Abstract
Mammalian genomes are populated with thousands of transcriptional enhancers that orchestrate cell type-specific gene expression programs1-4, but how those enhancers are exploited to institute alternative, signal-dependent transcriptional responses remains poorly understood. Here we present evidence that cell lineage-specific factors, such as FoxA1, can simultaneously facilitate and restrict key regulated transcription factors, exemplified by the androgen receptor (AR), to act on structurally- and functionally-distinct classes of enhancers. Consequently, FoxA1 down-regulation, an unfavorable prognostic sign in certain advanced prostate tumors, triggers dramatic reprogramming of the hormonal response by causing a massive switch in AR binding to a distinct cohort of pre-established enhancers. These enhancers are functional, as evidenced by the production of enhancer-templated non-coding RNA (eRNA5) based on global nuclear-on (GRO-seq) analysis6, with a unique class apparently requiring no nucleosome remodeling to induce specific enhancer-promoter looping and gene activation. GRO-seq data also suggest that liganded AR induces both transcription initiation and elongation. Together, these findings reveal a large repository of active enhancers that can be dynamically tuned to elicit alternative gene expression programs, which may underlie many sequential gene expression events in development, cell differentiation and disease progression.
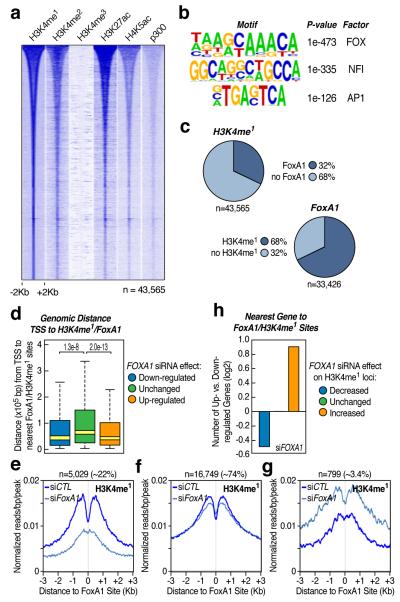
The wide diversity of mammalian cells is determined by a large repertoire of constitutive and inducible genes, which are regulated by general and cell-type specific transcription factors and cofactors through regulatory genomic elements7,8. Recent studies reveal that gene promoters are marked by tri-methylated H3K4 (H3K4me3) and distal regulatory elements are often associated with mono-methylated H3K4 (H3K4me1)1,2. Because these H3K4me1-positive, H3K4me3-negative regions exhibit striking cell type specificity1,2, we used this signature to characterize potential enhancers in prostatic LNCaP cells in which one of key regulatory transcriptional programs is mediated by the androgen receptor (AR). We identified by ChIP-seq 14,283 H3K4me3-marked and 51,544 H3K4me1-marked loci in androgen (5α-dihydrotestosterone, DHT)-treated LNCaP cells, among which 43,565 loci are uniquely marked by H3K4me1, largely localized distal to annotated TSSs (94%), and associated with other marks linked to enhancer activities (Fig. 1a).
Figure 1. FoxA1 contributes to the enhancer code in prostate cancer cells.
(a) Distribution of histone marks within ±2Kb windows around distinct genomic regions (n=43,565) marked by H3K4me1, but not H3K4me3, in androgen (DHT)-stimulated LNCaP cells. The ChIP-seq datasets for H3K4me1, H3K4me2, H3K4me3, H3K27ac, H4K5ac and p300 were each aligned with respect to the center of theH3K4me1 signal and sorted by the length of H3K4me1-marked regions. (b) Top-enriched DNA motifs with significant P-value and prospective families of DNA binding transcription factors identified by de novo motif analysis of non-promoter regions marked by H3K4me1. (c) Percentage of H3K4me1-marked regions that show FoxA1 binding events (top panel) and percentage of FoxA1 binding sites that are marked by H3K4me1 (bottom panel). Note that H3K4me1-marked regions tend to be broad, but FoxA1 binding sites are discrete, and as a result, many H3K4me1-positive regions may contain more than one FoxA1 binding site. (d) Genomic distance from FoxA1/H3K4me1-positive loci to the nearest TSS of genes in response to FOXA1 knockdown. Outliers were omitted from box plots. P-values indicate the significance in pair-wise comparisons. (e-g) Three classes of FoxA1/H3K4me1-positive loci according to the response in levels of H3K4me1 to FOXA1 knockdown: >1.5-fold decrease (e), no significant change (f), and >1.5-fold increase (g). (h) Ratio (log2) of up- and down-regulated genes in each H3K4me1 responsive category in e-g.
De novo DNA motif analysis revealed several highly enriched motifs, particularly the forkhead motif (Fig. 1b). Using a specific antibody against FoxA1, a major FOX family member expressed in LNCaP cells and normal prostate gland9-11 (Supplementary Fig. 1), we identified 33,426 FoxA1-bound sites, which extensively overlap with distal H3K4me1-marked regions (Fig. 1c and Supplementary Fig. 2a; see on KLK3 enhancer12 in Supplementary Fig. 2b). RNA profiling supports the functional relevance of these FoxA1/H3K4me1 loci, as genes responsive to FOXA1 siRNA are located more proximally to FoxA1/H3K4me1-marked loci than non-responsive genes (Fig. 1d and Supplementary Fig. 3).
FoxA1 has been characterized as a “pioneer” factor to facilitate DNA binding by other sequence-specific transcription factors9,13-16 and “translate” H3K4me1/me2 into AR-mediated gene expression9. Comparing the profile of H3K4me1 and H3K27ac before and after FOXA1 knockdown, we detected three classes of FoxA1 binding sites based on the H3K4me1 signal exhibiting reduced (~22%), relatively unaffected (~74%), or even increased (~3.4%) levels over candidate enhancers (Fig. 1e-g and Supplementary Fig. 4). RNA profiling analysis agrees with the functional significance of these selective FoxA1 effects, revealing more down-regulated genes in the first class, roughly equal numbers of up- or down-regulated genes in the second, and more up-regulated genes in the third (Fig. 1h), suggesting a contribution of FoxA1 to “writing” and “reading” the “histone code” on different enhancer cohorts, in line with its critical function in prostate gland development10,11.
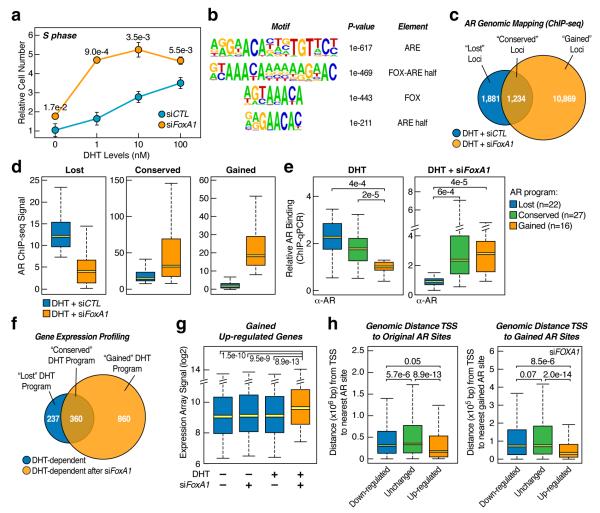
The rationale for our experimental strategy to use RNAi to study FoxA1-regulated enhancer network is the association of decreased FOXA1 expression with castration-resistant, poor prognostic prostate tumors (Supplementary Fig. 5). In LNCaP cells, FOXA1 RNAi enhanced cell entrance to S phase with reduced hormone (Fig. 2a). To understand the mechanistic basis for elevated hormone responsiveness, we mapped AR binding sites, identifying 3,115 high confident loci with ~65% co-incident with H3K4me1. De novo motif analysis revealed highly enriched elements for both AR and FoxA1, including a composite motif consisting of a FOX motif and AR regulatory element (ARE) half site, suggesting ternary complex formation on these sites (Fig. 2b). Indeed, 1,684 AR-bound loci (54% of total) are co-occupied by FoxA1 in DHT-treated LNCaP cells and FoxA1 appears to bind to most of these sites (~70%) before hormone treatment (Supplementary Fig. 6).
Figure 2. AR reprogramming and induced alternative hormonal response.
(a)FOXA1 siRNA-induced cell progression to S phase. Relative numbers of propidium iodide (PI)-labeled cells in S-phase at different DH) concentrations were determined by FASCan. The P-value for the difference detected at each hormonal level is indicated; mean±s.e.m. is based on three independent experiments. (b) Top-enriched motifs associated with AR-occupied loci (n=3,115). (c) Comparison between genome-wide AR binding programs before and after FOXA1 knockdown in DHT-treated LNCaP cells. (d) Quantitative levels of AR binding in the “lost”, “conserved”, and “gained” programs. Outliers were omitted from box plots. (e) ChIP-qPCR validated AR binding events on randomly selected loci from the lost (n=22), conserved (n=27), and gained (n=16) programs. (f) Microarray analysis of DHT-induced genes before and after FOXA1 knockdown. (g) Quantitative analysis of gained androgen up-regulated genes based on microarray analysis in f. Outliers were omitted from box plots. (h) Genomic distance of androgen-responsive genes from TSS to the nearest AR binding site in the original and gained AR binding programs.
The conundrum is that, while FoxA1 is known to facilitate AR binding on several DHT-responsive genes9, FOXA1 RNAi actually markedly elevated, rather than diminished, the DHT response (Fig. 2a). We found that ~60% of the original AR binding events was “expectedly” lost in response to FOXA1 RNAi, which we refer to as the “lost” AR program (Fig. 2c,d). We refer the remaining ~40% of AR binding events to as the “conserved” AR program, which often exhibited enhanced AR binding. Strikingly, we detected a massive gain of 10,869 new AR binding loci, referred to as the “gained” AR program (Fig. 2c,d). We extensively validated each of these AR programs by conventional ChIP-qPCR (Fig. 2e). This induced AR reprogramming appears to be qualitatively and quantitatively distinct from reported AR re-targeting on androgen-resistant LNCaP-abl cells compared to parental LNCaP cells17 and is also in sharp contrast to FoxA1-dependent genomic targeting of the estrogen receptor α (ERα) in breast cancer MCF7 cells18. In concert with such massive AR reprogramming, we observed corresponding changes in gene expression in each of three AR programs (Fig. 2f,g, Supplementary Fig. 7). The newly induced AR expression program is also linked to AR binding events (Fig. 2h), suggesting a direct gain-of-function on DHT-responsive genes, as illustrated on SOX9 and other genes (Supplementary Fig. 8), which have been previously documented to play critical roles in cancer progression19,20. Because we also observed ~3-fold elevation of AR expression in FOXA1 RNAi-treated cells (Supplementary Fig. 9a), we tested the possibility that increased AR expression might trigger these effects, finding that AR overexpression alone was insufficient to induce AR reprogramming (Supplementary Fig. 9b).
To explore the mechanism for AR reprogramming, we determined FoxA1 binding on different AR programs, finding that the gained AR program is largely devoid of FoxA1, while FoxA1 is present in more than half of the lost and conserved AR programs (Supplementary Fig. 10). This raises the possibility that FoxA1 may facilitate AR binding to its original binding program, but trans-repress AR from binding to other genomic regions that lack FoxA1 binding sites in the gained program, a strategy frequently used by other transcription activators21. Indeed, as previously reported22, FoxA1 overexpression squelched AR element (ARE)-driven transcription in transfected HEK293 cells (Supplementary Fig. 11a), which is consistent with the ability of AR to directly interact with FoxA123. This mechanism appears to be exploited during tumor progression because an AR mutation identified in advanced prostate tumors lacks part of the hinge domain important for interactions with FoxA123, its ability to interact with FoxA1, and became resistant to FoxA1-mediated trans-repression (Supplementary Fig. 11b,c). Furthermore, our functional analysis indicates that the missing AR ligand-binding domain (LBD) also contributes to AR:FoxA1 interactions (Supplementary Fig. 12). Interestingly, similar AR truncations have also been reported to result from alternative splicing, gene rearrangement, and/or calpain-mediated cleavage (Supplementary Fig. 13). Based on these findings, we propose that FoxA1 regulates AR genomic targeting by simultaneously anchoring AR to cognate loci and restricting AR from other ARE-containing loci in the human genome.
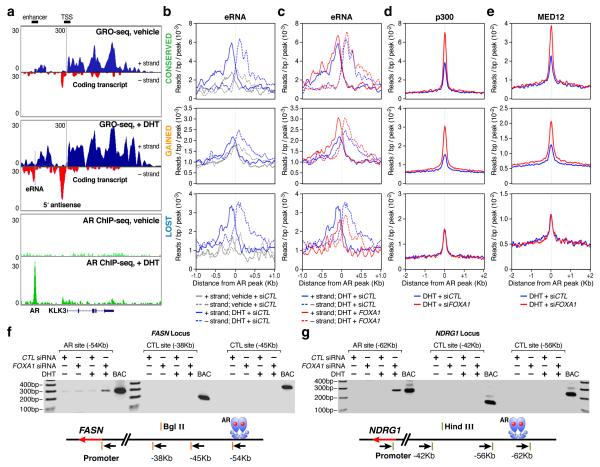
To understand how reprogrammed AR binding is translated to altered hormonal response, we took advantage of the recently established global run-on strategy (GRO-seq6) to detect the functional relationship between AR binding and hormone-induced gene expression. This powerful genome-wide interrogation of on-going transcription detected a broad scope of nascent RNAs. We uncovered 28,318 transcripts with 15,656 annotated and 12,662 unannotated transcripts, among which 450 coding and 347 unannotated transcripts were induced >1.5-fold with even just 1 hr DHT treatment (Supplementary Fig. 14). The TSS of GRO-seq defined transcripts are typically marked by H3K4me3 and H3K27Ac (Supplementary Fig.15a,b). Importantly, GRO-seq also detected non-coding RNAs from a subset of H3K4me1/2 positive, H3K4me3 negative regions (Supplementary Fig. 15c). As illustrated on the enhancer of the KLK3 transcription unit (Fig. 3a), these enhancer-derived RNAs (or eRNAs)5 are largely symmetrical and bidirectional (see additional examples on other well-known hormone regulated genes, such as PMEPA1 and KLK2 in Supplementary Fig. 16). Interestingly, we often detected a large amount of nascent RNA before DHT treatment, particularly near their TSS (e.g. KLK3); DHT not only enhanced the expression of these nascent RNAs, but also permitted the extension of transcription toward the end of the gene (Fig. 3a, Supplementary Fig. 16). We estimated that ~79% of the transcription units induced by liganded AR are regulated at the level of transcriptional initiation, while ~21% appear to be primarily regulated at the level of elongation (Supplementary Fig. 17).
Figure 3. Transcriptional response on individual enhancer programs to FOXA1 down-regulation.
(a) Display of nascent RNA detected by GRO-seq on the KLK3 locus. The DHT-induced AR binding is shown at bottom as a reference. (b,c) Induction of eRNA by DHT (b) or FOXA1 knockdown in DHT-treated LNCaP cells (c). The eRNA levels under different conditions (indicated at bottom) are separately displayed on three AR binding programs. (d,e) Effects of FoxA1 on binding of p300 (d) and Med12 (e) in each AR program in DHT-treated LNCaP cells. (f, g) Long-distance interaction between gene promoter and AR bound site (at ~50Kb) was determined by the 3C assay on two representative gene loci selected from the conserved and gained AR programs. Negative controls at shorter distances (~30 and ~40Kb) and a positive control with the corresponding BAC in the region are included in each case.
The ability to detect regulated eRNA expression permitted us to analyze different AR programs during transcriptional reprogramming. In the presence of FoxA1, DHT enhanced eRNA expression from AR-bound enhancers in both the lost and conserved AR programs. In contrast, a basal level of eRNAs was detectable on the gained program, but is independent of the hormone treatment, indicating that these are pre-established enhancers (Fig. 3b). In response to FOXA1 RNAi, the expression of eRNAs was diminished from the lost program, but modestly or dramatically enhanced from the conserved and gained programs, respectively (Fig. 3c). The DHT-induced nascent transcripts (detected by GRO-seq) and steady-state RNAs (detected by microarrays) best predicts direct target genes by liganded AR, as they show the shortest distance (<50kb) to nearby AR binding sites compared to genes identified by either criterion alone (Supplementary Fig. 18), indicating that AR-activated enhancers marked by increased eRNA are responsible for activation of nearby coding transcription units.
In concert with differential eRNA expression, we also observed corresponding changes in levels of another mark in the final step of enhancer activation4, specifically p300, on both conserved and gained AR programs (Fig. 3d). Interestingly, enhancers in the lost AR program continued to exhibit significant p300 binding even after AR binding and eRNA expression were diminished in FOXA1 knockdown cells (Fig. 3c,d). The transcription mediator Med12 has recently been suggested to mediate enhancer-promoter looping24. We tested Med12 binding on individual AR programs, finding that it exhibited an identical binding pattern to p300 (Fig. 3e). Enhanced Med12 binding on the conserved and gained programs after FOXA1 knockdown suggests elevated or newly activated enhancer:promoter interactions. This was demonstrated by the 3C assay on two representative genes where FOXA1 knockdown either enhanced (on the FASN locus from the conserved AR program) or create new (on the NDRG1 locus in the gained AR program) long-range interactions between AR-bound enhancers and specific gene promoters in DHT-treated cells (Fig. 3f,g and Supplementary Fig.19). These data strongly suggest that the induction of eRNAs, rather than binding of either p300 or Med12, is the most precise mark of the final, functional looping between an activated enhancer and its regulated gene promoter.
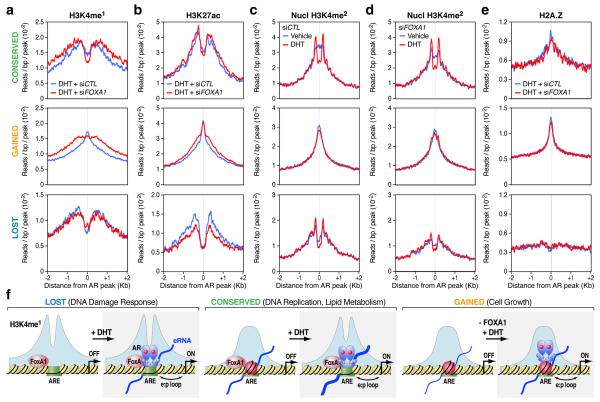
Addressing the structural basis for different functional classes of AR enhancers, we note that the distinct profiles of H3K4me1 and H3K27ac on the lost, conserved, and gained AR programs and FOXA1 RNAi had little effect on these profiles (Fig. 4a,b and Supplementary Fig. 20). The histone marks H3K4me1 and H3K27ac around the lost and conserved AR programs exhibit a bimodal distribution, which is particularly pronounced on the lost program (Fig. 4a, bottom panel). The DNA binding sites in the lost AR program are actually significantly less enriched in canonical AREs, which may render AR binding on these sites particularly dependent on FoxA1, whereas both the conserved and gained AR programs are associated with nearly perfect palindromic, canonical AREs (Supplementary Fig. 21), explaining why AR is able to target those sites in a FoxA1-independent manner. Strikingly, the gained AR binding sites are coincident with sharp H3K4me1 and H3K27ac peaks (Fig. 4a,b, middle panels), suggesting a distinct nucleosome architecture underlying the gained AR program.
Figure 4. Distinct classes of AR enhancers in the human genome.
(a,b) Profiles of H3K4me1 (a) and H3K27ac (b) associated with the lost (bottom panels), conserved (top panels), and gained (middle panels) AR programs in DHT-treated LNCaP cells in response to FOXA1 knockdown. (c, d) Profiles of H3K4me2 around AR binding loci at the nucleosomal resolution in response to DHT stimulation in control siRNA-treated (c) or FOXA1 siRNA-treated (d) LNCaP cells. (e) Profiles of the histone variant H2A.Z on the three different AR programs. (f) Model for FoxA1-mediated AR targeting and reprogramming in LNCaP cells. In Class I (the lost AR program), FoxA1 licenses liganded AR to bind to ARE in relatively nucleosome-free regions. AR binding does not induce nucleosome remodeling in this class of enhancers. In Class II (the conserved AR program), AR binds independently of FoxA1 to ARE, inducing nucleosome remodeling. In Class III (the gained AR program), FoxA1 restricts AR binding, despite the presence of strong AREs. Although pre-established, these gained loci exhibit a strong central nucleosome and are associated with H2AZ, which is not affected by AR binding. FOXA1 knockdown converted these sites to androgen-responsive sites. In all these three classes, eRNAs were generated or increased after AR binding.
A recent study suggests that AR binding leads to dynamic dismissal of a central, H2A.Z-containing nucleosome, replacing by two flanking H3K4me2-marked nucleosomes25. We found that the lost AR program was largely devoid of a “central” nucleosome even prior to AR binding (Fig. 4c, bottom panel). The conserved AR program exhibited DHT-induced switch from the central H3K4me2-marked nucleosome to two flanking H3K4me2-marked nucleosomes, which is largely independent of FoxA1 (Fig. 4c,d top panels). The gained program showed a strong H3K4me2-marked central nucleosome both before and after AR binding (Fig. 4c,d, middle panel). Thus, this gained AR program represents a new type of enhancer topography that requires no nucleosome remodeling for enhancer recognition and subsequent enhancer-promoter interactions. H2A.Z is prevalently associated with the gained AR program, modestly with the conserved AR program, and absent in the lost AR program (Fig. 4e). Together, these findings establish distinct chromatin structures underlying functionally distinct classes of AR enhancers.
In summary, our findings infer a general principle for establishing cell type-specific transcription programs. Cell lineage-specific factors (such as FoxA1) coupled with other general transcriptional factors “create” a cell type-specific enhancer network, allowing other regulated factors (such as AR) to “activate” these pre-established enhancers (Fig. 4f). The enhancer activation process is tightly linked to eRNA production, which appear to serve as a more robust indicator of enhancer activities than any enhancer-bound transcription activators or chromatin marks. On the current biology model, AR reprogramming dramatically altered the androgen responsive pathway, which, according to GO analysis (Supplementary Fig. 22 and Fig.23), may contribute to enhanced cell growth and the establishment of appropriate microenvironment in advanced prostate cancer26-28. Together, these findings provide a conceptual framework to understand complex gene expression switching events, as occurred during disease progression and development.
METHODS SUMMARY
Experiments were performed on LNCaP cells, LNCaP-AR cells (generous gift of Dr. C. Sawyers), and HEK293 cells. Chromatin immunoprecipitations (ChIPs) were done as previously described29 and global run-on (GRO) was performed as described6,30. Control siRNA was purchased from Qiagen (Cat# 1027280). FOXA1 siRNA #1 (M-010319) and #2 (sense 5′-GAGAGAAAAAAUCAACAGC-3′; antisense 5′-GCUGUUGAUUUUUUCUCUC-3′) (ref9) were purchased or synthesized from Dharmacon. Full Methods and associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
The authors are grateful to Hsing-Jien Kung for providing the PC3-AR cell lines, C. Sawyers for providing the AR-LNCaP cell line, and to J. Hightower, D. Benson, and M. Fisher for assistance with figure and manuscript preparation. This work was supported by grants from NIH (DK01847, DK37949, NS34934), DoD, NCI (CA97134) to M.G.R; (GM049369 and HG004659) to X.D.F. and Prostate Cancer Foundation to M.G.R. and X.D.F. M.G.R. is an investigator of the Howard Hughes Medical Institute.
Appendix
SUPPLEMENTARY EXPERIMENTAL PROCEDURES
Antibodies
Specific antibodies were purchased from the following commercial sources: anti-FoxA1 (ab5089), anti-H3K4me1 (ab8899), anti-H3K27ac (ab4729), anti-H3K36me3 (ab9050), and H2A.Z (ab4174) from Abcam; anti-AR (N-20), anti-FoxA1 (C-20), p300 (C-20, generous gift of Dr. Bing Ren) from Santa Cruz Biotechnology; anti-H3K4me2 (07-030), anti-H3K4me3 (07-473), anti-H4K5ac (07-327), and anti-H3K27me3 (07-449) from Upstate Biotechnology; anti-Med12 (A300-774A) from Bethyl Laboratories; and anti-beta Actin (AC74) from Sigma.
siRNA transfection
One day prior to transfection, LNCaP cells were seeded in RPMI 1640 medium with 10% FBS. 6 hrs after siRNA transfection (20 pmol/ml) with Lipofectamine 2000 (Invitrogen), cells were washed twice with PBS and then maintained in hormone-deprived phenol-free RPMI 1640 media. For gene expression profiling and Western blotting, cells were cultured for 3 days following transfection and then treated with DHT for 20 hrs; for ChIP-qPCR and ChIP-seq, cells were cultured for 4 days following transfection and then treated with DHT for 1 hr. ChIP-seq analysis at the nucleosome resolution was based on cells treated with DHT for 4 hrs.
3C assay
Cells were crosslinked with 1% formaldehyde for 20min at room temperature and processed according to the standard 3C protocol31. For the study on the FASN locus, fixed chromatin from 5×106 cells was digested with 400 units of Bgl II and EcoR I (NEB). For the NDRG1 locus, fixed chromatin from 5×106 cells was digested with 400 units of Hind III (NEB). Ligation was done with 800 units of T4 DNA ligase (NEB) for 4 hrs. The 3C product was quantified by qPCR after diluting the template 10-fold in comparison with purified genomic DNA of known concentration. For each semi-quantitative PCR, the amount of template was titrated to determine the linear range in which the PCR product was amplified. PCR primers were designed next to Bgl II and Hind III restriction sites, respectively, for the FASN (all in minus strand) promoter (5′-AAGCTGTGAGTCAGCATGGTAG-3′) and three upstream sites (−38Kb, 5′-TGTCTTCTGATGTGTCTGCTTAGAG-3′; −45Kb, 5′-AATCCTGCTCAGGAATCTGTATGT-3′; −54Kb, 5′-GGACACTACTGCTTTTTCCTGTG-3′) and for the NDRG1 (all in plus strand) promoter (5′-ATAGGTTCTGCCTTATTAGGG-3′) and three upstream sties (−42Kb, 5′-ATAGAGTTAGAGAAACGGAGGCAGT-3′; −56Kb: 5′-GCCGTGAAGAATAAACAAGATGAG-3′; −62Kb: 5′-ACACATTTTGTTCCCAGTGCAG-3′).
Co-IP and Western blotting analysis
HEK293 cells were seeded for one day, transfected with the expression plasmids expressing wt, mutant AR and FoxA1 using Lipofectamine2000 (Invitrogen) and then changed to hormone-depleted, phenol-free DMEM medium. One day after plasmid transfection, cells were treated with 100 nM DHT for another day. Cells were washed by cold PBS twice and treated with 1 ml of lysis buffer (50 mM Tris-pH 8.0, 150mM NaCl, 1% NP-40) supplemented with a cocktail of proteinase inhibitors (Sigma) for 5 min at 4°C. Harvested lysed cells were collected, rotated for 1 hr at 4°C, and cell debris removed by centrifugation at 14000 rpm for 30 min in cold room. The supernatant was incubated with anti-AR, anti-FoxA1 or IgG overnight at 4°C followed by the addition of 50 μl of 50% protein G beads to each tube. After rotating for another 2 hrs at 4°C, the beads were washed 5 times with the lysis buffer, twice with cold PBS, and boiled for 6 min in 40 μl of 2×SDS loading buffer. Western blotting analysis was carried out with anti-AR or anti-FoxA1.
Luciferase reporter assay
PC3-AR and HEK293 cells were seeded into 24-well plates in hormone-depleted and phenol-free RMPI 1640 medium and DMEM 1 day before transfection. Transfection was performed according to manufacture’s recommendations (DOTAP Liposomal Transfection Reagent from Roche or Lipofectamin 2000 from Invitrogen). One day after transfection, these cells were treated with DHT for an additional day. After washing with cold PBS twice, cells were treated with the lysis buffer (Promega) and the Luciferase signal was recorded.
Cell proliferation assay
The assay was based on the published protocol32. Briefly, LNCaP cells were transfected with control siRNA and FoxA1 siRNA (sequences listed in Methods Summary) and cultured at hormone-depleted medium for 3 days. The cells were treated with different amount of DHT for another day. After the treatment, cells were washed by PBS, fixed by 70% EtOH, stored at −20°C for at least 2 hrs. Before analysis, cells were washed with cold PBS, resuspended at the PI/Triton X-100 staining solution, and incubated at 37°C for 15 min. After removing cell clumps, stained cells were sorted on a Beckman FASCan, and the percent of S phase cells were calculated.
ChIP and ChIP-seq analyses
ChIP was as previously described29. Briefly, ~107 treated cells were crosslinked with 1% formaldehyde at room temperature for 15 min. After sonication, the soluble chromatin was incubated with 1-5μg of antibody. Specific immunocomplexes were precipitated with Protein A/G beads (Sigma-Aldrich). Complexes were washed, DNA extracted and purified by QIAquick Spin columns (Qiagen). 1μl from 60μl extracted DNA was used for qPCR with specific PCR primers listed in Supplementary Fig. 24, each of which was designed surrounding a specific region of 150-250bp on target DNA. PCR products were detected with SYBR Green on a MX3000P System (Stratagene) and the percentage of immunoprecipitated chromatin was calculated from ΔCt relative to IgG control after normalizing against input chromatin. For ChIP-seq, extracted DNA was ligated to specific adaptors followed by deep sequencing in the Illumina GAII system according to manufacture’s instruction (Illumina). The first 25 bp for each sequence tag returned by the Illumina Pipeline was aligned to the hg18 assembly (NCBI Build 36.1) using Bowtie, allowing up to 2 mismatches. Only tags uniquely mapped to the genome were used for further analysis. The data where visualized by preparing custom tracks for the UCSC Genome browser using HOMER33 (http://biowhat.ucsd.edu/homer/introduction/programs.html). The total number of mappable reads was normalized to 107 for each experiment presented in this study. ChIP-seq at nucleosome resolution was performed as previously reported34. A summary of ChIP experiments is provided in Supplementary Fig. 25.
Identification of ChIP-seq peaks
The identification of ChIP-seq peaks (bound regions) was performed using HOMER (http://biowhat.ucsd.edu/homer). For transcription factors, peaks were identified by searching locations of high read density using a 200bp sliding window. Regions of maximal density exceeding a given threshold were called as peaks, and we required adjacent peaks to be at least 500bp away to avoid redundant detection. Only one tag from each unique position was considered to avoid clonal artifacts from the sequencing. The threshold for the number of tags that determine a valid peak was selected at a false discovery rate of 0.001 determined by peak finding using randomized tag positions in a genome with an effective size of 2×109bp. We also required peaks to have at least 4-fold more tags (normalized to total count) than input control samples. In addition, we required 4-fold more tags relative to the local background region (10kb) to avoid identifying regions with genomic duplications or non-localized binding.
The peak finding procedure was modified to identify regions harboring specific histone modifications, as these experiments tend to yield broad areas of enrichment over several hundreds or thousands of base pairs. Seed regions were initially found using a peak size of 500bp at the false discovery rate of 0.001 to identify enriched loci. Enriched loci found within 1kb of one another were then merged to yield variable-length regions. Transcription factor peaks and histone modification regions were associated with gene products by identifying the nearest RefSeq TSS. Annotated positions for promoters, exons, introns, and other features were based on RefSeq transcripts and repeat annotations from UCSC. Peaks from separate experiments were considered equivalent/co-bound if their peak centers were located within 200bp of each other. Read density heat maps were created by first using HOMER to generate read densities and then visualized using Java TreeView (http://jtreeview.sourceforge.net/).
HOMER for de novo motif discovery and known motif enrichment
Motif discovery was performed using a comparative algorithm similar to those previously described35 and an in depth description will be published elsewhere (Benner et al., in preparation). Motif finding for transcription factors was performed on sequence from +/− 100bp relative to the peak center, while motif finding for histone modification regions was performed on sequence from +/− 500bp relative to the region center. Briefly, sequences were divided into target and background sets for each application of the algorithm. Background sequences were then selectively weighted to equalize the distributions of G+C content in target and background sequences to avoid comparing sequences of different general sequence content. Motifs of length 8, 10, 12, 14, 16, and 18bp were identified separately by first exhaustively screening all oligos for enrichment in the target set compared to the background set using the cumulative hypergeometric distribution to score enrichment. Up to three mismatches were allowed in each oligonucleotide sequence to increase the sensitivity of the method. The top 200 oligonucleotides of each length with the lowest p-values were then converted into probability matrices and heuristically optimized to maximize hypergeometric enrichment of each motif in the given data set. As optimized motifs were found they were removed from the data set to facilitate the identification of additional motifs in subsequent rounds. HOMER also screens the enrichment of known motifs previously identified through the analysis of published ChIP-ChIP and ChIP-Seq data sets by calculating the known motifs’ hypergeometric enrichment in the same set of G+C normalized sequences used for de novo analysis. Sequence logos were generated using WebLOGO (http://weblogo.berkeley.edu/). Motif enrichment heatmaps and dendrograms were created by clustering hypergeometric log p-values using Cluster (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/software.htm#ctv) and Java TreeView (http://jtreeview.sourceforge.net/).
Global run-on sequencing analysis (GRO-seq)
Global run-on was done as described6 and library preparation for sequencing as described30. Briefly, four 10cm plates of confluent LNCaP cells per treatment were washed 3 times with cold PBS buffer. Cells were then swelled in swelling buffer (10mM Tris-pH7.5, 2mM MgCl2, 3mM CaCl2) for 5min on ice. Harvested cells were resupended in 1ml of the lysis buffer (swelling buffer with 0.5% IGEPAL and 10% glycerol) with gentle vortex and brought to 10ml with the same buffer for nuclei extraction. Nuclei were washed with 10ml of lysis buffer and resuspended in 1ml of freezing buffer (50mM Tris-pH8.3, 40% glycerol, 5mM MgCl2, 0.1mM EDTA), pelleted down again, and finally resuspended in 100μl of freezing buffer.
For run-on assay, resuspended nuclei were mixed with an equal volume of reaction buffer (10mM Tris-pH 8.0, 5mM MgCl2, 1mM DTT, 300mM KCl, 20 units of SUPERase-In, 1% Sarkosyl, 500μM ATP, GTP, and Br-UTP, 2μM CTP) and incubated for 5 min at 30°C. Nuclei RNA were extracted with TRIzol LS reagent (Invitogen) following manufacturer’s instructions. RNA was then resuspended in 20μl of DEPC-water and subjected to base hydrolysis by addition of 5μl of 1M NaOH and incubated on ice for 40min. Then, 25μl of 1M Tris-pH 6.8 was added to neutralize the reaction. RNA was purified through a p-30 RNAse-free spin column (BioRad), according to the manufacturer’s instructions and treated with 6.7μl of DNase buffer and 10μl of RQ1 RNase-free DNase (Promega), purified again through a p-30 column. A volume of 8.5μl 10×antarctic phosphatase buffer, 1μl of SUPERase-In, and 5μl of antarctic phosphatase was added to the run-on RNA and treated for 1hr at 37°C. Before proceeding to immuno-purification, RNA was heated to 65°C for 5min and kept on ice.
Anti-BrdU argarose beads (Santa Cruz Biotech) were blocked in blocking buffer (0.5×SSPE, 1mM EDTA, 0.05% Tween-20, 0.1% PVP, and 1mg/ml BSA) for 1 hr at 4°C. Heated run-on RNA (~85μl) was added to 60μl beads in 500μl binding buffer (0.5×SSPE, 1mM EDTA, 0.05% Tween-20) and allowed to bind for 1hr at 4°C with rotation. After binding, beads were washed once in low salt buffer (0.2×SSPE, 1mM EDTA, 0.05% Tween-20), twice in high salt buffer (0.5% SSPE, 1mM EDTA, 0.05% Tween-20, 150mM NaCl), and twice in TET buffer (TE pH7.4, 0.05% Tween-20). BrU-incorporated RNA was eluted with 4×125μl elution buffer (20mM DTT, 300mM NaCl, 5mM Tris-pH 7.5, 1mM EDTA, and 0.1% SDS). RNA was then extracted with acidic phenol/chloroform once, chloroform once and precipitated with ethanol overnight. The precipitated RNA was resuspended in 50μl reaction (45μl of DEPC water, 5.2μl of T4 PNK buffer, 1μl of SUPERase_In and 1μl of T4 PNK (NEB)) and incubated at 37°C for 1 hr. The RNA was extracted and precipitated again as above.
cDNA synthesis was performed basically as described30 with minor modifications. First, RNA fragments were subjected to poly-A tailing reaction in 8.0μl volume containing 0.8μl poly-A polymerase buffer, 1μl 1mM ATP, 0.5μl of SUPERase-In, and 0.75μl poly-A polymerase (NEB). The reaction was carried out for 30 min at 37°C. Subsequently, reverse transcription was carried out using oNTI223 primer (5′-pGATCGTCGGACTGTAGAACTCT;CAAGCAGAAGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN-3′) where the (p) indicates 5′ phosphorylation, (;) indicates the abasic dSpacer furan and (V, N) indicate degenerate nucleotides.
Tailed RNA (8.0μl) was mixed with 1μl dNTP (10mM each) and 2.5μl 12.5μM oNTI223, heated for 3 min at 75°C, and chilled briefly on ice. Then, the tube was added 0.5μl SUPERase-In, 3μl 0.1M DTT, 2μl 25mM MgCl2, 2μl 10× reverse transcription buffer, and 1μl superscript III reverse transcriptase (Invitrogen). The tube was incubated for 30 min at 48°C. After that, 4μl of Exonuclease I (Fermentas) was added into the reaction and the tube was incubated for 1 hr at 37°C to eliminate extra oNTI223. Then, RNA was eliminated by adding 1.8μl 1M NaOH and incubated for 20 min at 98°C. The reaction was neutralized with 1.8μl of 1M HCl. After running on a 10% polyacrylamide TBE-urea gel, the extended first-strand cDNA product was excised and recovered by soaking the grinded gel in DNA gel elution buffer (TE with 0.1% Tween-20 and 150mM NaCl) overnight and then precipitated with ethanol.
Circularization of first-strand cDNA was carried out by resuspending cDNA in 9.5μl reaction solution (7.5μl water, 1μl CircLigase buffer, 0.5μl 1mM ATP, and 0.5μl 50mM MnCl2) and then adding 0.5μl CircLigase (Epicentre). The reaction went for 1 hr at 60°C and then was heat-inactivated for 20 min at 80°C. Circularized ssDNA was relinearized by adding 3.8μl of 4×relinearization supplement (100mM KCl, 2 mM DTT) followed by 1.5μl of Ape1 (15u, NEB). The reaction was incubated for 1 hr at 37°C. Relinearized ssDNA was separated in a 10% polyacrylamide TBE-urea gel (Invitrogen) as described above. The relinearized product band was excised (~120-300bp) and the DNA was recovered as described above.
The ssDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB) according to the manufacturer’s instructions. The oligonucleotide primers oNTI200 (5′-CAAGCAGAAGACGGCATA-3 ′) and oNTI201 (5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGACG-3′) were used to generate DNA for sequencing. PCR was carried out with an initial 30s denaturation at 98°C, followed by 13 cycles of 10s denaturation at 98°C, 15s annealing at 60°C, and 15s extension at 72°C. The PCR product was run on a non-denaturing 8% polyacrylamide TBE gel and recovered as mentioned before. DNA was then sequenced on the Illumina Genome Analyzer II according to the manufacturer’s instructions with small RNA sequencing primer 5′-CGACAGGTTCAGAGTTCTACAGTCCGACGATC-3′.
De novo identification of GRO-Seq transcripts
To identify transcription units in an unbiased manner, GRO-Seq read densities were analyzed to classify genomic regions into contiguous transcripts using HOMER. GRO-Seq read densities were initially normalized using the GC-content of individual reads to remove any systematic bias introduced by overall GC-content variation between read libraries. To maximize read depth for transcript identification, all GRO-Seq libraries were merged to perform the initial transcript discovery, and later considered separately to identify regulated transcripts. For each strand of each chromosome, GRO-Seq read densities were calculated using a sliding window of 250bp. Regions for which GRO-Seq reads could not be uniquely mapped (i.e. repeats) were first identified and then read densities in these regions were estimated using upstream read densities from mappable regions to avoid ending predicted transcripts prematurely. Transcript initiation sites were identified as regions where the GRO-Seq read density increases 3-fold relative to the previous 1kb region. Transcript termination sites were defined by either a reduction in reads below 10% of the start of the transcript or when another transcript’s start site occurs on the same strand. Single spikes in read density covering a span less than 250bp were considered artifacts and discarded. Identified transcripts were strand-specifically compared to RefSeq transcripts by looking for overlap in the transcribed region. Transcripts were defined as putative eRNAs if their TSS was located distal to RefSeq TSS (>3 kb) and were associated with H3K4me1 regions. To identify differentially regulated transcripts, strand specific read counts from each GRO-Seq experiment were determined for each transcript using HOMER33. EdgeR (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) was then used to calculate differential expression (>1.5 fold, < 0.01 False discovery rate).
Microarray and RT-qPCR analysis of gene expression
Total RNA was isolated with the RNeasy® Mini Kit (Qiagen) and treated by RNase-free DNase I. For RT-qPCR, first-strand cDNA synthesis from total RNA was performed with the Superscript III cDNA Synthesis System (Invitrogen). Microarray was performed on Human V2 Chips (Illumina). The published gene expression profiling data GDS2545 (ref.36,37) and GDS1439 (ref.38) were extracted from NCBI, normalized, and P-values calculated by T-test (2 tails). For validation by RT-PCR, cDNA was analyzed with SYBR Green (Stratagene) on the Mcx300P System (Stratagene). The relative mRNA level was calculated by comparing with non-treatment control, after normalization with GAPDH or ACTB mRNA.
Survival analysis
Two independent sets of gene expression data were used to check the association between FoxA1 and clinical outcome of patients by the Kaplan-Meier analysis. One data set comes from 78 patients with prostate tumors (age<70)39, and the other 131 patients40. Significant association with outcome was determined by log-rank test for survival. Hazard ratios were calculated by the Cox proportional model. All statistical analyses were performed with the use of the statistical software R (Version 2.6.2), available from The R Project for Statistical Computing website (http://www.r-project.org). The cutoff was determined so that the log-rank P-value was the smallest one in the cutoffs that go through 5th ~ 95th percentiles of signals.
References
- 31.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbro T, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to derive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandran UR, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 38.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceraline J, et al. Constitutive activation of the androgen receptor by a point mutation in the hinge region: a new mechanism for androgen-independent growth in prostate cancer. Int J Cancer. 2004;108:152–157. doi: 10.1002/ijc.11404. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 3.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 4.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 9.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao N, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–43. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 11.Mirosevich J, Gao N, Matusik RJ. Expression of Foxa transcription factors in the developing and adult murine prostate. Prostate. 2005;62:339–52. doi: 10.1002/pros.20131. [DOI] [PubMed] [Google Scholar]
- 12.Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol. 2004;18:2633–48. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 13.Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmqvist PH, Belikov S, Zaret KS, Wrange O. FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp Cell Res. 2005;304:593–603. doi: 10.1016/j.yexcr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Crowe AJ, et al. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the alpha-fetoprotein gene. J Biol Chem. 1999;274:25113–20. doi: 10.1074/jbc.274.35.25113. [DOI] [PubMed] [Google Scholar]
- 16.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomsen MK, et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70:979–87. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68:1625–30. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–21. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, Hwang M, Chattopadhyay S, Choi HS, Lee K. Hepatocyte nuclear factor-3 alpha (HNF-3alpha) negatively regulates androgen receptor transactivation in prostate cancer cells. Biochem Biophys Res Commun. 2008;367:481–6. doi: 10.1016/j.bbrc.2007.12.162. [DOI] [PubMed] [Google Scholar]
- 23.Gao N, et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17:1484–507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 24.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laitinen S, Karhu R, Sawyers CL, Vessella RL, Visakorpi T. Chromosomal aberrations in prostate cancer xenografts detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2002;35:66–73. doi: 10.1002/gcc.10097. [DOI] [PubMed] [Google Scholar]
- 27.Legrier ME, et al. Hormone escape is associated with genomic instability in a human prostate cancer model. Int J Cancer. 2009;124:1103–11. doi: 10.1002/ijc.24073. [DOI] [PubMed] [Google Scholar]
- 28.Wolf M, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004;6:240–7. doi: 10.1593/neo.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.