Abstract
Focal adhesions are large multi-protein assemblies that form at the basal surface of cells on planar dishes, which mediate cell signaling, force transduction, and adhesion to the substratum. While much is known about focal adhesion components in 2-D systems, their role in migrating cells within a more physiological three-dimensional (3-D) matrix is largely unknown. Live-cell microscopy shows that for cells fully embedded in a 3-D matrix, focal adhesion proteins, including vinculin, paxillin, talin, α-actinin, zyxin, VASP, FAK, and p130Cas, do not form aggregates but are diffusively distributed throughout the cytoplasm. Despite the absence of detectable focal adhesions, focal adhesion proteins still modulate cell motility but in a manner distinct from cells on planar substrates. Rather, focal adhesion proteins in matrix-embedded cells regulate cell speed and persistence by affecting protrusion activity and matrix deformation, two processes that play no direct role in controlling 2-D cell speed. This study shows that membrane protrusions constitute a critical motility/matrix-traction module that drives cell motility in a 3-D matrix.
Introduction
Two-dimensional (2-D) cell motility depends upon forces generated from the dynamic remodeling of the acto-myosin cytoskeleton as transmitted through focal adhesions (FAs) to the extracellular matrix. FAs, which contain > 100 different proteins and play both mechanosensory and signaling functions1, 2, are observed at the basal surface of cells in 2-D cultures 3, 4. When cells are partially embedded in a 3-D matrix, FAs become smaller and their composition changes compared to the conventional 2-D case 5-8. However, when the cell is completely buried inside a 3-D matrix – the in vivo case – FAs are not readily detected 9, 10. FAs also disappear when cells are placed on soft substrates 11-13. This suggests an important question: Since FAs are not apparent in matrix-embedded cells, what is the role of key components of FAs in cells in a 3-D matrix that more closely mimics the physiological condition? This is particularly important since expression levels of several FA proteins—focal adhesion kinase (FAK) 14, paxillin15, and zyxin 16—correlate with metastatic potential in vivo.
Little is known about the function(s) of FAs for cells in matrix or in vivo. Understanding this is important since the physiological environment of most cells in vivo is essentially soft and 3-D. Even endothelial and single-layered epithelial cells, which form 2-D structures, begin to move within 3-D extracellular matrices in the context of wound healing and cancer metastasis.
The architecture of adhesion complexes in 3-D matrices are remarkably different from those of cells in 2-D cultures5, 6. However, in previous work, cells were only partially embedded in the matrix, i.e. the apical surface of the cell was not in contact with the matrix. This is an important distinction from cells that are completely embedded inside a matrix away from all stiff walls. Herein, we determined the properties conferred by FA components to the migration of cells fully embedded in a 3-D matrix. Despite the absence of any detectable FA structures, FA components were still found to regulate cell speed, predominantly by modulating pseudopodia activity and matrix deformation – two cellular processes that play little role in controlling 2-D cell speed 17.
Results and Discussion
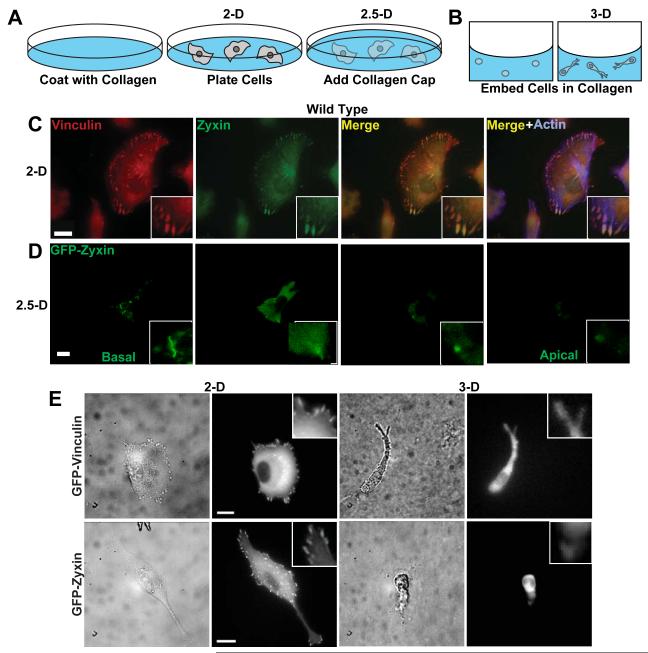
Confocal microscopy of FA proteins confirmed the formation of FAs at the basal surface of wild type (WT) HT-1080 cells, a human fibrosarcoma cell line commonly used to study cell migration18-21, on collagen-coated 2-D glass substrates (Fig. 1, A and C). When these cells were sandwiched between the same collagen-coated substrate and a thick collagen gel deposited on the apical surface of the cell (“2.5-D”, Fig. 1A), FAs still formed, but were greatly decreased in size and number (Fig. 1, A and D) and protein clusters did not appear on the apical surface facing the top gel (Fig. 1D).
Fig. 1. Regulated formation of FAs in 2-dimensional, 2.5-dimensional, and 3-dimensional collagen matrix microenvironments.
A and B. Schematics of type I collagen microenvironments studied here, including cells on conventional flat type I collagen-coated glass substrates (“2-D”, A), cells sandwiched between a collagen-coated substrate and coated with a thick layer of collagen (“2.5-D”, A), and cells fully immersed inside a 3-D collagen matrix (“3-D”, B). C and D. Confocal fluorescence micrographs of vinculin and zyxin, two major constituents of standard FAs in 2-D, in wild type (WT) HT-1080 human fibrosarcoma cells, which were either plated on conventional substrates (2-D case, C) or partially embedded in a matrix (2.5-D case, D). Scale bar, 10 μm. As a control, we verified that we were able to visualize the microtubule network using antibodies against tubulin and the same secondary antibodies used to stain FA proteins (data not shown). E. Phase contrast and fluorescence micrographs of live wild-type cells stably expressing either EGFP-vinculin (top panels) or EGFP-zyxin (bottom panels) on flat 2-D substrates (left panels) and inside a 3-D matrix (right panels). Insets show large vinculin or zyxin-containing FAs in the 2-D case, and diffuse staining in the 3-D case. Scale bar, 10 μm.
When cells were embedded in a 3-D matrix, and only cells inside the matrix and well removed from the bottom glass were analyzed (Fig. 1B), no FAs were detected by confocal microscopy in fixed cells. In an alternative strategy, we established cells stably transfected with EGFP-tagged proteins (Fig. 1E). FAs were again not observed. Given the resolutions of our light microscopes, we estimate that, if FAs exist for cells in a 3-D matrix, their size is smaller than 0.3 μm and their lifetime shorter than a second. In contrast, the size of FAs for cells on flat substrates is as large as 15 μm and can last > 15 min 22.
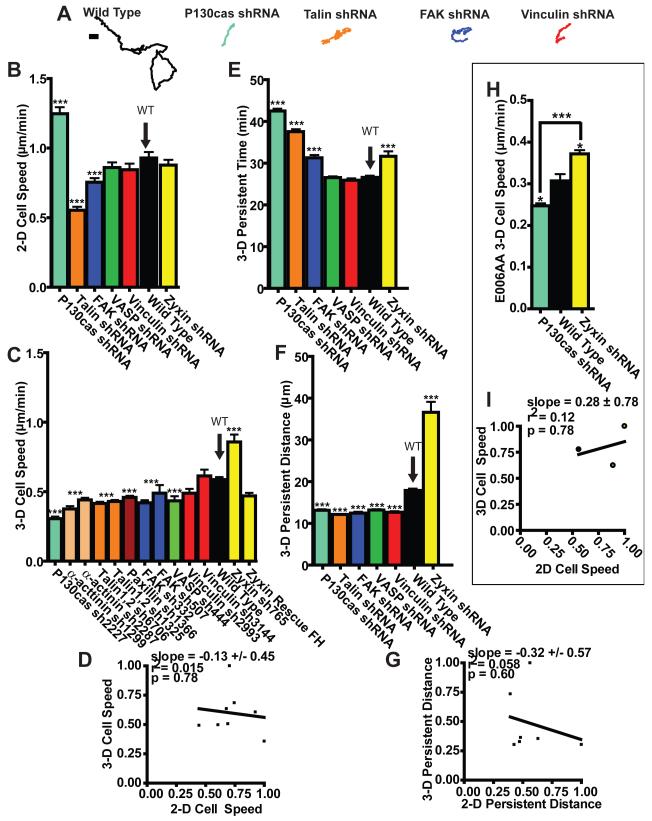
These results suggest that cells completely, as opposed to partially, embedded inside a matrix do not display observable FAs. To determine whether and how FA proteins play a role in cell motility in a 3-D matrix, we systematically RNAi-depleted (Fig. S1) major FA proteins, including structural proteins talin, vinculin, α-actinin, zyxin, paxilin, and vasodilator-stimulated phosphoprotein (VASP), and enzymes and adaptor proteins FAK and p130Cas. We measured the speed of individual cells during random migration inside a type I collagen matrix (Fig. 2, A and C) and compared this to speed on a collagen I-coated flat substrate (Fig. 2B). Strikingly, changes in 3-D cell speed resulting from the depletion of FA proteins did not correlate at all with changes in 2-D cell speed (Fig. 2, B and C), i.e. 2-D cell speed was a poor predictor of 3-D cell speed (Fig. 2D and ST1). For instance, the depletion of p130Cas uniquely enhanced cell speed in 2-D compared to control cells (Fig. 2B), but reduced cell speed in a 3-D matrix, more than any other tested FA protein (Fig. 2C). In contrast, the depletion of zyxin induced higher cell motility inside a 3-D matrix (Fig. 2C), while it had no significant effect on 2-D cell motility (Fig. 2B). The specificity of the role of FA proteins in both 2-D and 3-D cell motility was verified by showing that concurrent re-expression of an RNAi-resistant isoform of the depleted FA proteins rescued the 3-D cell motility phenotypes or by using multiple RNAis targeting different positions in mRNA (Fig. 2C). When a β1 integrin antibody was used, cell speed was drastically reduced, indicating that cell motility in a 3-D matrix, as on 2-D substrates, depends on β1 integrin-ECM binding (Fig. S2 A). Acto-myosin contractility also plays a significant role in 3-D cell migration23-25.
Fig. 2. Regulation of 2-D cell motility by FA proteins is not predictive of regulation of 3-D cell motility in matrix.
A. Typical trajectories of individual matrix-embedded WT HT-1080 cells and HT-1080 cells RNAi-depleted of major FA proteins p130Cas, talin, FAK, and vinculin. Scale bar, 10 μm. B and C. Average random-motility speed of WT cells and multiple cells stably depleted of major FA proteins on 2-D substratum (B) and inside a 3-D collagen matrix (C). D. Lack of correlation between 2-D cell speed and 3-D cell speed. Cell speeds were normalized by the maximum mean value in each data set (here zyxin-depleted cells in 3-D and p130Cas-depleted cells in 2-D). Slope evaluated from a linear fit of the data, R squared value, and p value of correlation are indicated. E and F. Persistence time (E) and persistence distance of migration (F) of cells in matrix for WT cells and FA protein-depleted cells. G. Correlation function between the 2-D and the 3-D persistence distances normalized by maximum persistence distance mean value in each data set. H. 3-D cell speed of WT E006AA human prostate cancer cells and E006AA cells depleted of either p130Cas or zyxin. I. Lack of correlation between 3-D cell speed and 2-D cell speed of WT, p130Cas-depleted, and zyxin-depleted E006AA cells. 2-D and 3-D cell speeds were normalized by the maximum mean value in each data set. *** in panels B, C, E, F and H indicate p values <0.001 between the type of cell considered and WT cells. The speed, persistence time, and persistence distance of at least 35 cells were measured on three different days for each condition. Bar graphs show mean and SEM values of three independent experiments. Arrows point to the WT case.
Measurements of the persistent time and distance, which represent the time and curvilinear length a cell travels before significantly deviating from a straight trajectory in the matrix (e.g. Fig. 2A), indicated that FA proteins regulate the persistence of migration of cells inside a matrix (Fig. 2, E and F, and ST1, red box). However, similar to cell speed, we found a complete absence of correlation between 2-D and 3-D persistence distances and times (Figs. 2, E-G, S2 B, and ST1).
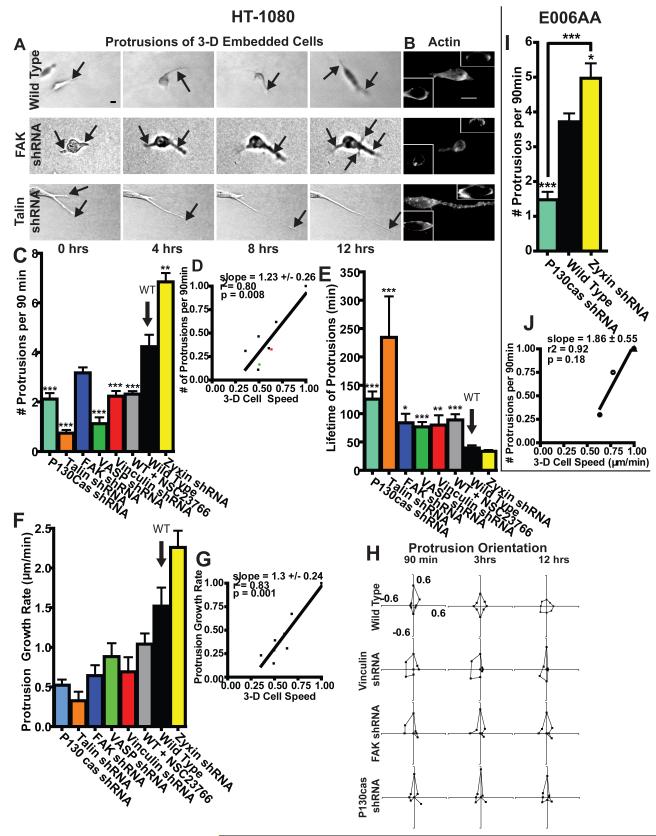
The striking absence of correlation between 2-D and 3-D motility suggests that FA proteins regulate motility in a matrix in a manner fundamentally different from planar cell motility. 2-D cell speed is controlled by the regulated assembly/disassembly of FA complexes at the basal cellular surface, cell-matrix adhesion or traction force, and the assembly and turnover of actin structures that advance lamellipodium or filopodia protrusions at the cell’s leading edge, but not by the rate of membrane protrusion 26-28. Moreover, there is no correlation between the location of filopodial protrusions at the edge of the lamella and the location of maximum traction17. Wild type cells inside a matrix featured neither wide lamella (Figs. 1E and 3A) nor classical FAs (Fig. 1), but displayed long-lived (> 30 min) protrusions, typically much wider and longer (>5 μm) than filopodia and much thinner than the lamella displayed by cells on substrates, which we shall call pseudopodia. Smaller and thinner filipodia were observed, but did not correlate with the formation of pseudopodia and cell speed.
Fig. 3. Extent of focal adhesion protein-mediated protrusion activity predicts 3-D cell speed.
A. Typical time-dependent morphological changes of WT, FAK-depleted and talin-depleted cells embedded in a 3-D matrix showing actively growing protrusions (indicated by arrows). Scale bar, 10 μm. B. Actin filament organization in WT, FAK-depleted and talin-depleted cells in a 3-D matrix. Insets show cross-sectional view. Scale bar, 10 μm. C. Averaged number of actively growing protrusions per 90 min (i.e. protrusion activity) for matrix-embedded WT cells and cells RNAi-depleted of major FA proteins. D. Correlation function between 3-D cell speed and cellular protrusion activity. Values are normalized by corresponding maximum mean values. Slope evaluated from a linear fit of the data, R squared value, and p value of correlation are indicated. E. Averaged lifetime of actively growing protrusions. F. Averaged growth rate of individual protrusions. G. Correlation between 3-D cell speed and growth rate of protrusions. Values were normalized by maximum mean value in each data set. H. Time-dependent angular distributions of actively growing protrusions along the matrix-embedded cell periphery after 90 min, 3h, and 12 h. The largest protrusion at time 0 was arbitrarily taken as being pointing in the 0 degree direction. I. Averaged number of actively growing protrusions per 90 min for WT E006AA cells and E006AA cells depleted of either p130Cas or zyxin. J. Correlation between 3-D cell speed and protrusion activity for the cells characterized in panel I and in Fig. 2H. *, **, and *** in panels C, E, and I indicate p values <0.05, <0.01, and <0.001, respectively, between the KD cells considered and WT cells unless indicated. The pseudopodial protrusions of at least 35 cells were characterized on three different days for each condition. Bar graphs show mean and SEM values of three independent experiments.
To move within a crosslinked network of mesh size smaller than the cell and its nucleus, cells in a matrix may exploit a different motility/traction “module” from that used on substrates. In particular, pseudopodial activity at the cellular periphery could constitute a critical component of the module required to efficiently migrate and negotiate the dense collagen matrix. We first asked whether FA protein depletion affected the number, lifetime, orientation, rate of growth, and length of pseudopodial protrusions generated by 3-D matrix-embedded cells (Fig. 3). With the exception of zyxin, the depletion of FA proteins decreased the number of protrusive processes generated per unit time (Fig. 3C). Changes in 3-D cell speed correlated strongly with changes in the number and growth rate of pseudopodial protrusions (Fig. 3, C-G and ST1, red boxes). For instance, similarly to their opposed effects on the regulation of 3-D cell speed, p130Cas and zyxin regulated the rate of formation of membrane protrusions in diametrically opposite ways (Fig. 3C).
To assess the predictive power of protrusion activity in determining 3-D cell speed, we compared the speed of α-actinin and vinculin KD cells, estimated using the model in Fig. 3D, to direct measurements of cell speed. We found that the measured speeds (red and green dots in Fig. 3D) were predicted within <20% error. The relevance of these findings to other cancer cell lines was verified with E006AA human prostate cancer cells (Fig. S1, I and J, 2, H and I, 3, I and J, and S3).
FA proteins also influenced the lifetime (Fig. 3E) and length of protrusions (Figs. 3A and S4 A and B), as well as the rate at which the direction of protrusions became uniformly distributed (Fig. 3H). However, lifetime and length did not correlate with 3-D cell speed or persistence of migration (Fig. S4, C-F and ST1). These results indicate that extended, long-lived protrusions did not necessarily drive fast or persistent migration in a 3-D matrix. Moreover, 2-D and 3-D cell speed did not correlate with biophysical parameters that should not influence them. For instance, 3-D cell speed did not correlate with 2-D persistence time (ST1). Staining for F-actin in WT cells showed regions of accumulation located mostly at the cell periphery, which was not significantly affected by the depletion of FA proteins (Fig. 3B).
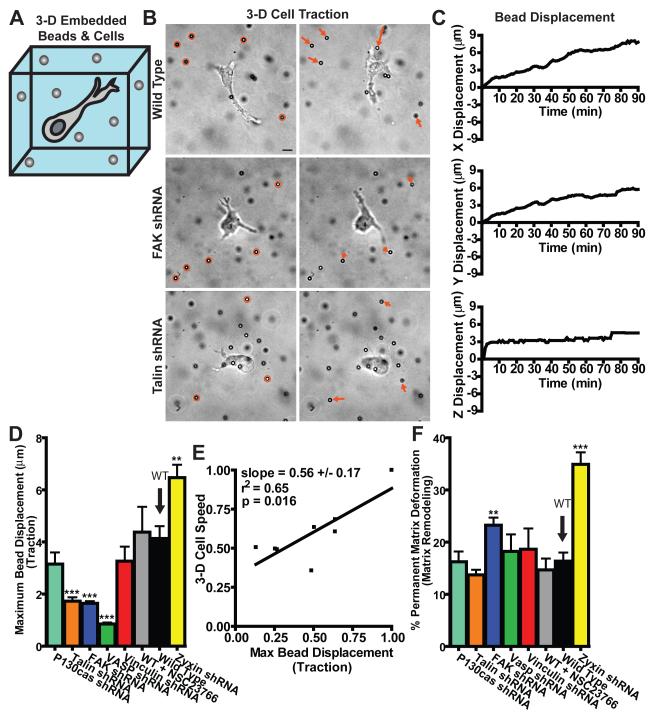
Next we asked whether FA proteins influenced the magnitude and location of the adhesive traction forces exerted by cells on the matrix. 3-D tracking of large carboxylated beads tightly embedded in the matrix (Fig. 4, A-C) revealed that cells only pulled and did not push their surrounding matrix (Fig. 4B). Despite their lack of clustering within cells in 3-D matrix (Fig. 1E), VASP, talin, vinculin, p130Cas, and FAK, but not zyxin, contributed to high traction forces—max bead displacement—on the surrounding matrix (Fig. 4D). Apart from FAK and zyxin, FA proteins individually did not significantly affect the mechanical character of the matrix, which was typically much more elastic (fully reversible deformation) than irreversible (Fig. 4F, where 0% corresponds to a purely elastic deformation of the matrix and 100% corresponds to an irreversible deformation). This result indicates that most FA proteins played no significant role in matrix remodeling, defined here as the quantitative ratio of final-to-total matrix deformation. Cell motility in the matrix was moderately correlated with cell-mediated traction—max bead displacement—(Fig. 4E; ST1), but not with the total deformation of the matrix—total distance travelled by beads—or the extent of matrix remodeling—percent matrix deformation—(Fig. S5, ST1).
Fig. 4. Regulation of 3-D cell-matrix interactions by FA proteins.
A. Schematic of the method used for the measurements of local matrix traction mediated by embedded cells, whereby large fiduciary beads are tightly embedded in the matrix and are monitored by high-resolution 3-D multiple-particle tracking. B. Typical movements of fiduciary beads in the vicinity of a WT cell and cells depleted of talin and FAK, as indicated. Left and right micrographs respectively show the initial and final positions of the beads after 90 min. Arrows indicate the magnitude and direction of the displacements of the matrix-bound beads, which were magnified three times for ease of visualization. Scale bar, 10 μm. C. Typical x, y, and z displacements of an individual matrix-bound bead in the vicinity of a WT cell in the 3-D matrix. D. Maximum displacements of the fiduciary beads in the matrix (i.e. traction). E. Correlation function between 3-D cell speed and the maximum bead displacement (i.e. traction). Values were normalized by maximum mean value in each data set. Slope evaluated from a linear fit of the data, R squared value, and p value of the correlation are indicated. F. Percentage deformation (i.e. matrix remodeling) calculated as the ratio of the final distance between initial and final bead position and the total bead displacement. This percentage is 0 when the matrix deformation is purely elastic and 100 when the matrix deformation is irreversible. See more details in the Materials and Methods section. ** and *** in panels D and F indicate p values <0.01 and <0.001, respectively, between the KD cell considered and WT cells. The local matrix traction in the vicinity of at least 5 cells (~30 beads per cell) was measured on three different days for each condition. Bar graphs show mean and SEM values of three independent experiments.
Matrix traction always occurred in the vicinity of an actively pulling protrusion (data not shown). FA proteins could regulate matrix traction per pseudopodium by modulating the adhesive strength of protrusions to collagen and the connection between integrins and the actin network. However, the traction per pseudopodium—max bead displacement per cell—did not significantly correlate with cell speed (Fig. S5E and ST1). We conclude that the ability of FA proteins to regulate matrix traction stems mainly from their differential ability to regulate the number of protrusions (Fig. 3C), but not the length or lifetime of protrusions (Fig. S4 and S2, C and D).
Together these results suggest that to move and negotiate their matrix environment, cells launch adhesive protrusion processes (Fig. 3), as often as possible, as opposed to producing long-lasting, elongated extensions. Rather than producing matrix traction through a single prolonged protrusion, cells generate multiple pseudopodial protrusions that mediate motility by more effectively probing, then selecting, and pulling collagen fibers in the cellular vicinity for a short time before generating another protrusion to engage another fiber. A WT cell generates on average one major protrusion before using a new protrusion to move in a new direction. Following a relatively short-lived (compared to the time scale of migration) tug on a fiber, a protrusion either releases the fiber or runs into a denser meshwork. The cell then sends off new protrusions in new directions to explore paths of migration in the matrix. Matrix-embedded cells establish a tightly regulated (low) number of major protrusions, which is controlled by FA proteins: too few protrusions and the cell cannot efficiently explore its surroundings, too many protrusions and the cell cannot move because its protrusions pull in too many directions simultaneously. This optimum number of protrusions lies between zero, for which cells would not be able to move at all, and ≤ 2, a number above which cells would not be able to move persistently. These results obtained in a matrix contrast with the small role played by filopodia in controlling 2-D cell speed and persistence 17, 27, 29 with the central role played by FAs, which mediate both adhesion to and traction onto the underlying substrate in conventional planar migration 13, 30, 31. For matrix-embedded cells, protrusion dynamics plays a central role in driving motility, and organized FAs, if they exist, are too small and short-lived compared to the length and lifetime of pseudopodia (~60 μm over 50 min; Fig. 3) or the amplitude and time scale associated with matrix deformation (4 μm over 50 min; Fig. 4) to likely play a significant role.
Moving cells from a hard 2-D substrate (i.e. glass) into a relatively soft 3-D matrix subjects cells to changes in dimensionality (2-D vs. 3-D) and extracellular mechanical compliance. To investigate whether we could reproduce the 3-D cell motility phenotype in 2-D, we compared the speed of cells placed on collagen I substrates (elasticity, >70 kPa) to that of cells placed on substrates covered with a soft, lightly crosslinked polacrylamide gels coated with collagen I (elasticity, 1 kPa). We found that FA proteins regulated cell speed the same way on (hard) glass and on soft substrates (Fig. 5). This suggests that the regulation of cell speed by FAs in a 3-D matrix is not primarily caused by the compliance of the matrix, but by the change in geometry of the microenvironment. In conclusion, modulation of cell speed and persistence on planar substrates by FA proteins, even compliant substrates, is not predictive of their regulation of cell speed in a matrix, which highlights limitations of traditional planar migration studies in understanding 3-D cell motility and the role of FA proteins.
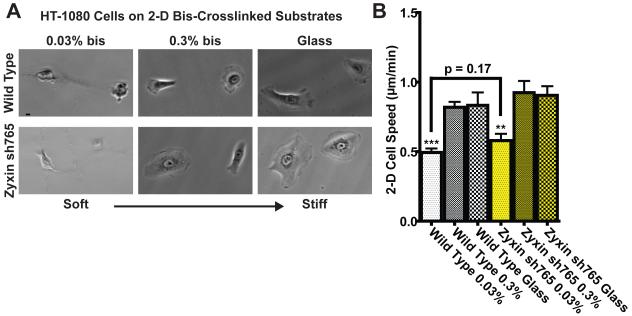
Fig. 5. Regulation of cell motility on compliant substrates by FA proteins.
A. Wild-type and zyxin-depleted cells on soft collagen-coated bis-crosslinked polyacrylamide gels and stiff collagen-coated glass substrates. Scale bar, 10 μm. B. Averaged cell speed of wild-type and zyxin-depleted cells on collagen-coated glass vs. soft substrates. ** and *** indicate p value <0.01 and <0.001 respectively comparing cells on the softest substrates to the same cells on glass. The speed of at least 35 cells were measured on three different days for each condition. Bar graphs show mean and SEM values of three independent experiments.
Supplementary Material
Figure S1. shRNA depletion of FA proteins in HT0180 and E006AA cells. Western blots of fibrosarcoma HT1080 (A-H) or prostate E006AA (I-K) cells infected with lentivirus expressing the indicated shRNA (see methods for a list of shRNAs) directed against indicated FA proteins, or mock infected, or infected with lentivirus expressing a nonspecific shRNA against luciferase. Loading controls are either α-tubulin or α-tubulin. A) Talin 1,2. B) Vinculin. C) Paxillin. D) FAK. E) α-actinin 1,4. In lane 3 is sh822 that did not result in efficient knockdown and was not used further. Only sh1299 and sh2287 were used to knockdown α-actinin in experiments. F) P130Cas. G) VASP. In lane 5 cells were transduced with a lentivirus that expressed both VASP sh444 and an RNAi-resistant isoform of VASP (rrhVASP-FH), containing a Flag and His epitope tag. Knockdown of endogenous VASP was rescued by co-transduction with RNAi-resistant VASP. H) Zyxin and concurrent endogenous Zyxin knockdown with sh756 and rescue with RNAi-resistant rrhZyxin-FH (right panel). I) E006AA cells and Zyxin. J) E006AA cells and P130Cas.
Figure S2. Dependence of 3-D cell speed on β 1 integrin, and correlations between 2-D and 3-D persistent time and protrusion number and growth rate. A. 3-D cell speed of control HT-1080 cells, HT-1080 cells in the presence of an anti-β1 integrin function-blocking antibody, and HT-1080 cells in the presence of a non-specific mouse IgG antibody. NS: P > 0.05. ***: P < 0.001. Bar graphs show mean and SEM values of three independent experiments. B. Absence of correlation between the persistence time of HT-1080 cells placed on conventional 2-D collagen I-coated substrates and cells placed inside 3-D collagen matrix. C and D. Correlation function between the number of actively growing protrusions per 90 min per cell and persistence distance during random cell motility (C) and between the growth rate of protrusions and the persistence distance for HT-1080 cells inside a 3-D collagen matrix (D).
Figure S3. Regulation of cell speed on a 2-D substrate by FA proteins in E006AA cells. 2-D cell speed of WT E006AA human prostate cancer cells and E006AA cells depleted of either p130Cas or zyxin on collagen coated glass. Bar graphs show mean and SEM values of three independent experiments.
Figure S4. Regulation of length and lifetime of protrusions by FA proteins. A and B. Maximum length and average protrusion length of protrusions for various HT-1080 cells depleted of FA proteins. C. Correlation function between 3-D cell speed and the lifetime of protrusions. D-F. Correlations between (D) 3-D cell speed and averaged protrusion length, (E) averaged protrusion length and 3-D persistence distance, and (F) lifetime of protrusions and 3-D persistence distance for HT-1080 cells inside a 3-D collagen matrix. Bar graphs show mean and SEM values of three independent experiments.
Figure S5. Correlation between cell motility in a 3-D matrix and cell-matrix interaction parameters. A-E. Correlations between (A) 3-D HT-1080 cell speed and total bead movement in the matrix (total matrix deformation), (B) 3-D cell speed and % permanent matrix deformation (matrix remodeling), (C) growth rate of protrusions and maximum bead displacement (traction), (D) number of protrusions per 90 min per cell and maximum bead displacement (traction), (E) 3-D cell speed and maximum bead displacement per cell (traction per pseudopodia).
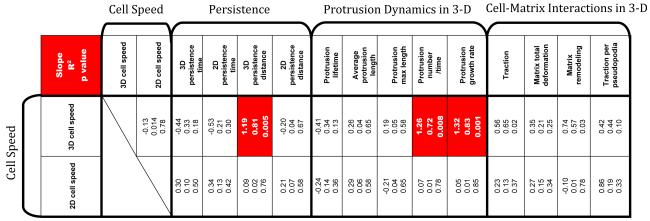
Table ST1. Table of correlations among cell motility parameters, protrusion dynamics parameters, and cell-matrix interaction parameters. The three numbers in each box represent the slope of a linear fit between the two parameters considered, the R squared value of the fit, and the p value. Boxes in red show high statistical significance. Cell motility parameters include: speed, persistence time, and persistence distance of cells both on 2-D substrates and inside a 3-D matrix. Protrusion dynamics parameters include: lifetime, length, maximum length, number of protrusions per 90 min, and growth rate of individual protrusions. Cell-matrix interaction parameters include: traction (averaged maximum bead displacements), total matrix deformation (averaged total bead movement), matrix remodeling (percent permanent matrix deformation), and traction per pseudopodia (averaged maximum bead displacements per cell). A strong correlation is defined as having slope ≥ 0.80 and R2 ≥ 0.70 and p ≤ 0.009.
table.
Acknowledgements
The authors acknowledge support from NIH (CA143868, GM084204, GM080673, and CA85839). SIF was supported by ARCS and NSF-GRFP. We thank Michael McCaffery and Ned Perkins for help with confocal microscopy, members of the Wirtz and Longmore groups for helpful discussions, and Dr. John Isaacs of JHMI and his group for generously providing E006AA prostate cancer cells.
Appendix
Material and Methods
Cell culture
HT-1080 cells (ATCC, Manassas, VA) were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% (v/v) fetal bovine serum (ATCC) and 0.1% gentimicin (Sigma, St. Louis, MO). E006AA cells (provided by Dr. John Isaacs, JHMI) were cultured in RPMI (Roswell Park Memorial Institute) 1640 medium supplemented with 10% (v/v) fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Invitrogen). Medium for HT-1080 and E006AA cells transfected with shRNA constructs (see below) also included 1.5-μg/ml and 3.0-μg/ml puromycin respectively for selection. Cells were maintained at 37°C and 5% CO2 in a humidified environment during culture and imaging. The cells were passaged every 2-3 days.
Depletion of FA proteins with shRNAs
The RNAi sequences targeting mRNA of each FA component or isoforms were selected with RNAi design online program from Dharmacon (Waltham, MA) (with advanced options if targeting multiple isoforms or species). Three or four targeting sites were chosen for each gene. After testing in HT-1080 or E006AA cells with lentiviral mediated RNAi, we selected those that showed more than 90% knocking down efficiency for subsequent experiments. They include (the number after the sequence denotes the targeting position in mRNA):
mh-Talin1,2 GTGGATGAGAAGACCAAGGA (1372);
mh-Talin1,2 GCCAAGGTGATGGTGACCAA (6706);
mh-Vinculin GCCAAGCAGTGCACAGATA (2993);
mh-Vinculin GCCACAGAGATGCTGGTTCA (3144);
h-Paxillin GCAAGGACTACTTCGACATGT (1366);
mh-Paxillin gTCAAGGAGCAGAACGACAA (1770);
mh-FAK GCGAGTATTAAAGGTCTTTCA (332);
mh-FAK GGGCATCATTCAGAAGATA (507);
h-p130Cas GGTCGACAGTGGTGTGTAT (1336);
h-p130Cas GCAGTTTGAACGACTGGAACA (2227);
mh-ACTN1,4 GCAGAGAAGTTCCGGCAGA (1299);
h-ACTN1,4 GACAACAAGCACACCAACTA (2287);
h-Zyxin GGATGACATGACCAAGAAT (623);
h-Zyxin GCCTGTGTCTTTGGCTAACA (756);
mh-VASP GCGTCCAGATCTACCACAA (444);
mh-VASP GAGCCAAACTCAGGAAAGT (586).
A firefly luciferase shRNA was used as a control (5′- GCTTACGCTGAGTACTTCGA).
shRNA expression cassettes and lentiviral vectors were constructed as described 32. To rescue the depleted VASP or zyxin, RNAi resistant isoform of hVASP (rrhVASP) or rrhZyxin with four point mutations in the VASP or zyxin shRNA target sequence were generated. Full length rrhVASP and rrhZyxin were obtained by joint PCR and subcloned into pFLRu-VASP-shRNA(444) or pFLRu-hZyxin-shRNA(756) in-frame with C-terminal Flag-His tag. Following the method described in 32, lentiviruses were generated, HT1080 cells were transduced and selected in medium containing 2 μg/ml puromycin for three days. Western blots were conducted as described 32.
2-D Collagen I Substratum
Two-dimensional cell culture plates were coated with soluble rat tail type I collagen in acetic acid (BD Biosciences, San Jose, CA) to achieve a coverage of 33 μg/cm2 and incubated at room temperature for 2 h. Plates were then washed gently three times with PBS and seeded with a low density of cells. For 2.5-D experiments, collagen I gel (final concentration of 2 mg/ml, see below) was added on top of 2-D cell cultures after cells were allowed to spread for 2-3 h and incubated for 5 h.
3-D Collagen I Matrix
Cell-impregnated 3-D collagen matrices were prepared by mixing cells suspended in culture medium and 10X reconstitiution buffer, 1:1 (v/v), with soluble rat tail type I collagen in acetic acid (BD Biosciences) to achieve a final concentration of 2 mg/ml collagen. 1M NaOH was then added to normalize pH (pH 7.0, 10-20 μl 1M NaOH), and the mixture was placed in multiwell, coverslip-bottom culture plates (LabTek, Campbell, CA). In integrin blocking experiments, β1 integrin antibody (ms 4B4-FITC, Beckman Coulter) or a non-specific mouse IgG antibody (Sigma, St. Louis, MO) was added to cell suspension and incubated for 10min before mixing with collagen. For traction experiments, 3.6-μm diameter carboxylated polystyrene beads (Duke Scientific, Palo Alto, CA) were added in the first step. All ingredients were kept chilled and care was taken to avoid bubbles. Collagen gels solidified overnight in an incubator at 37°C and 5% CO2, then 500 μl of cell culture medium was added. The concentration of the collagen matrix was chosen to be 2 mg/ml so that the matrix pore size (<1 μm) was significantly smaller than cell body and nucleus. Cell density was kept low to ensure that single cell motility measurements were accurate. We verified that cells continued to proliferate normally inside the 3-D matrix over >48 h.
Immunofluorescence microscopy and confocal microscopy
To visualize vinculin, zyxin, and actin in cells plated on 2-D or in 2.5-D, cells were fixed, permeablized, then incubated with primary anibodies against vinculin and zyxin (Sigma) and Alexa Fluor 568 phalloidin (Invitrogen), respectively, for 1 h. Images of cells were collected using a Cascade 1K CCD campera (Roper Scientific, Trenton, NJ) mounted on a Nikon TE2000E epiflorescence microscope equipped with a 60x oil-immersion objective (Nikon, Melville, NY) and controlled by Metamorph imaging software (Universal Imaging, Sunnyvale, CA) 23, 24. Mouse monoclonal antibodies against talin, vinculin, α-ACTN1, α-tubulin, α-tubulin, and rabbit polyclonal antibody against zyxin were purchased from Sigma; rabbit polyclonal antibody against p130Cas was from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit polyclonal antibody against VASP, paxillin, FAK were from Cell Signaling (Danvers, MA); rabbit polyclonal antibody against α-ACTN4 was from LifeSpan Biosciences (Seattle, WA). To visualize vinculin and zyxin in live cells embedded in 3-D collagen matrices, HT-1080 cells transfected with EGFP-vinculin or EGFP-zyxin were imaged using a Zeiss Confocor II Confocal Microscope (Integrated Imaging Center, JHU).
Cell speed and persistence
Cells embedded in 3-D collagen matrices were imaged at low magnification (10X) for 16.5 h. Speed was determined by tracking single cells using image recognition software (Metamorph/Metavue), with distance/time measurements taken every 2 min then averaged for each cell. An excel macro was written which uses tracking data (x, y coordinates, distance, and time) obtained by tracking individual cells to calculate persistent time, persistent distance, and angles explored between persistent moves. A persistent move is defined as the length (≥10 μm) traveled by a cell before it makes a significant change in direction (angle between previous direction and new direction <70°). These definitions were deduced so as to exclude noise incorporated by microscope stage drift, etc. Persistent distance is the distance traveled by a cell during a persistent move, and persistent time is the duration of a persistent move. The speed, persistence time, and persistence distance of at least 35 cells were computed on three different days for each condition.
Cell protrusion dynamics
Protrusions at least 5 μm in diameter and length were monitored using a Roper Scientific Cascade 1K CCD camera mounted on a Nikon TE2000E microscope. Low magnification images were taken 2 min apart for at least 12 h. The length, lifetime, position along the cell periphery, and number per cell were computed by hand using Metamorph. The growth rate of protrusion was taken as the ratio of length and lifetime. The protrusions of at least 10 cells were characterized on three different days for each condition.
Local 3-D traction of the matrix
To asses the matrix-remodeling capabilities of the wild type and shRNA transfected cells, mechanical traction and relaxation of the collagen matrix induced by cell migration was measured by tracking 3.6-μm carboxylated polystyrene beads in the x, y, and z, directions as previously described 25. Four measurements were obtained: total distance traveled by the beads, maximum displacement of all beads from their original location, maximum bead displacement of beads per cell, and final bead displacement divided by total distance to measure percent of permanent deformation of the collagen gel. The local matrix traction in the vicinity of at least 5 cells (~30 beads per cell) was measured on three different days for each condition.
Statistics
The number of cells examined for each experiment is indicated in the figure captions. Mean values, standard error of measurement (SEM), and statistical analysis were calculated and plotted using Graphpad Prism (Graphpad Software, San Diego, CA). Two-tailed unpaired t tests were conducted to determine significance, which was indicated using standard Michelin Guide scale (*** for P < 0.001, ** for P < 0.01, and * for P < 0.05).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Sastry SK, Burridge K. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp Cell Res. 2000;261:25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- 2.Riveline D, et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyamoto S, et al. Integrin function - molecular hierarchies of cytoskeletal and signaling molecules. J. Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381:531–5. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- 5.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 6.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–9. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 7.Mao Y, Schwarzbauer JE. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of alpha5beta1 versus alphavbeta3 integrin receptors. Cell Commun Adhes. 2006;13:267–77. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–99. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Friedl P, Entschladen F, Conrad C, Niggemann B, Zanker KS. CD4+ T lymphocytes migrating in three-dimensional collagen lattices lack focal adhesions and utilize beta1 integrin-independent strategies for polarization, interaction with collagen fibers and locomotion. Eur J Immunol. 1998;28:2331–43. doi: 10.1002/(SICI)1521-4141(199808)28:08<2331::AID-IMMU2331>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 10.Petroll WM, Ma L, Jester JV. Direct correlation of collagen matrix deformation with focal adhesion dynamics in living corneal fibroblasts. J Cell Sci. 2003;116:1481–91. doi: 10.1242/jcs.00357. [DOI] [PubMed] [Google Scholar]
- 11.Wehrle-Haller B, Imhof B. The inner lives of focal adhesions. Trends Cell Biol. 2002;12:382–9. doi: 10.1016/s0962-8924(02)02321-8. [DOI] [PubMed] [Google Scholar]
- 12.Pelham RJ, Jr., Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shemesh T, Geiger B, Bershadsky AD, Kozlov MM. Focal adhesions as mechanosensors: a physical mechanism. Proc Natl Acad Sci U S A. 2005;102:12383–8. doi: 10.1073/pnas.0500254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanada M, et al. Focal adhesion kinase is activated in invading fibrosarcoma cells and regulates metastasis. Clin Exp Metastasis. 2005;22:485–94. doi: 10.1007/s10585-005-3733-6. [DOI] [PubMed] [Google Scholar]
- 15.Salgia R, et al. Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene. 1999;18:67–77. doi: 10.1038/sj.onc.1202273. [DOI] [PubMed] [Google Scholar]
- 16.Yu YP, Luo JH. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–9. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]
- 17.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf K, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–77. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, et al. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 2008;22:1231–43. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–9. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren XD, et al. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J Cell Sci. 2000;113(Pt 20):3673–8. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
- 23.Panorchan P, Lee JS, Kole TP, Tseng Y, Wirtz D. Microrheology and ROCK signaling of human endothelial cells embedded in a 3D matrix. Biophys J. 2006;91:3499–507. doi: 10.1529/biophysj.106.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JS, et al. Ballistic intracellular nanorheology reveals ROCK-hard cytoplasmic stiffening response to fluid flow. J Cell Sci. 2006;119:1760–1768. doi: 10.1242/jcs.02899. [DOI] [PubMed] [Google Scholar]
- 25.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys J. 2008;95:4077–88. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–61. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 27.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 28.Kole TP, Tseng Y, Jiang I, Katz JL, Wirtz D. Intracellular mechanics of migrating fibroblasts. Mol Biol Cell. 2005;16:328–38. doi: 10.1091/mbc.E04-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munevar S, Wang YL, Dembo M. Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Mol Biol Cell. 2001;12:3947–54. doi: 10.1091/mbc.12.12.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 31.Tilghman RW, Parsons JT. Focal adhesion kinase as a regulator of cell tension in the progression of cancer. Semin Cancer Biol. 2008;18:45–52. doi: 10.1016/j.semcancer.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ngu H, et al. Effect of focal adhesion proteins on endothelial cell adhesion, motility and orientation response to cyclic strain. Ann Biomed Eng. 2010;38:208–22. doi: 10.1007/s10439-009-9826-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. shRNA depletion of FA proteins in HT0180 and E006AA cells. Western blots of fibrosarcoma HT1080 (A-H) or prostate E006AA (I-K) cells infected with lentivirus expressing the indicated shRNA (see methods for a list of shRNAs) directed against indicated FA proteins, or mock infected, or infected with lentivirus expressing a nonspecific shRNA against luciferase. Loading controls are either α-tubulin or α-tubulin. A) Talin 1,2. B) Vinculin. C) Paxillin. D) FAK. E) α-actinin 1,4. In lane 3 is sh822 that did not result in efficient knockdown and was not used further. Only sh1299 and sh2287 were used to knockdown α-actinin in experiments. F) P130Cas. G) VASP. In lane 5 cells were transduced with a lentivirus that expressed both VASP sh444 and an RNAi-resistant isoform of VASP (rrhVASP-FH), containing a Flag and His epitope tag. Knockdown of endogenous VASP was rescued by co-transduction with RNAi-resistant VASP. H) Zyxin and concurrent endogenous Zyxin knockdown with sh756 and rescue with RNAi-resistant rrhZyxin-FH (right panel). I) E006AA cells and Zyxin. J) E006AA cells and P130Cas.
Figure S2. Dependence of 3-D cell speed on β 1 integrin, and correlations between 2-D and 3-D persistent time and protrusion number and growth rate. A. 3-D cell speed of control HT-1080 cells, HT-1080 cells in the presence of an anti-β1 integrin function-blocking antibody, and HT-1080 cells in the presence of a non-specific mouse IgG antibody. NS: P > 0.05. ***: P < 0.001. Bar graphs show mean and SEM values of three independent experiments. B. Absence of correlation between the persistence time of HT-1080 cells placed on conventional 2-D collagen I-coated substrates and cells placed inside 3-D collagen matrix. C and D. Correlation function between the number of actively growing protrusions per 90 min per cell and persistence distance during random cell motility (C) and between the growth rate of protrusions and the persistence distance for HT-1080 cells inside a 3-D collagen matrix (D).
Figure S3. Regulation of cell speed on a 2-D substrate by FA proteins in E006AA cells. 2-D cell speed of WT E006AA human prostate cancer cells and E006AA cells depleted of either p130Cas or zyxin on collagen coated glass. Bar graphs show mean and SEM values of three independent experiments.
Figure S4. Regulation of length and lifetime of protrusions by FA proteins. A and B. Maximum length and average protrusion length of protrusions for various HT-1080 cells depleted of FA proteins. C. Correlation function between 3-D cell speed and the lifetime of protrusions. D-F. Correlations between (D) 3-D cell speed and averaged protrusion length, (E) averaged protrusion length and 3-D persistence distance, and (F) lifetime of protrusions and 3-D persistence distance for HT-1080 cells inside a 3-D collagen matrix. Bar graphs show mean and SEM values of three independent experiments.
Figure S5. Correlation between cell motility in a 3-D matrix and cell-matrix interaction parameters. A-E. Correlations between (A) 3-D HT-1080 cell speed and total bead movement in the matrix (total matrix deformation), (B) 3-D cell speed and % permanent matrix deformation (matrix remodeling), (C) growth rate of protrusions and maximum bead displacement (traction), (D) number of protrusions per 90 min per cell and maximum bead displacement (traction), (E) 3-D cell speed and maximum bead displacement per cell (traction per pseudopodia).
Table ST1. Table of correlations among cell motility parameters, protrusion dynamics parameters, and cell-matrix interaction parameters. The three numbers in each box represent the slope of a linear fit between the two parameters considered, the R squared value of the fit, and the p value. Boxes in red show high statistical significance. Cell motility parameters include: speed, persistence time, and persistence distance of cells both on 2-D substrates and inside a 3-D matrix. Protrusion dynamics parameters include: lifetime, length, maximum length, number of protrusions per 90 min, and growth rate of individual protrusions. Cell-matrix interaction parameters include: traction (averaged maximum bead displacements), total matrix deformation (averaged total bead movement), matrix remodeling (percent permanent matrix deformation), and traction per pseudopodia (averaged maximum bead displacements per cell). A strong correlation is defined as having slope ≥ 0.80 and R2 ≥ 0.70 and p ≤ 0.009.