Abstract
Haplotype information is essential to the complete description and interpretation of genomes1, genetic diversity2 and genetic ancestry3. Although individual human genome sequencing is increasingly routine4, nearly all such genomes are unresolved with respect to haplotype. Here we combine the throughput of massively parallel sequencing5 with the contiguity information provided by large-insert cloning6 to experimentally determine the haplotype-resolved genome of a South Asian individual. A single fosmid library was split into a modest number of pools, each providing ~3% physical coverage of the diploid genome. Sequencing of each pool yielded reads overwhelmingly derived from only one homologous chromosome at any given location. These data were combined with whole-genome shotgun sequence to directly phase 94% of ascertained heterozygous single nucleotide polymorphisms (SNPs) into long haplotype blocks (N50 of 386 kilobases (kbp)). This method also facilitates the analysis of structural variation, for example, to anchor novel insertions7,8 to specific locations and haplotypes.
The high quality of the human reference genome derives from the hierarchical sequencing of large-insert clones, such that the assembly corresponding to each clone represents a single haplotype9. One of the first ‘personal genomes’ exploited clone-based mate pairing and long, accurate Sanger reads to resolve variants into haplotype blocks (N50 of 350 kbp; that is, 50% of resolved sequence is within blocks of at least 350 kbp)1. Although new technologies5 have subsequently enabled >1,000-fold reduction in genome sequencing costs, the short read-lengths and paucity of contiguity information are such that it remains challenging to determine haplotypes at a genome-wide scale. Genomic phase, the assignment of alleles to homologous chromosomes, was determined for SNPs using mate-paired reads on the SOLiD (sequencing by oligonucleotide ligation and detection) platform10 for an individual genome, but only 43% of heterozygous variants were phased, and nearly all in blocks no greater than the insert size, that is, <3.5 kbp10. Experimental limitations on the size and complexity of mate-pair libraries based on in vitro circularization11 make it difficult to improve upon this approach.
An alternative is to infer haplotypes from population-based linkage disequilibrium data or from pedigree analysis. For example, haplotypes were successfully inferred in the YH (YanHuang) genome for variants at which phased CHB/JPT HapMap data were available (CHB, Han Chinese from Beijing, China; JPT, Japanese from Tokyo, Japan)12. The genomes of a family of four have been sequenced and these relationships used to infer inheritance blocks13. Although they can be successful, inferential methods have limitations. Statistical phasing, whether based on genotyping2 or sequencing14, performs poorly when linkage disequilibrium is not high, and for rare variants. Phasing by pedigree analysis requires genome sequencing of many related individuals, increasing costs and limiting practical application.
We describe a cost-effective method for determining long-range haplotypes at a genome-wide scale by massively parallel sequencing of complex, haploid subsets of an individual genome (Fig. 1). We apply this method to the first reported whole-genome sequencing of a human of South Asian ancestry. The Indian subcontinent is home to myriad culturally and genetically diverse groups with distinct population histories15. We selected a female from the HapMap panel of ‘Gujarati Indians in Houston’ (GIH; NA20847) for sequencing. Notably, the imputation of genotypes for GIH was the least effective of all non-African populations in HapMap2.
Figure 1.
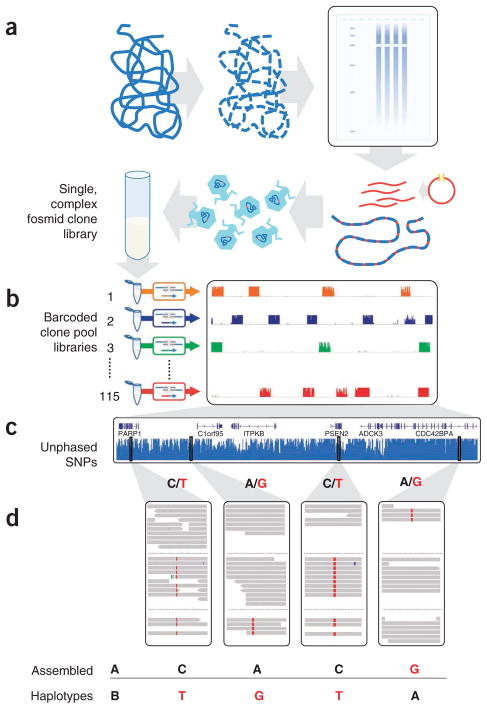
Haplotype-resolved genome sequencing. (a,b) A single, highly complex fosmid library was constructed (a) and split into 115 pools (b), each representing ~3% physical coverage of the diploid human genome. Barcoded shotgun libraries from each pool were constructed, then combined and sequenced. As expected, reads from each library map to ~5,000 × ~37 kbp blocks, minimally redundant within each library. (c) Whole-genome shotgun sequencing of the same individual generated unphased variant calls. (d) Unphased variant calls were combined with haploid genotype calls to assemble haplotype blocks using a maximum parsimony approach19 (reference allele in black, nonreference allele in red).
Genomic DNA from NA20847 was used to construct a single, complex fosmid library, containing clones packaged in phage for infecting Escherichia coli cells (>2 × 106 clones with ~37 kbp inserts) (Fig. 1a and Supplementary Methods). We then split a portion of this library to 115 pools, at a density such that each pool contained ~5,000 independent clones. Each pool was expanded by either scraping a single plate of infected cells and inoculating outgrowth culture, or by direct liquid outgrowth after infection. However, at no point does this method require the isolation of individual colonies. We next constructed 115 barcoded, shotgun sequencing libraries from fosmid DNA isolated from each of the 115 pools16. Libraries indexed with barcodes were combined and sequenced (Illumina GAIIx; PE76 or PE101 reads) to a mean 2.4× depth per haploid clone (Fig. 1b).
Because each pool captures an essentially random ~3% of the 6-gigabase (Gb) diploid genome (that is, ~5,000 fosmids × ~37 kbp inserts) sequence reads from each pool are overwhelmingly (99.1%) derived from only one homologous chromosome or the other at any single location. Upon mapping reads from each pool to the reference assembly, the approximate boundaries of 538,009 individual clones (37.2 ± 4.7 kbp) were identified by read depth (4,678 ± 1,229 clones per pool). Coverage was uniform across the genome (98.6% covered by one or more clones) and within each pool (82% of clones with mean read depth within a tenfold range) (Supplementary Fig. 1).
For unphased variation discovery, we performed conventional whole-genome resequencing to 15× depth (Illumina HiSeq; PE50) (Supplementary Table 1 and Supplementary Fig. 2). After alignment to the reference, we called 3.3 × 106 SNPs and 3.4 × 105 short indels17,18 (Fig. 1c). Nonreference sensitivity for SNPs was 91%, that is, HapMap variant genotypes at positions also called in our data, and genotype concordance to high-quality HapMap 3 genotypes2 at called positions was 99.2% (n = 1,436,495). Other bulk statistics, including the heterozygous-to-homozygous call ratio, the fraction of called variants previously ascertained in the NCBI SNP database (dbSNP), the transition-to-transversion ratio, and the numbers and classes of coding variants, were consistent with expectations based on previously sequenced non-African genomes (Supplementary Table 2).
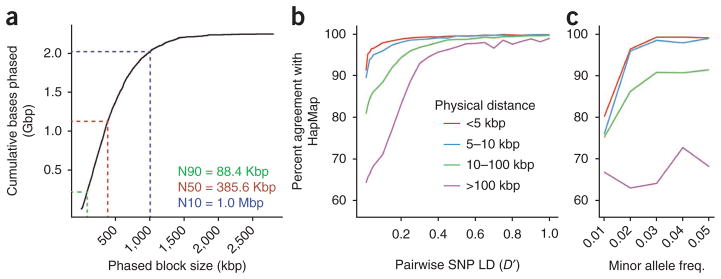
Several methods have been described for assembling haplotypes from sequence data1,19–21. We adopted a maximum parsimony approach19 to combine the unphased variants from shotgun whole-genome sequencing with haploid genotype calls from sequencing of the 115 pools (Fig. 1d). The resulting assembly incorporated 94% of ascertained heterozygous SNPs into haplotype-resolved blocks, with an N90 of 89 kbp, an N50 of 386 kbp and an N10 of 1 megabase (Mbp) (Fig. 2a). Sixty-two percent of genes were fully encompassed by single blocks, and 73% were covered for over half their length.
Figure 2.
Haplotype assembly results. (a) Size distribution of blocks within the haplotype assembly up to a maximum block size of 2.79 Mbp. Half of the assembly comprised blocks longer than 386 kbp (N50). (b) Comparison of experimental phasing with HapMap population-based inference2 for NA20847, with agreement of pairwise haplotype predictions as a function of physical distance and linkage disequilibrium. (c) Agreement of pairwise haplotype predictions as a function of physical distance and minor allele frequency (defined as the lower allele frequency of the pair in GIH). Key is the same as for b.
To evaluate accuracy, we compared our haplotype assembly with HapMap phase predictions for NA20847 (Fig. 2b)2. For pairs of SNPs in exceptionally high-linkage disequilibrium (D′ > 0.90 among GIH), we observed nearly perfect concordance (>99.7%). Because NA20847 was not part of a trio, HapMap predictions rely upon linkage disequilibrium between alleles to predict phase from genotypes. Correspondingly, concordance was reduced to ~71% when D; < 0.10, which is the case for most (66%) pairwise SNP combinations. Concordance is also reduced when one or both alleles in the pair is rare in GIH (Fig. 2c). Note that our haplotype assembly is experimental and specific to an individual, and therefore completely independent of population-based phenomena such as linkage disequilibrium and allele frequency. Consequently, these trends likely reflect errors in HapMap phasing1.
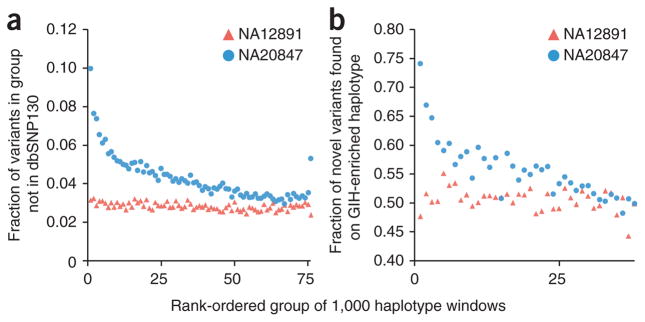
South Asian history includes admixture between two ancestral groups, one genetically close to Europeans (ANI) and another more highly diverged from well-ascertained populations (ASI)15. Furthermore, principal components analysis revealed a distinct subgroup of Indian populations in general and GIH in particular, including NA20847, that may harbor substantial genetic ancestry from a third population distinct from ANI and ASI15. We compared haplotype blocks for this individual to HapMap allele frequencies in the GIH and CEPH European (CEU) populations to distinguish ‘GIH-like’ from ‘CEU-like’ haplotypes. Notably, novel SNPs are markedly enriched on the most GIH-like haplotypes (Fig. 3). We also scored haplotype blocks against allele frequencies from the 1000 Genomes Project14 (Supplementary Fig. 3). Haplotypes that least resembled all three populations in that study (CEU, CHB/JPT and Yoruba) were also markedly enriched for novel SNPs. We propose that GIH-like blocks and other well-differentiated haplotypes may be derived from more poorly ascertained ancestral populations, and therefore enriched for novel variants. Such haplotypes may represent a valuable source of information about human history on the South Asian subcontinent.
Figure 3.
Enrichment of novel variants on ‘GIH-like’ haplotypes. (a) Haplotypes were scored and rank ordered within sliding windows of 20 HapMap variants2 for greater similarity to GIH or CEU on the basis of population allele frequencies (left on x axis: more similar to GIH). Plotted is the fraction of novel variants (not in dbSNP v130) in rank-ordered groups of haplotype windows, demonstrating that the most ‘GIH-like’ haplotype windows are enriched for novel variants. Values from trio-phased14 CEU individual NA12891 are shown for comparison (red). (b) Scores calculated in a for haplotype windows were compared between homologous chromosomes, and haplotypes were ranked based on the extent to which they scored as ‘GIH-like’ relative to their homolog. Plotted is the fraction of novel variants found on the more ‘GIH-like’ haplotype in rank-ordered groups of homologous haplotype windows. As above, the analysis was also performed for individual NA12891 using the rank ordering from individual NA20847. Haplotype blocks that are most differentiated relative to their homolog (higher ranked) with respect to GIH versus CEU similarity are enriched for novel variants relative to their homolog, consistent with the pattern observed in a.
A substantial fraction of the human genome consists of gene-rich segmental duplications and otherwise structurally complex regions that continue to defy accurate diploid consensus assembly within individual genomes. We sought to evaluate whether haplotype-resolved sequencing is useful for the fine-mapping and haplotype-assignment of deletions, inversions and novel contigs.
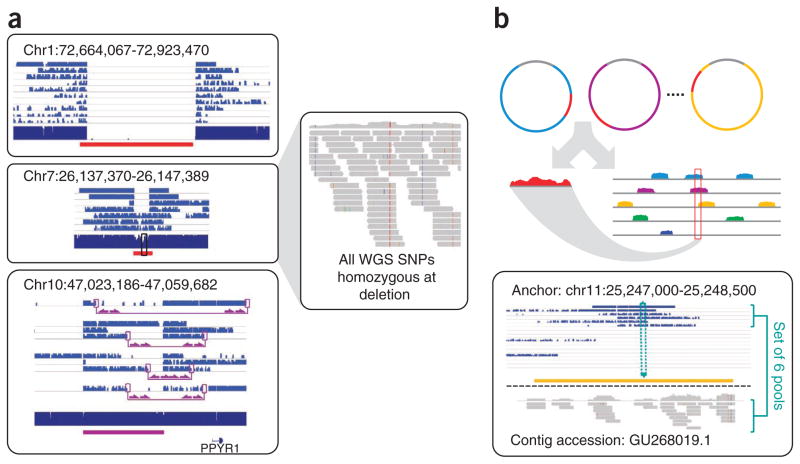
We used shotgun read depth22, discordant pairing in shotgun data23 and array-based SNP calls2 to estimate copy number and detect 58 deletions (>8 kbp), 15 of which were flanked by segmental duplications. Of these, 48 deletions (83%) were unambiguously confirmed by sequenced fosmid clones spanning the breakpoints, providing fine-scale resolution and confirming 30 as hemizygous (Fig. 4a and Supplementary Table 3). Heterozygous variants in flanking clones allowed for unambiguous incorporation of these deletions into haplotype-resolved blocks.
Figure 4.
Insertion anchoring and structural variation detection. (a) Homozygous deletion (top), hemizygous deletion (middle) and inversion (bottom) with fosmid clone support. Deletion calls were made using read depth and paired-read discordance. Inversions were called by paired-read discordance. SNPs within hemizygous deletions appear as stretches of hemizygosity by whole-genome shotgun sequencing. Purple connections indicate the additional support of strand discordance of read pairs spanning genomic DNA and the vector backbone. (b) Novel contigs not present in the reference assembly (red) but detected among clone pool–derived reads (light blue, purple, yellow) are anchored by searching for positions in the reference common to those pools but missing from most or all other pools. This approach anchors 1,733 recently reported insertion sequences7,8 including contig GU268019.
Inversions are challenging to detect because they are copy-number neutral and frequently mediated by repetitive sequences. As even fosmid end-sequencing tends to overcall inversions6, the added information from interrogating full ~37-kbp inserts may be useful for discriminating true inversions from false positives (Supplementary Fig. 4). Indeed, we observed a number of unambiguous inversions by means of breakpoint-spanning clones (Supplementary Fig. 5). However, larger clones (>100 kbp) may be required to span the large duplication blocks where inversion breakpoints typically map6. NA20847 is heterozygous for the inversion-containing H2 haplotype at the MAPT locus (17q21) (Supplementary Fig. 6). Of note, we properly phased all 287 SNPs that tag the H2 haplotype across a 588-kbp span24.
We also detected common human sequences unrepresented in the reference, that is, the ‘pan-genome’ (Supplementary Table 4)7,8. Of 16,904 contigs (total 12.8 Mbp) reported by two recent studies7,8, we identified 8,993 in NA20847. We exploited the contiguity of fosmids to anchor ~30% of these (Fig. 4b), with 73% agreement (±50 kbp) with a previously anchored subset8. De novo assembly of remaining unmapped reads yielded 2,242 additional contigs after filtering, of which we anchored 396. To validate anchoring accuracy, we simulated novel insertions by deleting 600 intervals (250 bp–10 kbp) in silico from the reference and remapping reads to the modified reference. Unmapped reads were de novo assembled into 5,435 contigs that covered ~61% of simulated insertions. Of these, we predicted anchoring locations for 2,184 with an accuracy of 87%, with the remaining contigs unassigned because of limited clone coverage. The sensitivity and specificity with which novel contigs can be anchored by this approach is likely to improve with increased clone and shotgun coverage.
We recently demonstrated exome sequencing as a strategy for identifying causal variants in Mendelian disorders25, for example, implicating compound heterozygote variants in DHODH in Miller syndrome26. In such studies, phasing reduces the number of candidate genes consistent with a recessive, compound heterozygous model13. For example, in this Gujarati Indian individual, unphased variant data included 44 genes consistent with compound heterozygosity (that is, two or more heterozygous, novel, nonsynonymous or splice-site variants that altered the same gene). But after phase was taken into account, only ten were validated as trans heterozygous, with the remainder having both variants on the same haplotype.
This method requires significantly greater expertise and sample preparation than the haplotype-blind shotgun sequencing of an individual genome—specifically, the construction of a single fosmid library and >100 in vitro shotgun libraries, as compared with constructing one or a few in vitro shotgun libraries. A detailed consideration of the added effort and cost are provided in Supplementary Table 5. In summary, sample preparation can be completed in <2 weeks by a single technician at a cost (~$4,000) that is much greater than that of preparing a single shotgun library, but low relative to the overall cost of whole-genome sequencing. We use an unconventional method based on in vitro transposition16 to significantly reduce the time and effort for producing >100 shotgun libraries. Current costs are primarily driven by commercial reagents for fosmid and shotgun library construction, and may therefore be amenable to optimization16. Furthermore, most steps are compatible with manual scaling and/or automation.
We also note that the total bases sequenced here (~87 Gb shotgun, ~110 Gb clone-based) is only modestly higher than for other individual human genomes sequenced to date. To estimate the minimal amount of clone sequencing required, we subsampled our data for either the number of independent clones or the depth of clone library sequencing (Supplementary Fig. 7). The primary effect was a reduction in the length of assembled haplotype blocks, rather than any decay in accuracy. For example, at 80% of clones and 60% of sequencing depth (which is 48% as much clone-based sequencing), the N50 dropped from 386 kbp to 238 kbp. However, most ascertained heterozygous variants remained phased (85.4%), and phasing remained highly concordant with HapMap (>99% at D′ > 0.9). Other optimizations, for example, switching from plate-scraping to direct liquid outgrowth to improve clone uniformity (Supplementary Fig. 1), may further reduce sequencing requirements.
Haplotypes are essential to the information content that defines a diploid human genome, but have heretofore been intractable to genome-wide, experimental determination in the context of massively parallel sequencing. We anticipate that haplotype-resolved genome sequencing will be valuable in a broad range of scenarios, including the following. (i) Population genetics. Haplotype-resolved genome sequencing eliminates the need for population or pedigree-based haplotype inference. This will be most useful in populations that are poorly ascertained (e.g., South Asians) or have low linkage disequilibrium (e.g., Africans), and more generally for rare variants. (ii) Genetic anthropology. For example, the availability of the haplotype-resolved reference and Venter genomes was critical to the observation of a Neanderthal contribution to some modern humans3. (iii) Medical genetics of rare and common phenotypes. Haplotype information can facilitate the analysis of recessive Mendelian disorders13, the determination of the parent of origin for de novo mutations, and the study of complex interactions among multiple SNPs27. (iv) Structural variation in both germline and cancer genomes. Our approach is more comprehensive than long-insert mate-pairing (whether by fosmids6 or in vitro circularization28), as these methods determine the ends of large molecules but are blind to their internal contents. Also, the intermediate level of partitioning provided by fosmids may be more useful than whole chromosome amplification29, as many germline and somatic structural events are intrachromosomal. (v) Allele-specific phenomena. Haplotype information may be essential for understanding the genetic basis of phenomena such as allele-specific expression and methylation30. (vi) De novo genome assembly. Massively parallel sequencing of highly complex pools of minimally redundant haploid clones may facilitate the high-quality de novo assembly of new genomes, an area that continues to be a major challenge for the genomics field despite the falling costs of DNA sequencing11.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
Acknowledgments
We thank C. Lee and M. Malig for technical assistance, J. Akey, T. O’Connor and P. Green for helpful discussions, D. Reich for ancestry information on NA20847, the U.W. Genome Sciences Genomics Resource Center (GS-GRC) for sequencing and the 1000 Genomes Project for early data release. This work was supported by National Institutes of Health grants AG039173 (J.B.H.) and HG002385 (E.E.E.), a National Science Foundation Graduate Research Fellowship (J.O.K.), a Natural Sciences and Engineering Research Council of Canada Fellowship (P.H.S.) and a fellowship from the Achievement Rewards for College Scientists Foundation (J.B.H.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
The project was conceived and experiments planned by J.O.K., E.E.E. and J.S. J.O.K., A.P.M. and R.Q. carried out all experiments. J.O.K., A.A., J.B.H., R.P.P., P.H.S., S.B.N. and C.A. performed data analysis. J.O.K., A.P.M., A.A., J.B.H., R.P.P. and J.S. wrote the manuscript, and all authors reviewed it. All aspects of the study were supervised by J.S.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Accession codes. Short read sequence data have been deposited at the NCBI Sequence Read Archive (SRA) under accession no. SRA026360. Assembled haplotype blocks and novel contigs are available from: http://krishna.gs.washington.edu/indianGenome/.
References
- 1.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. Human genome: Genomes by the thousand. Nature. 2010;467:1026–1027. doi: 10.1038/4671026a. [DOI] [PubMed] [Google Scholar]
- 5.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 6.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, et al. Building the sequence map of the human pan-genome. Nat Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 8.Kidd JM, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.McKernan KJ, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–i159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, Waterman MS, Li LM. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 2007;17:1101–1110. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal V, Halpern AL, Axelrod N, Bafna V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18:1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hormozdiari F, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;26:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drysdale CM, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L, et al. Direct determination of molecular haplotypes by chromosome microdissection. Nat Methods. 2010;7:299–301. doi: 10.1038/nmeth.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.