Abstract
The emergence of functional neuronal connectivity in the developing cerebral cortex depends on 1) neural progenitor differentiation, which leads to the generation of appropriate number and types of neurons, and 2) neuronal migration, which enables the appropriate positioning of neurons so that the correct patterns of functional synaptic connectivity between neurons can emerge. In this review, we discuss 1) currently available methods to study neural progenitor development and differentiation in the developing cerebral cortex and emerging technologies in this regard, 2) assays to study the migration of descendents of progenitors (i.e., neurons) in vitro and in vivo, and 3) the use of these assays to probe the molecular control of these events in the developing brain and evaluation of gene functions disrupted in human neurodevelopmental disorders.
Keywords: cortical development, cortical progenitors, neuronal migration
Introduction
The emergence of functional neuronal organization and connectivity in the developing cerebral cortex depends on 2 crucial early developmental events: 1) the differentiation and proliferation of neural progenitors, leading to the timely generation of appropriate numbers of cortical neuronal subtypes, and 2) migration of neurons to specific locations in the cortex where they establish functional synaptic connections with other neurons (Marin and Rubenstein 2003). Disruptions in these developmental events can have severe consequences, including mental retardation, lissencephaly, tumorigenesis, and neuropsychiatric disorders like autism and schizophrenia (Ayala et al. 2007; Kerjan and Gleeson 2007; Jackson and Alvarez-Buylla 2008). Furthermore, analogous events occur in the postnatal brain, where new neurons are born in neurogenic niches and targeted to where they are needed as part of either normal maintenance of the neural circuitry or in response to disease or injury (Falk and Frisen 2005; Ghashghaei et al. 2007; Zhao et al. 2008; Kriegstein and Alvarez-Buylla 2009). Thus, our ability to study the complexities of neural progenitor differentiation and neuronal migration is essential to understanding normal cerebral cortical formation, the prevention and treatment of neurodevelopmental disorders, and the regeneration of functional connectivity in damaged or diseased sites in the adult brain.
Since neural progenitor differentiation and neuronal migration depend on complex intercellular and intracellular interactions, the most effective methods to study these processes should combine an ability to genetically target and visualize relevant cells in their native environment with an analysis of global patterns of progenitor formation and neuronal movement. Traditional techniques for studying progenitor differentiation and neuronal migration have relied on either analysis of fixed tissue or in vitro culture of individual cells. But the recent advances in live imaging techniques and genetic labeling of distinct cell types in the developing brain have accelerated our ability to analyze the complexities of the fundamental processes of cerebral cortical formation. This review will describe the currently available and emerging methods for studying neural progenitor differentiation and neuronal migration, evaluating their uses and limitations, and will propose how these methodologies can be adapted and improved to resolve fundamental issues in cerebral cortical development and disease.
The first part of the review will focus on assays to study the differentiation of different types of neural progenitors (neuroepithelial cells, radial progenitors, and intermediate precursors) in the developing cortex. In the second part, we will survey neuronal migration assays, addressing 2 significant aspects of migration: 1) movement of neurons from their places of birth to positions where they form functional synaptic connections and 2) migratory pathway selection and migration mode differences among neuronal subtypes. Finally, we will describe how these methods can be adopted to test the functions of genes that are rapidly being identified as candidate susceptibility genes for human brain disorders in which progenitor development or neuronal migration is affected.
Progenitor Differentiation
During cerebral cortical development, after closure of the anterior neural tube, neuroepithelial cells proliferate within the walls of the ventricle, populating the ventricular zone (VZ) with a pseudostratified layer of bipolar cells (Fig. 1). At the onset of neurogenesis, neuroepithelial cells transform into radial glial progenitors (RGPs), which are characterized by a highly polarized morphology and a glial-like molecular identity (reviewed in Götz and Huttner 2005). Initially, RGPs expand their population by dividing symmetrically to produce 2 daughter RGPs. Then, as neurogenesis proceeds, an increasing number of RGPs begin to divide asymmetrically, giving rise to a daughter neuron or intermediate progenitor (IP) and a self-renewing daughter RGP (Malatesta et al. 2000; Miyata et al. 2001; Anthony et al. 2004; Noctor et al. 2004, 2008). IPs move to the subventricular zone (SVZ), where they form an additional neurogenic niche that supplies the majority of projection neurons to all layers of the cerebral cortex (Haubensak et al. 2004; Kowalczyk et al. 2009). As neurogenesis wanes, asymmetric neurogenic RGP divisions decrease in frequency due to the terminal differentiation of RGPs into glia and ependymal cells (Noctor et al. 2008) (Fig. 1). During neurogenesis, significant molecular and morphological heterogeneity among neural progenitors in the VZ has been characterized and this heterogeneity may be essential to create neuronal diversity in cerebral cortex (Gal et al. 2006; Mizutani et al. 2007; Anthony and Heintz 2008; Howard et al. 2008; Kawaguchi et al. 2008; Hansen et al. 2010; Stancik et al. 2010).
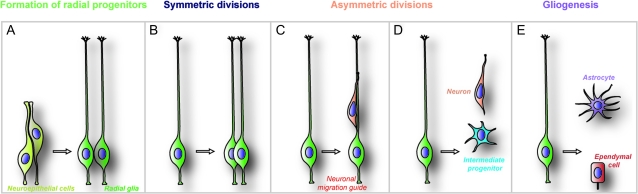
Figure 1.
Progression of radial progenitor development in embryonic cerebral cortex. Initially, RGPs are established from an undifferentiated sheet of neuroepithelial cells (A). Radial glia then divide symmetrically and generate more radial glia (B). Subsequent asymmetric division of radial glial cells gives rise to daughter neurons and other radial glial cells (C and D). Neurons either somally translocate or attach to radial glial guides (C) as they migrate to the cortical plate. Asymmetric divisions of a radial glial cell can also yield a daughter neuron and an IP (D). IPs divide and generate neurons. During late stages of corticogenesis, as neurogenesis and migration dwindle, radial progenitors give rise to astrocytes or become ependymal cells (E). These distinct phases of progenitor development can be studied using clonal analysis of isolated progenitors, neurosphere assays, real-time tracking of labeled progenitors in embryonic cortex, or in utero imaging. An important point to note is that significant molecular and morphological heterogeneity of radial progenitors exists within the VZ of the developing cerebral cortex and is not outlined in this figure.
In addition to their role as cortical progenitors, RGPs perform another important function as guides for neuron migration. Specifically, the pial-directed radial process of RGPs provides a permissive and instructive scaffolding for the oriented migration and placement of newly generated neurons (Ayala et al. 2007). Thus, as neuronal progenitors and migratory guides, RGPs perform multiple evolving functions as cortical development unfolds (Fig. 1), making it necessary to develop tools to analyze each of these functions.
Time-lapse Analysis of Adherent Neural Progenitor Clones
A fundamental question in radial progenitor development during corticogenesis is what governs the timing and mode of RGP division and the gradual restriction of daughter cell fate as development proceeds. Is the sequential production of RGPs, IPs and neurons, and then glia regulated by cell-intrinsic programs, extrinsic cues, or a combination of both? Similarly, what signals impinging on progenitors determine the orderly generation of layer-specific projection neurons?
In vitro assays using isolated, single cortical progenitors from different embryonic ages can be used to selectively study and manipulate RGP proliferation, differentiation, and cell fate in a defined environment. In this method, dissociated cortical progenitors are plated on an adherent substrate at clonal density, so that cell–cell contacts are minimized, and the only extracellular cues present are produced from the clones themselves or exogenously added to the defined culture medium (Shen et al. 2006). Over several days in vitro, single progenitors generate other progenitors, distinct neuronal subtypes, and glia, with a timing and order that parallels the timing and order observed in vivo (Shen et al. 2006). The identity of daughter cells generated from isolated progenitors can be determined using cell type–specific markers, thus defining the mode of progenitor division. For instance, if an isolated progenitor gives rise to only RGPs, it underwent symmetric self-renewing divisions (Fig. 1A). If the daughter cells are of different cell types (i.e., neurons, intermediate precursor, or astroglia), they are likely the result of asymmetric divisions (Fig. 1C,D). While this assay is ideal to test the cell-intrinsic capacities of cortical progenitors from different developmental stages (Fig. 1), extrinsic cues critical for distinct patterns of progenitor division can also be presented to the isolated clones and tested for their influence on the mode of progenitor division.
Time-lapse analysis of neural progenitor clones is a further extension of this technique where long-term live-cell imaging of isolated progenitors and their descendents (Fig. 2), followed by correlative staining for progenitor, neuronal, and glial subtypes, can be used to establish a comprehensive lineage tree (Al-Kofahi et al. 2006; Shen et al. 2006). The effects of extrinsic factors or cell–cell contacts on progenitor division can be examined by adding diffusible cues to the medium or culturing progenitors at higher density, respectively (Ravin et al. 2008; Shen et al. 2006). Additional refinements to the system might include methods to specify the identity of the initial progenitors. For example, one can use fluorescence activated cell sorting to purify progenitors from transgenic mice expressing XFP under the control of a promoter active in a specific progenitor subpopulation (e.g., BLBP-GFP to isolate RGPs, Tα1-GFP to isolate short neural precursors, or Tbr2-GFP to isolate IPs; Table 1).
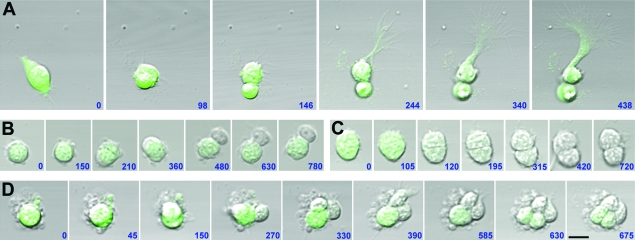
Figure 2.
Analysis of isolated cortical progenitor development. Radial progenitors micro dissected from the VZ of E14.5 BLBP-GFP embryonic cortices were plated at clonal density and isolated, and genetically defined radial progenitors (GFP+) were time-lapse imaged. (A) Division of GFP+ progenitor resulting in 2 other GFP+ cells, 1 of which extends a basal process. (B) Division of GFP+ progenitor resulting in 1 GFP+ cell and 1 GFP− cell. (C) Division of GFP+ progenitor resulting in 2 GFP− cells. (D) Generation of a mixed clone containing GFP+ and GFP− cells. The combined use of time-lapse analysis of genetically defined cortical progenitors with correlative immunolabeling of daughter cells at the end of imaging can be used to study patterns of normal and altered progenitor development. Time elapsed between observations are indicated in minutes. Simultaneous GFP and phase light images were obtained using a Zeiss inverted confocal microscope equipped with live-cell incubation chamber. Scale bar = 25 μm.
Table 1.
Progenitor-specific transgenic mouse lines
| Line | Time of initial expression | Labeled progenitor population | Reference |
| Rat α1-tubulin-YFP | E9.5 | Neuroepithelium | Sawamoto et al. (2001)a |
| IPs | |||
| Sox2-GFP | E10.5 | Neuroepithelium | Ellis et al. (2004) |
| RGPs | |||
| Postnatal dentate gyrus | |||
| Postnatal SVZ | |||
| Rat Nestin-GFP | E11.5 | Neuroepithelium | Yamaguchi et al. (2000) |
| RGPs | |||
| Postnatal dentate gyrus | |||
| Postnatal SVZ and RMS | |||
| GLAST-dsRed | E12.5 | RGPs | Glowatzki et al. (2006) |
| Postnatal dentate gyrus | |||
| Eomes/Tbr2-GFP | E12.5 | IPs | Arnold et al. (2009), Kowalczyk et al. (2009) |
| Human GFAP-GFP | E13.5 | RGPs | Malatesta et al. (2000), Zhuo et al. (1997) |
| Astrocytes | |||
| BLBP-GFP, YFP, dsRed | E14.5 | RGPs | Schmid et al. (2006) |
| Astrocytes | |||
| Postnatal dentate gyrus | |||
| ALDH1L1-GFP | E16.5 | RGPs | Anthony and Heintz (2007, 2008), Barres (2008) |
| Hes5-GFP | N.D. | RGPs | Akazawa et al. (1992), Gong et al. (2003) |
| Fezf2-GFP | E12.5 | Progenitors of deep layer projection neurons | Kwan et al. (2008), Gong et al. (2003) |
| Tis21-GFP | E10.5 | IPs and some RGPs | Haubensak et al. (2004), Attardo et al. (2008) |
Note: N.D., not determined.
Also expressed in early born neurons (Coksaygan et al. 2006).
The advantage of this technique over neurosphere assays, which are also used to evaluate progenitor proliferation, is that it permits complete registering of individual cell divisions and behavior and a clear analysis of intrinsic mechanisms at work in choosing cell fate. Neurospheres, which are essentially a collection of cells arising from a progenitor proliferating in suspension, do not permit such analysis. However, the neurosphere culture is a useful assay to expand progenitors (Fig. 1B) using extrinsic factors. Neurospheres thus generated can then be differentiated on an adhesive substratum to generate neurons and glia.
An obvious drawback of this method of clonal analysis of isolated cortical progenitors is that dissociated progenitors neither encounter the permissive microenvironments (i.e., progenitor niches) found in vivo nor do they maintain critical features such as polarity and orientation they would have in the intact brain. This limitation might have major consequences considering that coordinated patterns of symmetric and asymmetric progenitor divisions in vivo rely on appropriate cell–cell contacts (Lu et al. 2001) that are absent in isolated cultures. Cortical slice assays help overcome these limitations of isolated progenitor cultures.
Cerebral Cortical Slice Assays for Real-Time Analysis of Progenitor Development
Improvements in tissue culture methods and live-cell imaging in tissue slices have made possible the real-time tracking of individual progenitors and their progeny within tissue explants. Currently, this involves targeted expression of a fluorescent protein in progenitors followed by time-lapse confocal imaging of slice cultures. Depending on the objective of the experiment, single or multiple progenitors can be targeted in specific proliferative regions of both embryonic and postnatal cortices. The most common modes of fluorescent gene expression rely on electroporation (in utero and ex utero), viral transduction, and germ line genetic manipulation, each having unique advantages. For example, while viral transduction enables tracking of individual cells in isolation, electroporation and genetic manipulation allow targeting of cohorts of specific progenitor subpopulations.
Likely the most rapid method of progenitor transfection, DNA electroporation results in 1 or more regions of the cortex expressing a fluorescent transgene in multiple progenitors and their neuronal and glial progeny. In this method, concentrated (2–5 mg/mL) purified plasmid DNA containing the gene of interest is injected with a micropipette into the lateral ventricle of embryos, either removed from the uterus (ex vivo) (Gongidi et al. 2004) or left intact in the embryonic sac (in utero) (Tabata and Nakajima 2001). A low-voltage current is then passed through the cortex, and the DNA is incorporated into progenitors lining the lateral ventricles. The current's path can be oriented differently so that plasmid DNA will be taken up preferentially by progenitors that fall within the current's path. In this way, progenitors in the dorsal (Tabata and Nakajima 2001), ventral, and hippocampal (Navarro-Quiroga et al. 2007) anlages can be exclusively targeted. After ex utero electroporation, the brains are removed, sliced, and maintained in vitro. After sufficient time for transgene expression (24–48 h), long-term live-cell imaging can be performed on labeled progenitors in slices to analyze their patterns of differentiation (Xie et al. 2007; Yoon et al. 2008; Yokota et al. 2009) using a confocal microscope equipped with live-cell incubation system. With in utero electroporation, the embryos are returned to the mother, where they develop for the desired number of additional days. The advantage of the ex utero approach is that it avoids the potential for embryonic death that, at times, occurs from the insults associated with in utero electroporation and allows for rapid analysis of large numbers of cortical progenitors. In contrast, in utero electroporation allows the cortex to develop intact until the effect of transgene expression in progenitors is ready to be analyzed.
In its simplest form, electroporation followed by live-cell imaging is a powerful method for observing the dynamics of progenitors as they differentiate, divide, and generate neurons. With the development of brighter, stable fluorescent markers, critical features of progenitors such as interkinetic nuclear movement, interprogenitor interactions, and pial end-feet dynamics can be followed in RGPs in both normal cortex and models of neurodevelopmental disorders (Xie et al. 2007; Yokota et al. 2009). Moreover, combining fluorescent labeling with genetic manipulation of progenitors, by coelectroporating mutated genes or short hairpin RNA (shRNA) constructs, can be an extremely efficient way to analyze the molecular mechanisms that regulate progenitor morphology, proliferation, and neurogenesis (LoTurco et al. 2009; Loulier et al. 2009). For example, the efficiency and reproducibility of electroporation make this technique amenable to rapid screening with shRNA libraries or shRNA screens targeting specific susceptibility genes for neurodevelopmental disorders. Further, by varying the timing of electroporation and using progenitor-type specific promoters, one can target and study radial progenitors and their progeny at distinct stages of development (Mizutani et al. 2007; Anthony and Heintz 2008). Postnatally, electroporation can be used to label and study progenitors and new neurons in the neurogenic niches (e.g., SVZ) of the neonatal or mature brain (Barnabe-Heider et al. 2008) This approach can significantly facilitate our ability to study adult neurogenesis and the response of adult neural progenitors to injury and disease.
Viral transduction offers an alternative to electroporation. In this technique, high-titer retroviral particles expressing a gene of interest and a fluorescent marker are injected into the lateral ventricle 1–2 days prior to slice culture and imaging. The viral particles diffuse evenly through the ventricular space and infect single isolated progenitors, which can then be visualized and tracked as they divide and generate daughter neurons (Noctor et al. 2001, 2004, 2008). Because viral vectors stably integrate into the genome, both progenitors and their progeny will express the transgene at relatively consistent levels. This is in contrast to electroporated cells where the plasmid DNA does not always integrate, and expression levels can vary widely among cells. However, when analysis of large cohorts of progenitors and rapid gene manipulation are required, electroporation-based methods are optimal.
In contrast to electroporation or viral transduction, germ INS> line genetic manipulation provides a non-invasive technique for labeling progenitors. Recently, a number of transgenic mouse lines have been produced that express fluorescent protein tags under the control of different progenitor-specific promoters (Table 1). These lines label neuroepithelial precursors (Ellis et al. 2004), RGPs (Zhuo et al. 1997; Malatesta et al. 2000; Gong et al. 2003; Haubensak et al. 2004; Schmid et al. 2006; Regan et al. 2007; Attardo et al. 2008; Kwan et al. 2008), and IPs (Sawamoto et al. 2001; Gong et al. 2003; Haubensak et al. 2004; Attardo et al. 2008; Kwan et al. 2008; Arnold et al. 2009) and offer uniform labeling of a majority of cells in a specific progenitor population. These lines are powerful tools to investigate progenitor dysfunctions in neurodevelopmental disorders because when crossed with mutant models of neurodevelopmental diseases, they can reveal defects in large cohorts of progenitors in distinct cortical areas that might not be seen with more focal labeling of progenitors achieved with electroporation or retroviral vectors. A recent study demonstrating how defects in neurogenesis may lead to the increased inhibitory drive in the brains of Down's syndrome mouse model illustrates the insights that could be gained by combining neurodevelopment disease models with other relevant genetic tools (Chakrabarti et al. 2010).
Advanced Techniques to Study Progenitor Development in Intact Brains
In spite of many advantages, the isolated progenitor cell culture methods and the slice-based assays do not entirely preserve the neural progenitor cell niche. Neural stem cell niche in the developing brain, consisting of the extracellular matrix, inter-progenitor adhesive contacts, and the vasculature within the proliferative zone, can dynamically influence the appropriate unfolding of neural progenitor division and differentiation (Chenn and Walsh 2003; Klezovitch et al. 2004; Shen et al. 2004; Rasin et al. 2007; Loulier et al. 2009; Stubbs et al. 2009; Weimer et al. 2009). As such, it will be vital to develop methods to observe and study neural progenitor development in live embryonic brains in utero. Multiphoton microscopy-based methods can be employed to evaluate neural progenitor development in the intact developing cerebral cortex in utero (see Supplementary Figure 1 and Supplementary Movie 1). Embryonic mice expressing XFP in neural progenitors can be used to image specific types of neural progenitors at different stages of cortical development in vivo. Although technical issues such as mechanical stability of embryos necessary for repeated long-term imaging of an identical area of the proliferative zone and the increasing thickness of cerebral wall during development can present obstacles for this type of imaging, the ability to follow neural progenitor differentiation and behavior in utero holds great promise for our understanding of how patterns of progenitor differentiation influence the formation of cerebral cortex.
Neuronal Migration
Once generated from proliferating progenitors, postmitotic neurons migrate from the proliferative VZ and SVZ to their final sites of differentiation. During development, the waves of neuron migration that commence in distinct regions of the brain differ in their timing, mode of movement, cytoskeletal dynamics, and response to guidance cues. Developmental cascades triggered by migratory mode and pathway choice are thought to lead to the emergence of distinct neuronal subtypes. How these complex patterns of neuronal movement and identity development are orchestrated so that the right types and numbers of neurons are deposited into appropriate areas and layers of the cerebral cortex remains unclear.
In general, neurons utilize 2 modes of migration, radial and tangential, depending on their origin and destination. In the dorsal telencephalon, early generated neurons of the preplate and deeper layers migrate radially using somal translocation, whereas later generated neurons utilize elongated radial glial guides to attain their more distant locations at the interface between cortical plate and marginal zone (Rakic 1972, 2003; Sidman and Rakic 1973; Takahashi et al. 1990; Miyata et al. 2001; Nadarajah et al. 2001; Hatten 2002; Fig. 1). Meanwhile, in the ventral telencephalon, GABAergic cortical interneurons are generated in the ganglionic eminences, and they migrate tangentially into the cerebral wall (de Carlos et al. 1996; Anderson et al. 1997; Ang et al. 2003; Marin and Rubenstein 2003). Postnatally, radial migration continues in the dentate gyrus of the hippocampus as newly generated granule neurons migrate from the SVZ into the granular layer (Nowakowski and Rakic 1979). Tangential migration also occurs throughout life, exemplified by olfactory bulb neuroblasts that migrate from the anterior SVZ to the olfactory bulb, through the rostral migratory stream (Lois and Alvarez-Buylla 1994; Marin and Rubenstein 2003).
Several developmental brain disorders have been linked to defects in radial migration in humans, including periventricular heterotopia and lissencephaly (Kerjan and Gleeson 2007; Sarkisian et al. 2008). Also, aberrant migration, placement, and connectivity of the inhibitory GABAergic interneurons have been implicated in disorders such as schizophrenia, autism, and Tourette's syndrome (Di Cristo 2007). Thus, understanding the global orchestration of neuronal migration and the molecular mechanisms of cell movement will contribute toward delineating the biological basis of a plethora of neurodevelopmental disorders. Further, strategies to promote functional recovery in the injured adult brain depend on the generation of new neurons and the appropriate guidance of these neurons to where they are needed. Thus, an understanding of the process of neuronal migration could be of use in neural disease settings where there is a demand for neurogenesis and targeted guidance of neurons to repair and maintain functional neural circuitry.
In Vitro Migration Assay Using Dissociated Neuronal Cells
Migration assays using dissociated neurons and glial cells are highly suitable for the analysis of intracellular signaling activities (e.g., calcium transients or cytoskeletal dynamics), cell–cell and cell–substratum interactions during neuronal migration. In these assays, dissociated neurons and astroglia from embryonic cortical tissue can be recombined in vitro to promote neuronal attachment and migration on astroglial processes (Hatten and Liem 1981; Gregory et al. 1988; Elias et al. 2007). Tissue imprints of embryonic cortex made by peeling off adhered cortical slices can also yield neurons attached to radial glial processes (Barres et al. 1990; Anton et al. 1996). Alternately, dissociated neurons or micro explants from embryonic cortices can also be plated on specific adhesive substratum to analyze neuronal response to surface bound cues (Fishman and Hatten 1993; Lois et al. 1996; Wichterle et al. 1997; Tsai et al. 2007). Neuronal motility in all these different assays can be easily tracked using live-cell imaging. The advantage of these methods lies in the ability to simultaneously visualize individual isolated neurons and their intracellular cytoskeleton or signaling components (e.g., calcium, endosomes), making possible the study of cytoskeletal or signaling dynamics during neuronal migration. For example, using dissociated, migrating cerebellar granule neurons, it has been shown that the physical coupling of the nucleus and centrosome is a key feature of migrating neurons (Solecki et al. 2004; Tanaka et al. 2004; Bellion et al. 2005; Schaar and McConnell 2005) and that defects in this coupling are seen in neurons taken from mutant models of migration disorders (Tanaka et al. 2004; Koizumi et al. 2006; Tsai et al. 2007). These assays were instrumental in examining the role of neuron–glial contact and adhesion in neuronal migration, as they can be used to selectively perturb neuronal or glial components involved (Anton et al. 1997; Edmondson et al. 1988). Further, these techniques are also commonly used to study the role of guidance cues in directing migration, where neurons migrating from an explant are confronted with a choice of permissive/nonpermissive substratum or with a gradient of a soluble chemoattractant or repellant (Hu and Rutishauser 1996; Wu et al. 1999; Ward et al. 2003; Ward et al. 2005; Higginbotham et al. 2006).
Real-time Neuronal Migration in Embryonic Brain Slice Assay
A major deficit of migration assays using dissociated neurons is that they disrupt many of the endogenous signals critical for patterned migration within the developing cerebral cortex. Evaluating neuronal migration in living embryonic slices helps overcome this deficit. This method of tracking neurons as they migrate through their native environment has helped expand our understanding of aspects of migration that had only been postulated using fixed tissue, such as distinct phases and modes of projection or interneuron migration (Noctor et al. 2004), neuronal–RGP interactions (Gongidi et al. 2004), and the mechanisms of maintaining neuronal polarity during migration (Xie et al. 2003; Guerrier et al. 2009). Further, analysis of human neuronal migration disorders indicates that disruptions in selective stages of migration can have vastly different effects on cerebral cortical formation and function (e.g., disrupted initiation of migration linked to periventricular heterotopia, aberrant maintenance of migration associated with lissencephaly and cortical band heterotopia, and deregulated terminal phase of migration leading to over migration of neurons in Walker–Warburg syndrome (Gleeson and Walsh 2000). The cortical slice assay in which all different stages of neuronal migration can be visualized is highly amenable to the study of mechanisms underlying selective stages of neuronal migration. Migrating neurons in embryonic slices can be labeled with either fluorescent dyes or XFP transgenes. Lipophilic or vital dyes, such as DiI, DiO, CMTMR, and Oregon Green, can be introduced into the ventricles or on top of cortical slices to label large cohorts of migrating neurons in the embryonic cortex and postnatal RMS (Honig and Hume 1989; de Carlos et al. 1996; Doetsch and Alvarez-Buylla 1996; O'Rourke et al. 1997; Nadarajah et al. 2001; Anton et al. 2004; Miyata et al. 2004). Dye- or transgene-coated gold particles can also be directly introduced into migrating neurons using a biolistics delivery approach (Gan et al. 2000; O'Brien and Lummis 2006). In utero or ex vivo electroporation, as described above, will achieve similar results, without the risk of the lipophilic dyes diffusing from cell to cell (Tabata and Nakajima 2001).
A more reliable and exact method to label migrating neurons is to use transgenic mice, where fluorescent markers are expressed exclusively in distinct migrating neuronal populations in both embryonic brain and adult brain. Table 2 describes some of these transgenic lines and the neuronal populations where fluorescence is expressed. A useful feature of some of these lines is the cis-expression of a fluorescent protein and Cre under the control of a neuronal-specific promoter. When crossed to mice with a floxed allele, one can simultaneously delete expression of the floxed gene and visualize the mutant neurons, allowing imaging of the altered patterns of neuronal migration resulting from gene deletion (Stenman et al. 2003; Stanco et al. 2009). Electroporation of postmitotic neuron-specific promoters driving XFP (e.g., NeuroD-GFP) can also be used to selectively target and label migrating neurons in embryonic cortex (Yokota, Ring, et al. 2007; Heng et al. 2008; Guerrier et al. 2009). This approach, when used in transgenic mice expressing a different fluorescent protein under the control of a progenitor-specific promoter, can provide differential labeling of migrating neurons and radial progenitors and thus enable the investigation of complex interactions between radial glia and migrating neurons (Yokota, Gashghaei, et al. 2007). Similarly, generation of bitransgenic mice in which projection neurons and interneurons are labeled with different fluorescent tags (Table 2) will be highly useful in evaluating how interneurons and projections neurons coordinate their patterns of migration to generate cortical laminar organization.
Table 2.
Neuron-specific transgenic mouse indicator lines for migration assays
| Line | Time of initial expression | Labeled neuronal population | Reference |
| Dcx-GFP, dsRed | E11.5 | Projection neurons and interneurons | Gong et al. (2003), Koizumi et al. (2006), Wang et al. (2007) |
| Postnatal neuroblasts in SVZ and RMS | |||
| Cdk5-GFP | E11.5 | Postmitotic projection neurons | Gong et al. (2003), Tsai et al. (1993) |
| NeuroD6-GFP | E11.5 | IPs and postmitotic projection neurons | Gong et al. (2003), Shimizu et al. (1995) |
| p35-GFP | E12.5 | IPs and postmitotic projection neurons | Delalle et al. (1997), Gong et al. (2003) |
| Cx43-GFP | E12.5 | Postmitotic projection neurons | Gong et al. (2003) |
| β-3-tubulin-GFP | E12.5 | Postmitotic projection neurons | Gong et al. (2003) |
| Fezf2-GFP | E12.5 | Postmitotic deep layer projection neurons | Kwan et al. (2008) |
| Nkx2.1-GFP | E10.5 | Interneurons from MGE | Gong et al. (2003) |
| Lhx6-GFP | E11.5 | Interneurons from MGE | Cobos et al. (2006), Gong et al. (2003) |
| Dlx5/6-Cre-IRES-GFP | E12.5 | Interneurons from LGE and MGE | Stenman et al. (2003), Yokota, Gashghaei et al. (2007) |
| Dlx1-GFP | E12.5 | Interneurons from ventral telencephalon | Cobos et al. (2005), Gong et al. (2003) |
| CXCR4-GFP | E12.5 | Interneurons from ventral telencephalon | Gong et al. (2003), Tissir et al. (2004) |
| GAD65-GFP | E12.5 | Interneurons from ventral telencephalon | De Marchis et al. (2004), Lopez-Bendito et al. (2004), Tanaka et al. (2006) |
| Postnatal neuroblasts in SVZ and RMS |
Note: LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence.
4-D Neuronal Migration Analysis and Examination of Neuronal Migration in Living Embryos
Although the migration assays using embryonic cortical slices have yielded extensive insights into the modalities and molecular control of neuronal migration, other fundamental questions related to neuronal migration, such as how subtypes of interneurons and projection neurons coordinate their migration and placement in specific layers and areas of the developing cerebral cortex, remain unanswered. Simultaneous live imaging of genetically defined populations of interneurons, projection neurons, and radial glia in the entire rostral–caudal extent of developing cerebral cortex from different embryonic ages is essential to understand how specific subtypes of neurons achieve their laminar and areal positions within the developing cortex. Slices from the entire extent of embryonic cortex can be repeatedly scanned in real time, using a confocal microscope equipped with x–y–z–motorized stage and live-cell incubation chamber. The images collected can be compiled and analyzed to get a 4D view of neuronal movement within the cortex at a particular stage of embryonic development. Further, multiphoton microscopy provides a powerful tool to map neuronal movement and dynamics within the emerging neocortex of intact living embryos (Fig. 3; Ang et al. 2003). For example, specific analysis of interneuronal movement in living embryos using multiphoton imaging indicates that multidirectional migration within local areas of the developing cortex may facilitate the appropriate placement of interneurons within distinct cortical regions (Yokota, Gashghaei, et al. 2007). This type of combined large-scale neuronal imaging in the developing brain of normal and genetic models of neurodevelopmental disorders can yield unexpected and significant insights in to how different types of neurons get to where they have to go in the developing brain and how disruption of this process can cause a specific developmental disorder.
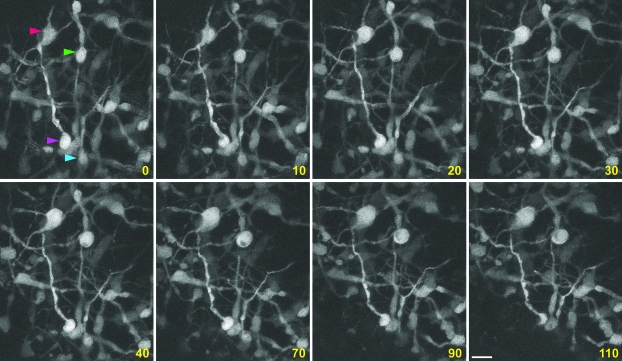
Figure 3.
In utero imaging of interneuronal dynamics. GFP+ interneurons in the developing cerebral cortex (parietal cortical area) of E16.5 Dlx5/6-CIE embryos were repeatedly imaged using 2-photon microscopy. Time-lapse imaging of these neurons in living embryos illustrates dynamic changes in their position, process growth, or neighbor relationships. Arrowheads indicate sample neurons undergoing such changes. Time elapsed is indicated in minutes. Scale bar = 30 μm.
Future Directions
The methods to study progenitor development or migration of their neuronal progeny are driven by the need to understand the molecular and cellular basis of these phenomena during normal brain development and in neurodevelopmental disorders. An emerging goal of these efforts is to map and study the behavior of large cohorts of developing progenitors and their progeny with high temporal, spatial, and molecular resolution across development. For example, the evolving functional status of critical polarity regulators during progenitor differentiation can be probed with functional state-specific fluorophores, such as bioprobes for CDC42 or Rac activity (Nalbant et al. 2004; Wu et al. 2009). Similarly, cell state–specific probes (e.g., Fucci to define different stages of cell cycle) can be developed and used to map changing differentiation states. Further, tagging critical signaling components with fluorescent timers (Subach et al. 2009), photoactivatable modifiers (Wu et al. 2009), and photoconvertible fluorophores such as Kaeda can be used to track the dynamics of critical molecular regulators (e.g., Wave1) (Yokota, Ring, et al. 2007) or organelles (Wang et al. 2009) during neuronal development. This approach of using biosensors (Miyawaki 2005; Miyawaki and Schnitzer 2007; Matsuda et al. 2008; Wu et al. 2009) specific to a particular functional state of key molecular determinants or particular cell state to query the dynamics of normal or altered development in mouse genetic models of neurodevelopmental disorders will be essential to fully understand how cerebral cortex forms and to define the neurodevelopmental pathways whose disruption is likely to lead to brain developmental disorders.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
National Institutes of Health grant (MH63660 to EA); NARSAD young investigator award (to HH); confocal imaging core of a National Institute of Neurological Disorders and Stroke institutional center core grant NS045892.
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
References
- Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–21885. [PubMed] [Google Scholar]
- Al-Kofahi O, Radke RJ, Goderie SK, Shen Q, Temple S, Roysam B. Automated cell lineage construction: a rapid method to analyze clonal development established with murine neural progenitor cells. Cell Cycle. 2006;5:327–335. doi: 10.4161/cc.5.3.2426. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Ang ES, Jr., Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500:368–383. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Heintz N. Genetic lineage tracing defines distinct neurogenic and gliogenic stages of ventral telencephalic radial glial development. Neural Dev. 2008;3:30. doi: 10.1186/1749-8104-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Anton ES, Cameron RS, Rakic P. Role of neuron-glial junctional domain proteins in the maintenance and termination of neuronal migration across the embryonic cerebral wall. J Neurosci. 1996;16:2283–2293. doi: 10.1523/JNEUROSCI.16-07-02283.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton ES, Ghashghaei HT, Weber JL, McCann C, Fischer TM, Cheung ID, Gassmann M, Messing A, Klein R, Schwab MH, et al. Receptor tyrosine kinase ErbB4 modulates neuroblast migration and placement in the adult forebrain. Nat Neurosci. 2004;7:1319–1328. doi: 10.1038/nn1345. [DOI] [PubMed] [Google Scholar]
- Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis. 2009;47:775–781. doi: 10.1002/dvg.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo A, Calegari F, Haubensak W, Wilsch-Brauninger M, Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS One. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider F, Meletis K, Eriksson M, Bergmann O, Sabelstrom H, Harvey MA, Mikkers H, Frisen J. Genetic manipulation of adult mouse neurogenic niches by in vivo electroporation. Nat Methods. 2008;5:189–196. doi: 10.1038/nmeth.1174. [DOI] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Barres BA, Koroshetz WJ, Chun LL, Corey DP. Ion channel expression by white matter glia: the type-1 astrocyte. Neuron. 1990;5:527–544. doi: 10.1016/0896-6273(90)90091-s. [DOI] [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, Best TK, Cramer NP, Carney RS, Isaac JT, Galdzicki Z. Haydar TF Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb Cortex. 2003;13:599–606. doi: 10.1093/cercor/13.6.599. [DOI] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Corte. 2006;16(Suppl 1):i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Coksaygan T, Magnus T, Cai J, Mughal M, Lepore A, Xue H, Fischer I, Rao MS. Neurogenesis in Talpha-1 tubulin transgenic mice during development and after injury. Exp Neurol. 2006;197:475–485. doi: 10.1016/j.expneurol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- de Carlos JA, Lopez-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchis S, Temoney S, Erdelyi F, Bovetti S, Bovolin P, Szabo G, Puche AC. GABAergic phenotypic differentiation of a subpopulation of subventricular derived migrating progenitors. Eur J Neurosci. 2004;20:1307–1317. doi: 10.1111/j.1460-9568.2004.03584.x. [DOI] [PubMed] [Google Scholar]
- Delalle I, Bhide PG, Caviness VS, Jr., Tsai LH. Temporal and spatial patterns of expression of p35, a regulatory subunit of cyclin-dependent kinase 5, in the nervous system of the mouse. J Neurocytol. 1997;26:283–296. doi: 10.1023/a:1018500617374. [DOI] [PubMed] [Google Scholar]
- Di Cristo G. Development of cortical GABAergic circuits and its implications for neurodevelopmental disorders. Clin Genet. 2007;72:1–8. doi: 10.1111/j.1399-0004.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Alvarez-Buylla A. Network of tangential pathways for neuronal migration in adult mammalian brain. Proc Natl Acad Sci U S A. 1996;93:14895–14900. doi: 10.1073/pnas.93.25.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson JC, Liem RK, Kuster JE, Hatten ME. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007;448:901–907. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Falk A, Frisen J. New neurons in old brains. Ann Med. 2005;37:480–486. doi: 10.1080/07853890500371890. [DOI] [PubMed] [Google Scholar]
- Fishman RB, Hatten ME. Multiple receptor systems promote CNS neural migration. J Neurosci. 1993;13:3485–3495. doi: 10.1523/JNEUROSCI.13-08-03485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB, Grutzendler J, Wong WT, Wong RO, Lichtman JW. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 2000;27:219–225. doi: 10.1016/s0896-6273(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Ghashghaei HT, Lai C, Anton ES. Neuronal migration in the adult brain: are we there yet? Nat Rev Neurosci. 2007;8:141–151. doi: 10.1038/nrn2074. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Walsh CA. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- Glowatzki E, Cheng N, Hiel H, Yi E, Tanaka K, Ellis-Davies GC, Rothstein JD, Bergles DE. The glutamate-aspartate transporter GLAST mediates glutamate uptake at inner hair cell afferent synapses in the mammalian cochlea. J Neurosci. 2006;26:7659–7664. doi: 10.1523/JNEUROSCI.1545-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gongidi V, Ring C, Moody M, Brekken R, Sage EH, Rakic P, Anton ES. SPARC-like 1 regulates the terminal phase of radial glia-guided migration in the cerebral cortex. Neuron. 2004;41:57–69. doi: 10.1016/s0896-6273(03)00818-3. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gregory WA, Edmondson JC, Hatten ME, Mason CA. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci. 1988;8:1728–1738. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Liem RK. Astroglial cells provide a template for the positioning of developing cerebellar neurons in vitro. J Cell Biol. 1981;90:622–630. doi: 10.1083/jcb.90.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Tanaka T, Brinkman BC, Gleeson JG. GSK3beta and PKCzeta function in centrosome localization and process stabilization during Slit-mediated neuronal repolarization. Mol Cell Neurosci. 2006;32:118–132. doi: 10.1016/j.mcn.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Honig MG, Hume RI. Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends Neurosci. 1989;12:333, 335–340. [PubMed] [Google Scholar]
- Howard BM, Zhicheng M, Filipovic R, Moore AR, Antic SD, Zecevic N. Radial glia cells in the developing human brain. Neuroscientist. 2008;14:459–473. doi: 10.1177/1073858407313512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Rutishauser U. A septum-derived chemorepulsive factor for migrating olfactory interneuron precursors. Neuron. 1996;16:933–940. doi: 10.1016/s0896-6273(00)80116-6. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Alvarez-Buylla A. Characterization of adult neural stem cells and their relation to brain tumors. Cells Tissues Organs. 2008;188:212–224. doi: 10.1159/000114541. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kerjan G, Gleeson JG. Genetic mechanisms underlying abnormal neuronal migration in classical lissencephaly. Trends Genet. 2007;23:623–630. doi: 10.1016/j.tig.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi H, Higginbotham H, Poon T, Tanaka T, Brinkman BC, Gleeson JG. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors. produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- LoTurco J, Manent JB, Sidiqi F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i120–i125. doi: 10.1093/cercor/bhp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulier K, Lathia JD, Marthiens V, Relucio J, Mughal MR, Tang SC, Coksaygan T, Hall PE, Chigurupati S, Patton B, et al. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Miyawaki A, Nagai T. Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nat Methods. 2008;5:339–345. doi: 10.1038/nmeth.1193. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Innovations in the imaging of brain functions using fluorescent proteins. Neuron. 2005;48:189–199. doi: 10.1016/j.neuron.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Schnitzer MJ. New technologies for neuroscience. Curr Opin Neurobiol. 2007;17:565–566. doi: 10.1016/j.conb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- Navarro-Quiroga I, Chittajallu R, Gallo V, Haydar TF. Long-term, selective gene expression in developing and adult hippocampal pyramidal neurons using focal in utero electroporation. J Neurosci. 2007;27:5007–5011. doi: 10.1523/JNEUROSCI.0867-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski RS, Rakic P. The mode of migration of neurons to the hippocampus: a Golgi and electron microscopic analysis in foetal rhesus monkey. J Neurocytol. 1979;8:697–718. doi: 10.1007/BF01206671. [DOI] [PubMed] [Google Scholar]
- O'Brien JA, Lummis SC. Diolistic labeling of neuronal cultures and intact tissue using a hand-held gene gun. Nat Protoc. 2006;1:1517–1521. doi: 10.1038/nprot.2006.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke NA, Chenn A, McConnell SK. Postmitotic neurons migrate tangentially in the cortical ventricular zone. Development. 1997;124:997–1005. doi: 10.1242/dev.124.5.997. [DOI] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Rakic P. Developmental and evolutionary adaptations of cortical radial glia. Cereb Cortex. 2003;13:541–549. doi: 10.1093/cercor/13.6.541. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Ravin R, Hoeppner DJ, Munno DM, Carmel L, Sullivan J, Levitt DL, Miller JL, Athaide C, Panchision DM, McKay RD. Potency and fate specification in CNS stem cell populations in vitro. Cell Stem Cell. 2008;3:670–680. doi: 10.1016/j.stem.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian MR, Bartley CM, Rakic P. Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex. Trends Neurosci. 2008;31:54–61. doi: 10.1016/j.tins.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Yamamoto A, Kawaguchi A, Yamaguchi M, Mori K, Goldman SA, Okano H. Direct isolation of committed neuronal progenitor cells from transgenic mice coexpressing spectrally distinct fluorescent proteins regulated by stage-specific neural promoters. J Neurosci Res. 2001;65:220–227. doi: 10.1002/jnr.1145. [DOI] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid RS, Yokota Y, Anton ES. Generation and characterization of brain lipid-binding protein promoter-based transgenic mouse models for the study of radial glia. Glia. 2006;53:345–351. doi: 10.1002/glia.20274. [DOI] [PubMed] [Google Scholar]
- Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shimizu C, Akazawa C, Nakanishi S, Kageyama R. MATH-2, a mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur J Biochem. 1995;229:239–248. doi: 10.1111/j.1432-1033.1995.tb20461.x. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanco A, Szekeres C, Patel N, Rao S, Campbell K, Kreidberg JA, Polleux F, Anton ES. Netrin-1-alpha3beta1 integrin interactions regulate the migration of interneurons through the cortical marginal zone. Proc Natl Acad Sci U S A. 2009;106:7595–7600. doi: 10.1073/pnas.0811343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs D, DeProto J, Nie K, Englund C, Mahmud I, Hevner R, Molnar Z. Neurovascular congruence during cerebral cortical development. Cereb Cortex. 2009;19(Suppl 1):i32–i41. doi: 10.1093/cercor/bhp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV. Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat Chem Biol. 2009;5:118–126. doi: 10.1038/nchembio.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Misson JP, Caviness VS., Jr. Glial process elongation and branching in the developing murine neocortex: a qualitative and quantitative immunohistochemical analysis. J Comp Neurol. 1990;302:15–28. doi: 10.1002/cne.903020103. [DOI] [PubMed] [Google Scholar]
- Tanaka DH, Maekawa K, Yanagawa Y, Obata K, Murakami F. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133:2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Wang CE, Goffinet AM. Expression of the chemokine receptor Cxcr4 mRNA during mouse brain development. Brain Res Dev Brain Res. 2004;149:63–71. doi: 10.1016/j.devbrainres.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai LH, Takahashi T, Caviness VS, Jr., Harlow E. Activity and expression pattern of cyclin-dependent kinase 5 in the embryonic mouse nervous system. Development. 1993;119:1029–1040. doi: 10.1242/dev.119.4.1029. [DOI] [PubMed] [Google Scholar]
- Wang X, Qiu R, Tsark W, Lu Q. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 2007;85:3567–3573. doi: 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M, McCann C, DeWulf M, Wu JY, Rao Y. Distinguishing between directional guidance and motility regulation in neuronal migration. J Neurosci. 2003;23:5170–5177. doi: 10.1523/JNEUROSCI.23-12-05170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ME, Jiang H, Rao Y. Regulated formation and selection of neuronal processes underlie directional guidance of neuronal migration. Mol Cell Neurosci. 2005;30:378–387. doi: 10.1016/j.mcn.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Weimer JM, Yokota Y, Stanco A, Stumpo DJ, Blackshear PJ, Anton ES. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development. 2009;136:2965–2975. doi: 10.1242/dev.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18:779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu JY, Rao Y. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Saito H, Suzuki M, Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Gashghaei HT, Han C, Watson H, Campbell KJ, Anton ES. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS One. 2007;2:e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Kim WY, Chen Y, Wang X, Stanco A, Komuro Y, Snider W, Anton ES. The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron. 2009;61:42–56. doi: 10.1016/j.neuron.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 2007;54:429–445. doi: 10.1016/j.neuron.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Sun B, Zhang CL, Fine A, Chiu SY, Messing A. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol. 1997;187:36–42. doi: 10.1006/dbio.1997.8601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.