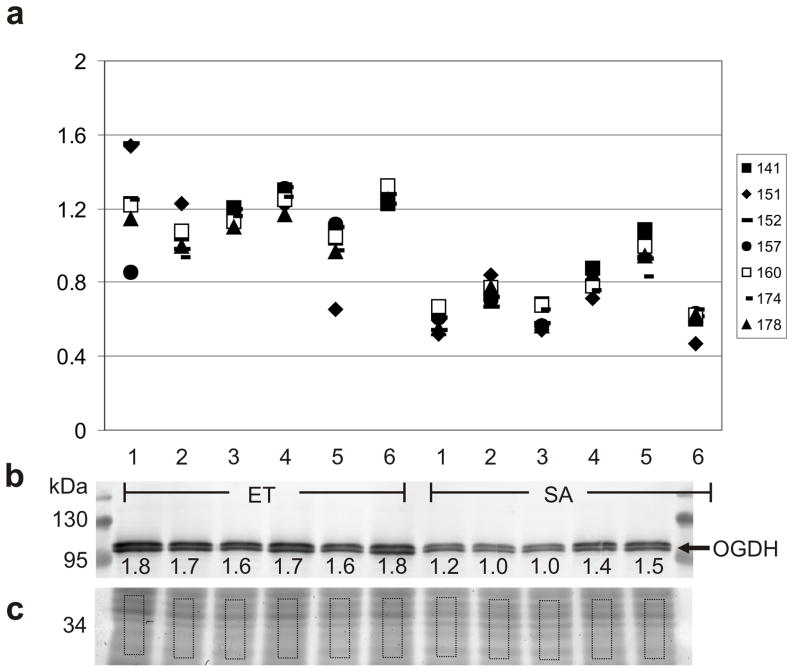
OGDH, uniquely identified in seven 2D gel spots (Table 1), was selected for verification of the DIGE results by western blotting. Two bands of ~114 kDa were detected with a combined increase in ET of 1.4 fold (Fig. 3). While the results of western blotting are consistent with the DIGE results, the 2D gel method is able to discriminate and independently quantify the significant fold changes for seven different isoforms of the same protein in contrast to the western blot which merges all of these variants of distinct charge (Fig. 2) into two poorly-resolved bands.
Fig. 3.
Comparison of DIGE and western quantitation for OGDH. (a) Isoform-specific quantitation using the normalized spot volumes obtained by DIGE of seven 2D gel spots for which OGDH was the uniquely identified protein; symbols represent protein spots as indicated; ET: n=6, SA: n=6; fold changes and Student’s t-test p values for each of the seven OGDH spots are in Table 1. DIGE values are aligned with western blot lanes, except for the depleted SA6. (b) Western blot distinguishes two isoforms of OGDH at ~114kDa; after electrophoretic separation, the protein gel was cut and proteins in the top portion were transferred for immunoblotting, the bottom (c) was stained with Sypro Ruby. Molecular mass is indicated in kDa, normalized volumes are indicated under each band after correction for protein loading to the volume of the corresponding rectangle in the Sypro Ruby-stained portion of gel. Fold change for OGDH (combined bands) as determined by western, 1.39X (ET>SA); ET: n=6, SA: n=5; p<0.0006, Student’s t-test.
The proteins that altered significantly between ET and SA demonstrated consistently small fold changes, with the majority less than two-fold. The largest fold change was spot 1189 at 2.31 fold. Small but significant fold changes might arise in two ways: either they reflect uniform small fold changes, or there are nonuniform large fold changes in a subset of cells due to the high functional heterogeneity of cells within the brainstem. To address the question of region-specific expression of the gene products revealed by our screen, we compared our list of ground squirrel proteins to mRNAs that show regional expression in mouse brain. The Allen Mouse Brain Reference Atlas comprises data from in situ hybridization experiments localizing the expression of ~20,000 mRNAs in mouse brain (Lein et al. 2007). This online resource (www.brain-map.org) was searched for mRNAs that correspond to the proteins listed in Tables 1 and 2 to determine whether expression was ubiquitous or cell type-specific. Of the 85 distinct proteins examined, 76 of the gene product names matched exactly to an entry in the Allen atlas. Serial coronal sections were available for 33 of these 76; 25 demonstrated regional enrichment of expression and the other eight were ubiquitous (Table 3). These results suggest that for many proteins identified in this proteomics screen, small fold changes found in the homogenized brainstem sample are actually “diluted” representations of larger, localized fold changes. Table 4 illustrates how this heterogeneity would affect protein quantification in brainstem homogenates.
Table 3.
Distribution in mouse brain of gene products that differ between SA and ET brains. Listed are official symbol (gene) from Tables 1 and 2 that exactly matched those of the mRNA in situ hybridization data in the Allen brain expression atlas for brainstem regions midbrain, pons and medulla for which a coronal section series was available. Specific areas of mRNA hybridization are listed (brainstem nuclei.)
Abbreviations: 3N: oculomotor nucleus, 6N: abducens nucleus, 5N: motor trigeminal nucleus, 12N: hypoglossal nucleus, 10N: dorsal motor nucleus of the vagus, AmbSC: subcompact nucleus ambiguus, AP: area postrema, ATg: anterior tegmental nucleus, CLi: caudal linear nucleus of the raphe, DR: dorsal raphe nucleus, Gi: gigantocellular reticular nucleus, LC: locus coeruleus, LDTg: laterodorsal tegmental nucleus, LPB: lateral parabrachial nucleus, LPNE: lateral parabrachial nucleus, external; LRt: lateral reticular nucleus, LSO: lateral superior olivary nucleus, IOB: inferior olive, subnucleus C of medial nucleus, IOC: inferior olive, subnucleus C of medial nucleus, IOM: inferior olive medial nucleus, me5: mesencephalic trigeminal tract, P5: peritrigeminal zone, Pn: pontine nuclei, PO: periolivary nuclei, RLi: raphe rostral linear nucleus, RMC: red nucleus, magnocellular part, ROb: raphe obscuris nucleus, RR: retrorubral nucleus, RtTg: reticulotegmental nucleus pons, scp: superior cerebellar peduncle, SNL: Substantia nigra, lateral part, Sp5C: spinal trigeminal nucleus caudal, SubB: Subbrachial nucleus.
| gene | brainstem nuclei |
|---|---|
| ACO1 | AmbSC |
| AKR1B3 | LC, 10N |
| ALDOC | AmbSC |
| ANXA5 | AmbSC, 10N, 12N |
| ARHGDIA | AmbSC, 10N, 12N |
| ATP5A1 | ubiquitous |
| BCAT1 | AP, 12N |
| CALB2 | Pn, LPNE, IOC, IOB |
| CCT2 | ubiquitous |
| CKMT1B | ubiquitous |
| CLIC4 | ubiquitous |
| CLTA | 5N, 12N |
| DPYSL3 | LPNE, me5, 10N, 12N |
| DPYSL5 | 10N |
| EIF5A | me5 |
| ERP29 | me5, AmbSC, 10N |
| GLO1 | 12N, LRt |
| GNAO1 | LDTg, light on 10N, 12N |
| HSPB1 | P5, LSO, AmbSC, 12N |
| NDRG2 | ROb |
| NDUFS1 | ubiquitous |
| NDUFV2 | Pn, Gi |
| NSF | ubiquitous |
| PEA15 | CLi, LPB, LC, AmbSC, 10N, 12N |
| PFN2 | ATg, RR, PO, 10N, 12N |
| PIR | 12N |
| PYGB | 6N, AmbSC, 10N, 12N |
| SERPINB1 | RtTg, scp, Sp5C |
| SNCG | RLi, SNL, RMC, 3N, SubB, DR, AmbSC, 12N, 10N, IOM |
| UCHL1 | neurons |
| UQCRH | ubiquitous |
| VDAC2 | ubiquitous |
| YWHAG | LC, AmbSC, 10N, 12N |
Table 4.
Illustration of fold change dilution in homogenate for a hypothetical protein expressed in glia and in neurons of brainstem nuclei a–i. Here, glial cells are 30 times more abundant than neurons, and the protein is present at a ten fold higher copy number in neurons than in glia. The neurons in nucleus “e” increase expression of this hypothetical protein 20 fold in ET, but this fold change is reduced to 1.33 in the homogenized sample.
| cell type | proportion of cells | SA | ET | scaled SA | scaled ET | fold change |
|---|---|---|---|---|---|---|
| glia | 30 | 0.01 | 0.01 | 0.3 | 0.3 | 1 |
| nuc. a | 0.08 | 0.1 | 0.1 | 0.008 | 0.008 | 1 |
| nuc. b | 0.1 | 0.1 | 0.1 | 0.01 | 0.01 | 1 |
| nuc. c | 0.08 | 0.1 | 0.1 | 0.008 | 0.008 | 1 |
| nuc. d | 0.11 | 0.1 | 0.1 | 0.011 | 0.011 | 1 |
| nuc. e | 0.07 | 0.1 | 2 | 0.007 | 0.14 | 20 |
| nuc. f | 0.15 | 0.1 | 0.1 | 0.015 | 0.015 | 1 |
| nuc. g | 0.13 | 0.1 | 0.1 | 0.013 | 0.013 | 1 |
| nuc. h | 0.08 | 0.1 | 0.1 | 0.008 | 0.008 | 1 |
| nuc. i | 0.2 | 0.1 | 0.1 | 0.02 | 0.02 | 1 |
|
| ||||||
| in homogenate: | 0.4 | 0.53 | 1.33 | |||
Accurate automated evaluation of large sets of biochemical data is still in its infancy. A new tool that enables statistically robust pathway searching is KEGG Spider (http://mips.gsf.de/proj/keggspider/); this was used to examine the uniquely identified proteins found to be elevated either in ET or in SA ground squirrels. By this method, using the feature to generate 200 random networks, two KEGG metabolic pathways in ET and two in SA were significantly elevated (p<0.05). Alanine/aspartate metabolism and the TCA cycle were elevated in ET, and pyruvate metabolism and glycolysis/gluconeogenesis were elevated in SA.
Other functional groups implicated in the present dataset include an upregulation in ET of the electron transport chain and oxidative phosphorylation, ATP synthesis machinery and components required for intracellular protein trafficking. Functional groups that contained both ET- and SA-elevated spots include protein anabolic and catabolic pathways, cytoskeleton and cell motility, redox balance and signal transduction. Changes in protein levels in all of these functional groups reveal a winter proteome that differs considerably from summer, and one which enables dynamic shifts during the torpor and arousal cycles in fuel consumption, cell survival, vesicular trafficking and cytoarchitecture (Table 5).
Table 5.
Functional groupings for proteins found to alter from SA to ET. The official symbols of the genes encoding proteins that differed between SA and hibernating (ET) ground squirrels are listed.
| higher in early torpor | higher in summer active |
|---|---|
|
glycolysis, sugar metabolism
| |
| PYGB | |
| MDH1-cytosolic (3 spots) | |
| ALDOC (2 spots) | |
| TPI1 | |
| ENO2 | |
| LDHB (2 spots) | |
| GALK2 | |
|
pyruvate dehydrogenase complex
| |
| DLD (2 spots) | |
| PDHB | |
| PDHX | |
|
TCA cycle
| |
| ACO2 | ACO2 (2 spots) |
| IDH3A | |
| DLD (2 spots) | |
| OGDH (7 spots) | |
|
intracellular trafficking
| |
| DNM1 | |
| NSF (2 spots) | |
| GSNAP | |
| CLTA (clathrin, 2 spots) | |
| NAPA (alpha SNAP) | |
|
signal transduction
| |
| GNAO1 (3 spots) | GNAO1 |
| GNB1 | YWHAG (14-3-3 gamma) |
| ANXA5 | ARHGDIA |
|
redox
| |
| ACO1 | |
| AKR1A1 | |
| DDAH1 | |
| AKR1B3 | |
| HDHD2 | |
| GSTM2 | |
| TKT (2 spots) | |
| PRDX2 | |
|
ion modulation
| |
| ASNA1 | CLIC4 |
| VDAC2 | CALB2 |
|
cytoskeleton/cell motility
| |
| TUBA1B (2 spots) | DPYSL2 (3 spots) |
| TUBA1C | DPYSL3 (2 spots) |
| TUBB2C | DPYSL5 |
| NDRG2 (2 spots) | CFL2 |
| RPSA | SNCG (3 spots) |
| TPM3 | PFN2 |
| HSPB1 | PDXP (2 spots) |
| CFL1 | |
|
ETC/oxidative phosphorylation
| |
| NDUFS1 | |
| ATP5A1 | |
| ATP5B (2 spots) | |
| UQCRC1 | |
| NDUFS3 | |
| NDUFV2 | |
| UQCRH | |
|
apoptosis/antiapoptosis
| |
| HSPA5 | GLO1 |
| PEA15 | |
| PDCD5 | |
|
protein synthesis and folding (anabolism)
| |
| CCT5 | HSPA4 (3 spots) |
| CCT2 | HSPA8 |
| TUFM | SERPINB1 |
| ERP29 | PCMT1 |
| EIF5A | PPIA (3 spots) |
|
protein and amino acid breakdown (catabolism)
| |
| USP5 | APEH |
| VCP | ALDH4A1 |
| PSMD14 | |
| BCAT1 | |
| HIBADH | |
| UCHL1 | |
|
transcription/nucleic acid processing
| |
| PURA (2 spots) | PIR |
| STRAP | HPRT1 |
| TCEB2 | |
Discussion
Hibernators display extraordinary physiological plasticity during the winter months. They sustain core temperature as well as cardiac, respiratory and metabolic rates at one to five percent of SA levels for more than ten days at a time, then spontaneously revert to euthermic conditions that resemble summer physiology before entering another bout of torpor (reviewed in Andrews 2007; Carey et al. 2003). While ischemia and reperfusion (I/R) leads to damage, stress response and cell death in homeotherms, the brains of hibernating ground squirrels are markedly resistant to oxidative damage in spite of measurable hypoxia (Ma et al. 2005). Cells in the brain exemplify the hibernators’ plasticity and resilience as they undergo transitions in activity (Kilduff et al. 1990; Krilowicz et al. 1988) and synaptic structure (Arendt et al. 2003; Ruediger et al. 2007; von der Ohe et al. 2007) during the torpor and arousal cycles of winter. As predicted, this physiological plasticity was reflected in a seasonal alteration in protein composition as measured by the changes in protein spots observed between summer and winter brainstem, a brain region containing neurons of the autonomic nervous system whose critical functions must be maintained in order to assure survival during hibernation.
This unbiased quantitative comparison of more than 2000 protein spots from 13-lined ground squirrel brainstem demonstrates that most of the proteomic complement remains unaltered between SA and ET, consistent with the maintenance of normal cellular function required throughout the year. Compared to proteomic studies in other tissues, the brainstem data revealed smaller fold changes in a larger percentage of the proteins surveyed. Thirteen percent of spots underwent a significant seasonal shift in brainstem, whereas other tissues demonstrated seasonal changes as follows: whole intestine, three percent (Martin et al. 2008); liver, 11%; lung, less than one percent; and plasma, three percent (unpublished results). Experiments using other 2DE methods also found seasonal changes in protein levels, e.g. liver, nine percent (Epperson et al. 2004), but most studies did not quantify the changes as a portion of the total. One protein in heart (Russeth et al. 2006), one protein in bat liver (Eddy et al. 2005) and six proteins in bat muscle (Lee et al. 2008) were reported to alter with hibernation state. In contrast to these other studies, almost all proteins here changed by less than two fold, likely a consequence of the extreme regional heterogeneity in the brainstem (Tables 3 and 4). Consistent with this explanation, the relative cell type homogeneity in liver may contribute to its greater fold changes (two to five fold) for a greater percentage of the proteins found there in a comparison of SA animals to hibernators (Epperson et al. 2004).
An advantage of 2D gel analysis over traditional SDS-PAGE is the possibility for separating and identifying multiple protein isoforms derived from the same gene (Fig. 3). In most cases, various isoforms of the same protein are elevated in the same season. For example, among significantly different spots in this study, all three spots identified as synuclein were elevated in SA over ET; the same was found for peptidyl-prolyl cis-trans isomerase A (three spots), and for several others. Previous results from studies in liver and intestine reveal similar patterns (Epperson et al. 2004; Martin et al. 2008). However, in the present study, two proteins were identified that were higher in SA in one spot, and higher in ET in a separate spot. These were aconitase 2 (ACO2) and guanine nucleotide binding protein (GNAO1.) Both proteins may be subject to shifts in charge (Bota and Davies 2002; Spickett et al. 2006). This spot pattern suggests a post-translational modification in hibernation (reviewed in Storey 1997), a strategy for controlling enzyme activity likely to be used along with alterations in small molecules (Nelson et al. 2009; Serkova et al. 2007) rather than de novo gene expression for the rapid winter switches required. Examples remain sparse, although several modified proteins have been identified that might play critical roles in hibernation (reviewed in Carey et al. 2003; Chen et al. 2001; Frerichs et al. 1998; Lee et al. 2007; van Breukelen et al. 2004). Investigation of multiple winter stages and identification of specific modifications is required to reveal the importance of post-translational modification in torpor and arousal cycles.
Highly significant changes between SA and ET in brainstem proteins illuminate pathways that are likely to be critical in enabling survival during hibernation (Table 5). The well-known shift away from predominantly carbohydrate metabolism in summer (Andrews et al. 2009) is indicated by the decrease in winter of many enzymes of glycolysis/gluconeogenesis as well as an increase in alanine and aspartate metabolism. The central role of energy homeostasis in hibernation shown previously in cortex metabolite shifts (Henry et al. 2007) is further revealed by the increased abundance of numerous key proteins in both the TCA cycle and oxidative phosphorylation.
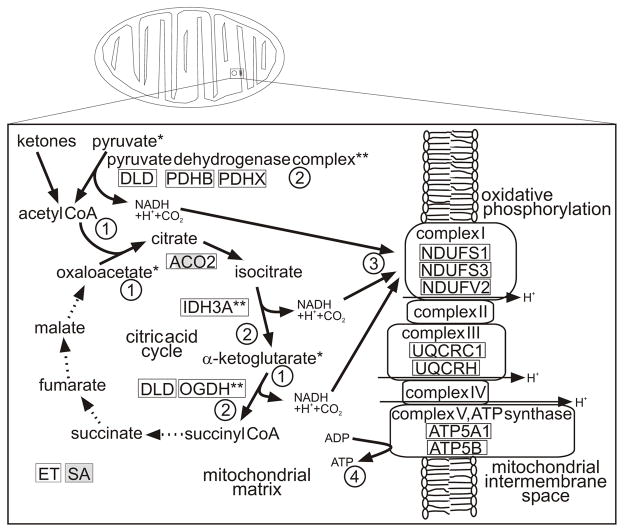
The collective winter upregulation of numerous mitochondrial proteins suggests a model for rapid ATP synthesis with selective use of a portion of the TCA cycle during hibernation, specifically the three steps that follow introduction of acetyl CoA, as depicted in Fig. 4. In this model, near the end of a euthermic interbout arousal when biochemical tasks that require higher temperatures are completed, demand for ATP tapers, allowing ATP to accumulate in the mitochondrial matrix. Elevated ATP would enable phosphorylation of key TCA cycle enzymes such as alpha-ketoglutarate dehydrogenase (OGDH), leading to inactivation by well-established mechanisms and a reduction in ATP synthesis. During torpor, TCA cycle precursors derived from ketone bodies or amino acids would accumulate in the mitochondrial matrix because the TCA enzymes are inactive. Near the end of a torpor bout after slow use and local depletion of ATP, OGDH would be dephosphorylated and thus reactivated. Isocitrate dehydrogenase (IDH3A) and pyruvate dehydrogenase (PDH) are similarly activated via dephosphorylation. A rapid shift in the activity of these three enzymes would be consistent with a predicted quick reversal of oxidative suppression at the end of torpor when Tb is still very low (reviewed in Staples and Brown 2008). The restored catalytic activity of these three enzymes would generate reducing power in the form of NADH to be taken up by the adjacent electron transport chain, a process that demonstrated a unilateral upregulation in brain in ET for several of its protein subunits. The higher winter copy number of proteins in the oxidative phosphorylation pathway would allow for rapid production of ATP during arousal from torpor when demand for energy is high and in the ensuing euthermic interbout arousal (Fig. 4).
Fig. 4.
A model for rapid ATP production in hibernation, specifically during arousal from torpor and in interbout arousal. Unshaded boxes, unique ids elevated in ET; shaded boxes, unique ids reduced in ET. Aconitase 2 (partially shaded) was found in 3 spots, one was elevated in ET and two were reduced in ET. *, derived from amino acids; **, activity reversible by phosphorylation where phosphorylated form is inactive and dephosphorylation activates the enzyme. Circled numbers represent the succession of events. 1) During a torpor bout, intermediates of the citric acid (TCA) cycle accumulate and ATP stores in the mitochondrial matrix are slowly consumed until 2) ATP concentrations are depleted to a threshold resulting in dephosphorylation and therefore activation of several key metabolic enzymes. 3) Catalysis of stockpiled intermediates rapidly creates reducing power in the form of NADH which is taken up by the adjacent electron transport chain and 4) converted to ATP.
Several additional pathways were identified as changing seasonally. Pathways of protein translation, folding and turnover were elevated in either ET or SA, indicating an elaborate and potentially protein-specific strategy for synthesis, salvage and degradation. Another of the largest functional groups represented in Table 5 is that of cytoskeleton and cell motility, consistent with dramatic cellular restructuring during hibernation. Finally, redox balance, signal transduction and clathrin-mediated vesicle formation all altered from SA to ET, with all identified proteins with roles in vesicle formation increased in winter.
The results of this study support the hypothesis that differential expression of genes common to the mammalian genome plays a major role in the biochemistry of hibernation rather than expression of novel genes not found in other, non-hibernating mammals (Srere et al. 1992). These specific protein changes provide new insights into the molecular events that orchestrate and enable survival of the remarkable physiological extremes of hibernation.
Supplementary Material
Acknowledgments
We thank T. Finger, K. Howell, J. Hooper and C. Nelson for helpful discussion. This work was supported by P30-DC004657 to the Rocky Mountain Taste and Smell Center, Defense Advanced Research Projects Agency W81XWH-05-2-0016 to SLM and HVC and National Institutes of Health HL089049 to SLM.
Abbreviations
- Tb
core body temperature
- SA
summer active
- ET
early torpor
References
- Andrews MT. Advances in molecular biology of hibernation in mammals. Bioessays. 2007;29:431–40. doi: 10.1002/bies.20560. [DOI] [PubMed] [Google Scholar]
- Andrews MT, Russeth KP, Drewes LR, Henry PG. Adaptive mechanisms regulate preferred utilization of ketones in the heart and brain of a hibernating mammal during arousal from torpor. Am J Physiol Regul Integr Comp Physiol. 2009;296:R383–93. doi: 10.1152/ajpregu.90795.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T, Stieler J, Strijkstra AM, Hut RA, Rudiger J, Van der Zee EA, Harkany T, Holzer M, Hartig W. Reversible paired helical filament-like phosphorylation of tau is an adaptive process associated with neuronal plasticity in hibernating animals. J Neurosci. 2003;23:6972–81. doi: 10.1523/JNEUROSCI.23-18-06972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bernard KR, Jonscher KR, Resing KA, Ahn NG. Methods in functional proteomics: two-dimensional polyacrylamide gel electrophoresis with immobilized pH gradients in-gel digestion and identification of proteins by mass spectrometry. Methods Mol Biol. 2004;250:263–82. doi: 10.1385/1-59259-671-1:263. [DOI] [PubMed] [Google Scholar]
- Bitting L, Sutin EL, Watson FL, Leard LE, O’Hara BF, Heller HC, Kilduff TS. C-fos mRNA increases in the ground squirrel suprachiasmatic nucleus during arousal from hibernation. Neurosci Lett. 1994 Jan 3;165:117–21. doi: 10.1016/0304-3940(94)90723-4. [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–80. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- Bratincsák A, McMullen D, Miyake S, Toth ZE, Hallenbeck JM, Palkovits M. Spatial and temporal activation of brain regions in hibernation: c-fos expression during the hibernation bout in thirteen-lined ground squirrel. J Comp Neurol. 2007;505:443–58. doi: 10.1002/cne.21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–81. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Chen Y, Matsushita M, Nairn AC, Damuni Z, Cai D, Frerichs KU, Hallenbeck JM. Mechanisms for increased levels of phosphorylation of elongation factor-2 during hibernation in ground squirrels. Biochemistry. 2001;40:11565–11570. doi: 10.1021/bi010649w. [DOI] [PubMed] [Google Scholar]
- Dave KR, Prado R, Raval AP, Drew KL, Perez-Pinzon MA. The arctic ground squirrel brain is resistant to injury from cardiac arrest during euthermia. Stroke. 2006;37:1261–5. doi: 10.1161/01.STR.0000217409.60731.38. [DOI] [PubMed] [Google Scholar]
- Eddy SF, McNally JD, Storey KB. Up-regulation of a thioredoxin peroxidase-like protein, proliferation-associated gene, in hibernating bats. Arch Biochem Biophys. 2005;435:103–11. doi: 10.1016/j.abb.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Dahl TA, Martin SL. Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol Cell Proteomics. 2004;3:920–33. doi: 10.1074/mcp.M400042-MCP200. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Martin SL. Quantitative assessment of ground squirrel mRNA levels in multiple stages of hibernation. Physiol Genomics. 2002;10:93–102. doi: 10.1152/physiolgenomics.00004.2002. [DOI] [PubMed] [Google Scholar]
- Fleck CC, Carey HV. Modulation of apoptotic pathways in intestinal mucosa during hibernation. Am J Physiol Regul Integr Comp Physiol. 2005;289:R586–R595. doi: 10.1152/ajpregu.00100.2005. [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Hallenbeck JM. Hibernation in ground squirrels induces state and species-specific tolerance to hypoxia and aglycemia: an in vitro study in hippocampal slices. Journal of Cerebral Blood Flow and Metabolism. 1998;18:168–175. doi: 10.1097/00004647-199802000-00007. [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Smith CB, Brenner M, DeGracia DJ, Krause GS, Marrone L, Dever TE, Hallenbeck JM. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc Natl Acad Sci, USA. 1998;95:14511–14516. doi: 10.1073/pnas.95.24.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgen KM, Cole FR, Helgen LE, Wilson DE. Generic Revision in the Holarctic Ground Squirrel Genus Spermophilus. Journal of Mammalogy. 2009;90:270–305. [Google Scholar]
- Henry PG, Russeth KP, Tkac I, Drewes LR, Andrews MT, Gruetter R. Brain energy metabolism and neurotransmission at near-freezing temperatures: in vivo (1)H MRS study of a hibernating mammal. J Neurochem. 2007;101:1505–15. doi: 10.1111/j.1471-4159.2007.04514.x. [DOI] [PubMed] [Google Scholar]
- Karp NA, Spencer M, Lindsay H, O’Dell K, Lilley KS. Impact of replicate types on proteomic expression analysis. J Proteome Res. 2005;4:1867–71. doi: 10.1021/pr050084g. [DOI] [PubMed] [Google Scholar]
- Kilduff TS, Miller JD, Radeke CM, Sharp FR, Heller HC. 14C-2-deoxyglucose uptake in the ground squirrel brain during entrance to and arousal from hibernation. J Neurosci. 1990;10:2463–75. doi: 10.1523/JNEUROSCI.10-07-02463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilduff TS, Sharp FR, Heller HC. [14C]2-deoxyglucose uptake in ground squirrel brain during hibernation. J Neurosci. 1982;2:143–57. doi: 10.1523/JNEUROSCI.02-02-00143.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krilowicz BL, Glotzbach SF, Heller HC. Neuronal activity during sleep and complete bouts of hibernation. Am J Physiol. 1988;255:R1008–19. doi: 10.1152/ajpregu.1988.255.6.R1008. [DOI] [PubMed] [Google Scholar]
- Lee K, Park JY, Yoo W, Gwag T, Lee JW, Byun MW, Choi I. Overcoming muscle atrophy in a hibernating mammal despite prolonged disuse in dormancy: proteomic and molecular assessment. J Cell Biochem. 2008;104:642–56. doi: 10.1002/jcb.21653. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Miyake S, Wakita H, McMullen DC, Azuma Y, Auh S, Hallenbeck JM. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J Cereb Blood Flow Metab. 2007;27:950–62. doi: 10.1038/sj.jcbfm.9600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lyman CP, Willis JS, Malan A, Wang LCH. Hibernation and Torpor in Mammals and Birds. New York: Academic Press; 1982. [Google Scholar]
- Ma Y, Wu S. Simultaneous measurement of brain tissue oxygen partial pressure, temperature, and global oxygen consumption during hibernation, arousal, and euthermy in non-sedated and non-anesthetized Arctic ground squirrels. J Neurosci Methods. 2008;174:237–44. doi: 10.1016/j.jneumeth.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YL, Zhu X, Rivera PM, Toien O, Barnes BM, LaManna JC, Smith MA, Drew KL. Absence of cellular stress in brain after hypoxia induced by arousal from hibernation in Arctic ground squirrels. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1297–306. doi: 10.1152/ajpregu.00260.2005. [DOI] [PubMed] [Google Scholar]
- Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–78. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- Martin SL, Dahl T, Epperson LE. Slow loss of protein integrity during torpor: a cause for arousal? In: BB, CHV, editors. Life in the Cold. Vol. 12. Fairbanks, AK: 2004. pp. 199–208. [Google Scholar]
- Martin SL, Epperson LE. A two-switch model for mammalian hibernation. In: Lovegrove BG, McKechnie AE, editors. Hypometabolism in Animals. Pietermaritzburg: Interpak Books; 2008. pp. 177–186. [Google Scholar]
- Martin SL, Epperson LE, Rose JC, Kurtz CC, Ane C, Carey HV. Proteomic analysis of the winter-protected phenotype of hibernating ground squirrel intestine. Am J Physiol Regul Integr Comp Physiol. 2008;295:R316–28. doi: 10.1152/ajpregu.00418.2007. [DOI] [PubMed] [Google Scholar]
- Milsom WK, Zimmer MB, Harris MB. Regulation of cardiac rhythm in hibernating mammals. Comp Biochem Physiol A Mol Integr Physiol. 1999;124:383–91. doi: 10.1016/s1095-6433(99)00130-0. [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Otis JP, Martin SL, Carey HV. Analysis of the hibernation cycle using LC-MS-based metabolomics in ground squirrel liver. Physiol Genomics. 2009;37:43–51. doi: 10.1152/physiolgenomics.90323.2008. [DOI] [PubMed] [Google Scholar]
- Ruediger J, Van der Zee EA, Strijkstra AM, Aschoff A, Daan S, Hut RA. Dynamics in the ultrastructure of asymmetric axospinous synapses in the frontal cortex of hibernating European ground squirrels (Spermophilus citellus) Synapse. 2007;61:343–52. doi: 10.1002/syn.20380. [DOI] [PubMed] [Google Scholar]
- Russeth KP, Higgins L, Andrews MT. Identification of proteins from non-model organisms using mass spectrometry: application to a hibernating mammal. J Proteome Res. 2006;5:829–39. doi: 10.1021/pr050306a. [DOI] [PubMed] [Google Scholar]
- Serkova NJ, Rose JC, Epperson LE, Carey HV, Martin SL. Quantitative analysis of liver metabolites in three stages of the circannual hibernation cycle in 13-lined ground squirrels by NMR. Physiol Genomics. 2007;31:15–24. doi: 10.1152/physiolgenomics.00028.2007. [DOI] [PubMed] [Google Scholar]
- Spickett CM, Pitt AR, Morrice N, Kolch W. Proteomic analysis of phosphorylation, oxidation and nitrosylation in signal transduction. Biochim Biophys Acta. 2006;1764:1823–41. doi: 10.1016/j.bbapap.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Srere HK, Wang LC, Martin SL. Central role for differential gene expression in mammalian hibernation. Proc Natl Acad Sci, USA. 1992;89:7119–7123. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples JF, Brown JC. Mitochondrial metabolism in hibernation and daily torpor: a review. J Comp Physiol [B] 2008;178:811–27. doi: 10.1007/s00360-008-0282-8. [DOI] [PubMed] [Google Scholar]
- Storey KB. Metabolic regulation in mammalian hibernation: enzyme and protein adaptations. Comp Biochem Physiol. 1997;118A:1115–1124. doi: 10.1016/s0300-9629(97)00238-7. [DOI] [PubMed] [Google Scholar]
- Tøien Ø, Drew KL, Chao ML, Rice ME. Ascorbate dynamics and oxygen consumption during arousal from hibernation in Arctic ground squirrels. Am J Physiol Regul Integr Comp Physiol. 2001;281:R572–83. doi: 10.1152/ajpregu.2001.281.2.R572. [DOI] [PubMed] [Google Scholar]
- van Breukelen F, Sonenberg N, Martin SL. Seasonal and state dependent changes of eIF4E and 4E-BP1 during mammalian hibernation: implications for the control of translation during torpor. Am J Physiol Regul Integr Comp Physiol. 2004;287:R349–53. doi: 10.1152/ajpregu.00728.2003. [DOI] [PubMed] [Google Scholar]
- von der Ohe CG, Garner CC, Darian-Smith C, Heller HC. Synaptic protein dynamics in hibernation. J Neurosci. 2007;27:84–92. doi: 10.1523/JNEUROSCI.4385-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics. 2008;32:170–81. doi: 10.1152/physiolgenomics.00075.2007. [DOI] [PubMed] [Google Scholar]
- Zhao HW, Christian SL, Castillo MR, Bult-Ito A, Drew KL. Distribution of NMDA receptor subunit NR1 in arctic ground squirrel central nervous system. J Chem Neuroanat. 2006;32:196–207. doi: 10.1016/j.jchemneu.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Zhu X, Castellani RJ, Stimmelmayr R, Perry G, Smith MA, Drew KL. Hibernation, a model of neuroprotection. Am J Pathol. 2001;158:2145–2151. doi: 10.1016/S0002-9440(10)64686-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.