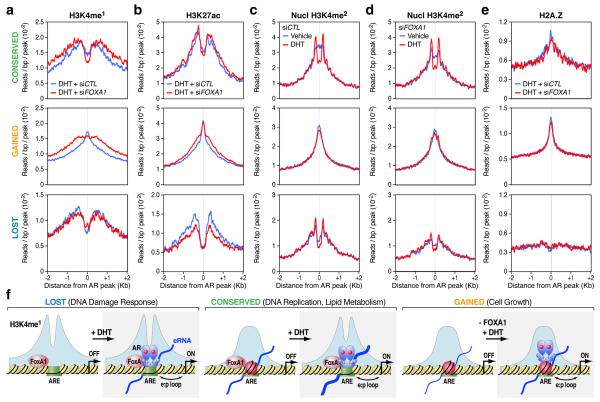
Figure 4. Distinct classes of AR enhancers in the human genome.
(a,b) Profiles of H3K4me1 (a) and H3K27ac (b) associated with the lost (bottom panels), conserved (top panels), and gained (middle panels) AR programs in DHT-treated LNCaP cells in response to FOXA1 knockdown. (c, d) Profiles of H3K4me2 around AR binding loci at the nucleosomal resolution in response to DHT stimulation in control siRNA-treated (c) or FOXA1 siRNA-treated (d) LNCaP cells. (e) Profiles of the histone variant H2A.Z on the three different AR programs. (f) Model for FoxA1-mediated AR targeting and reprogramming in LNCaP cells. In Class I (the lost AR program), FoxA1 licenses liganded AR to bind to ARE in relatively nucleosome-free regions. AR binding does not induce nucleosome remodeling in this class of enhancers. In Class II (the conserved AR program), AR binds independently of FoxA1 to ARE, inducing nucleosome remodeling. In Class III (the gained AR program), FoxA1 restricts AR binding, despite the presence of strong AREs. Although pre-established, these gained loci exhibit a strong central nucleosome and are associated with H2AZ, which is not affected by AR binding. FOXA1 knockdown converted these sites to androgen-responsive sites. In all these three classes, eRNAs were generated or increased after AR binding.