Abstract
Breast cancer is a major cause of cancer-related deaths in American women; therefore, the identification of novel breast-cancer related molecules for the discovery of new markers and drug targets remains essential. The human DEK gene, which encodes a chromatin-binding protein and DNA topology regulator, is up-regulated in many types of cancer. DEK has been implicated as an oncogene in breast cancer based on mRNA expression studies, but its functional significance in breast cancer growth and progression has not yet been tested directly. We demonstrate that DEK is highly expressed in breast cancer cells compared to normal tissue, and functionally important for cellular growth, invasion and mammosphere formation. DEK over-expression in non-tumorigenic MCF10A cells resulted in increased growth and motility with a concomitant down-regulation of E-cadherin. Conversely, DEK knockdown in MCF7 and MDA-MB-468 breast cancer cells resulted in decreased growth and motility with up-regulation of E-cadherin. The use of DEK-proficient and -deficient breast cancer cells in orthotopic xenografts provided further in vivo evidence that DEK contributes to tumor growth. Activation of the β-catenin signaling pathway is important for normal and cancer stem cell character, growth and metastasis. We show that DEK expression stimulated and DEK knockdown repressed β-catenin nuclear translocation and activity. Importantly, the expression of constitutively active β-catenin rescued breast cancer invasion defects of DEK knockdown cells. Together, our data indicate that DEK expression stimulates the growth, stem cell character, and motility of breast cancer cells, and that DEK-dependent cellular invasion occurs at least in part via β-catenin activation.
Keywords: DEK, breast cancer, metastasis, β-catenin, cancer stem cells
Introduction
Breast cancer is the second leading cause of cancer-related deaths among women in the United States (ACS, 2009). Early detection of breast cancer is crucial for successful treatment because advanced high grade disease correlates with increased metastasis and mortality (Tubiana and Koscielny, 1999). Therefore, the identification of new breast cancer mediators and biomarkers, particularly those associated with metastasis and growth, remains critical in order to combat mortality from recurrent disease. Recent reports and Oncomine data have shown that DEK mRNA expression is up-regulated in invasive ductal breast cancers with particularly strong gene expression in high grade and late stage breast cancers, making it a potential new target in the fight against recurrence (Abba et al., 2007, Rhodes et al., 2004, Wise-Draper et al., 2009a).
DEK is a unique protein expressed in the proliferating cells of all tissues (Waldmann et al., 2004). A fraction of the cellular DEK pool is detected in the cytoplasm bound to mRNA, but 90% of the protein is associated with chromatin (Kappes et al., 2001, Le Hir et al., 2001, McGarvey et al., 2000, Soares et al., 2006). Cell free assays showed that DEK can introduce constrained positive supercoils into DNA and facilitate the ligation of linear DNA molecules in vitro (Waldmann et al., 2003, Waldmann et al., 2002). Its ability to bind nucleic acids has led to functional associations with several cellular processes including mRNA splicing, transcriptional regulation, DNA replication, and DNA repair (Alexiadis et al., 2000, Campillos et al., 2003, Le Hir et al., 2001, McGarvey et al., 2000, Sammons et al., 2006, Soares et al., 2006).
DEK gene amplification and up-regulated mRNA expression have been described in multiple cancer types including hepatocellular carcinoma, bladder cancer, and melanoma (Abba et al., 2007, Carro et al., 2006, Evans et al., 2004, Khodadoust et al., 2009, Kondoh et al., 1999, Lu et al., 2005, Wise-Draper et al., 2009a). In particular, recent work has shown compelling evidence that DEK functions as an oncogene in lung neuroendocrine carcinomas and contributes to tumorigenicity, chemo-resistance, and the expression of stem cell markers (Shibata et al., 2010). DEK is an induced target of the human papillomavirus (HPV) E7 oncoprotein and an inhibitor of p53-dependent and –independent cellular senescence and apoptosis phenotypes (Johung et al., 2007, Kappes et al., 2008, Khodadoust et al., 2009, Wise-Draper et al., 2006, Wise-Draper et al., 2009a, Wise-Draper et al., 2009b). However, few studies have analyzed DEK with regard to breast cancer despite its description as one of the most deregulated transcripts involved in cellular proliferation in lymph node positive primary breast cancers (Abba et al., 2007, Bowles et al., 2007). This deregulation might occur because DEK is transcriptionally up-regulated by the Rb/E2F pathway, which is frequently perturbed in breast cancers (Bosco and Knudsen, 2007, Carro et al., 2006, Wise-Draper et al., 2005). Since DEK is an E2F target gene, its expression is strongly indicative of proliferation. Here we define specific oncogenic activities of DEK in breast cancer cells in vitro and in vivo, including roles in tumor growth, cancer stem cell characteristics, and the identification of a molecular mechanism through which DEK contributes to cell motility and invasion.
Results
DEK is highly expressed in breast cancer cells
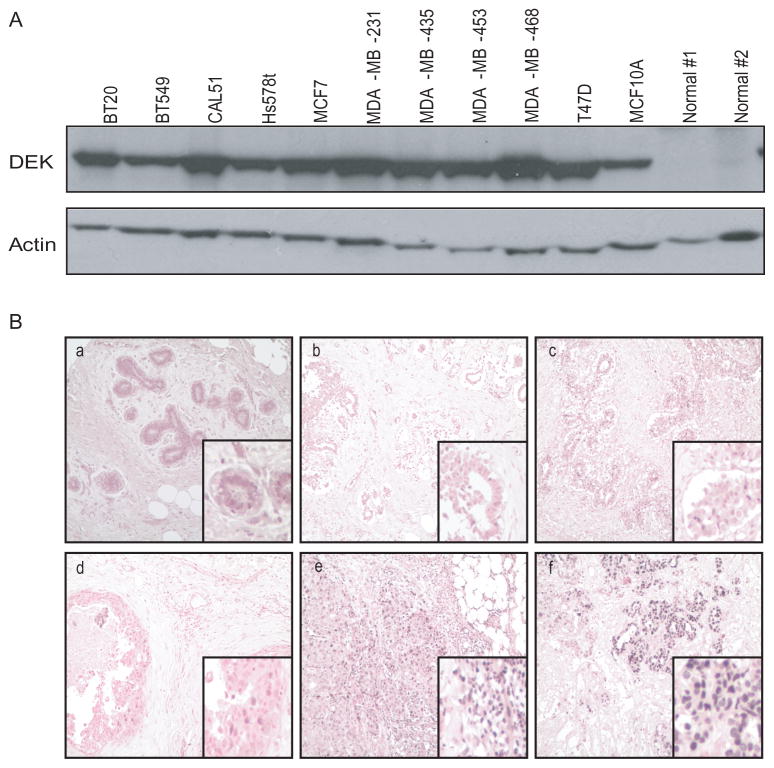
Meta-analysis of microarray data in Oncomine indicated that the DEK proto-oncogene may be important for breast cancer development and progression (Rhodes et al., 2004). In particular, DEK expression was higher in breast carcinomas compared to normal breast tissue and this correlated with tumor grade, tumor subtype, lymph node involvement, and disease-free survival (data not shown) (Miller et al., 2005, Richardson et al., 2006, van 't Veer et al., 2002, van de Vijver et al., 2002, Wang et al., 2005). To determine the frequency of DEK protein up-regulation in breast cancer cells, eleven distinct breast cell lines were analyzed by western blotting (Figure 1A; over-exposed to show expression in normal tissue). Consistent with previous reports on DEK mRNA levels (Miller et al., 2005, Richardson et al., 2006, van de Vijver et al., 2002, Wang et al., 2005), DEK protein was highly expressed in all eleven breast cancer cells compared to normal breast tissue, suggesting uniform DEK over-expression in breast cancer cell lines. To examine this further, immunohistochemistry was performed on a breast tissue microarray to assess DEK expression in primary tumors. DEK was detected in less than 50% of ductal epithelial cells in normal and non-malignant breast diseases, including hyperplasia and fibrocystic disease (Figure 1B, a-d). Consistent with prevalent DEK expression in breast cancer cell lines, 80% (24/30) of invasive carcinomas expressed DEK. Moreover, 20% (6 of 30, p=0.05 Chi Square) of the invasive carcinomas showed greater than 50% of the cancer cells staining positive for DEK (Figure 1B, e and f) demonstrating that DEK expression is more uniform in malignant tumors than in normal or hyperplastic breast epithelium and suggesting that DEK upregulation in cancer may result from an increase in DEK expressing cells. Together, these data demonstrate that DEK is highly expressed in breast cancer cell lines as well as primary breast cancers in vivo.
Figure 1. DEK is highly expressed in human breast cancer cell lines and primary breast cancers.
A) Western blotting for DEK revealed high expression in ten breast cancer cell lines and the non-tumorigenic MCF10A immortalized human mammary epithelial cell line compared to two primary normal breast samples from separate donors. Actin was used as a loading control. The blot is over-exposed to emphasize low expression in normal tissue (B) A tissue microarray was stained for the DEK protein (brown) by immunohistochemistry. Normal, abnormal, and cancerous tissue are shown: (a) normal tissue, (b) hyperplasia, (c) fibrocystic changes, (d) grade I carcinoma in situ, (e) grade II-III, and (f) grade III invasive ductal carcinomas. Low power images are at 100x total magnification.
DEK promotes breast cancer cell growth and survival
We next sought to determine the physiological role of DEK in regulating breast cancer cell growth. We chose three molecularly unique cell lines; non-tumorigenic estrogen receptor negative (ER-) MCF10A cells, tumorigenic ER- MDA-MB-468 cells, and tumorigenic ER+ MCF7 cells. These cells were retrovirally transduced with a DEK over-expression construct (R780:DEK) or empty vector (R780) as a control and expression was confirmed in the polyclonal population by western blotting (Figure 2A). The degree of over-expression was modest, as has been noted previously (Wise-Draper et al., 2005, Wise-Draper et al., 2009a). We found that the R780:DEK cells grew faster than their respective controls in each of the three lines. Interestingly, the observed differences manifested as a trend in tumorigenic MDA-MB-468 and MCF7 cells, but the strongest affect was observed in the non-tumorigenic, slower growing MCF10A cells (Figure 2B). Increased growth was further supported by the presence of elevated cyclin A levels in the DEK over-expressing compared to control cells (Figure 2A). The non-tumorigenic MCF10A cells demonstrated a more dramatic increase in cyclin A expression than the other two breast cancer cell lines, a finding which correlated well with the observed significant degree of growth stimulation by DEK.
Figure 2. DEK regulates breast cancer cell growth and survival.
(A) MDA-MB-468, MCF10A, and MCF7 cells were retrovirally transduced with either empty vector (R780) or a DEK over-expression construct (R780:DEK). MDA-MB-468 and MCF7 cells were transduced with lentiviral shRNA constructs for non-targeting shRNA (NTsh) or two distinct DEK shRNAs (DEKsh2 or DEKsh5). Lysates were analyzed by Western blotting for DEK, cyclin A and Actin expression. (B) DEK over-expression significantly increases cellular growth rates in non-tumorigenic MCF10A cells and moderately increases growth rates in cancer cells. Population growth was monitored by plating equal numbers of control and DEK over-expressing cells then counting cell numbers over the course of five to seven days (p<0.05 in MCF10A cells, p=0.06 for MCF7 cells). (C) Loss of DEK expression inhibits growth in breast cancer cells. Population growth was monitored as in (B) immediately after selection in puromycin.. (D) Loss of DEK expression results in increased rates of apoptosis. MCF7 and MDA-MB-468 NTsh or DEKsh2 cells were stained for cleaved caspase 3 and analyzed by flow cytometry.
Since MCF7 and MDA-MB-468 cells are already rapidly growing cancer cell lines and over-expression of DEK did not dramatically enhance this phenotype, we predicted that DEK knockdown might suppress cellular growth in cancer cells. Cells were transduced with non-targeting (NTsh) or two different DEK-specific lentiviral shRNA constructs (sh2 and sh5) and expression was confirmed by western blotting (Figure 2A). In both MCF7 and MDA-MB-468 cells, the most efficient DEK knockdown construct, DEKsh2, nearly abolished cellular growth. DEKsh5, which moderately decreased DEK expression, caused an intermediate phenotype (Figure 2C). The decreased cell growth induced by DEK knockdown was associated with decreased cyclin A levels in MDA-MB-468 cells, but not in MCF7 cells (Figure 2A). There is an overall correlation between cyclin A expression and DEK-mediated effects on cell growth. However, as indicated in MCF7 cells, it appears that DEK can promote cyclin A-independent cellular growth. To determine if impaired growth was due to changes in apoptosis, we analyzed active (cleaved) caspase 3 detection in MCF7 and MDA-MB-468 cells and both cell lines had an approximate 3-fold increase in cell death following DEK depletion (Figure 2D). To define a potential molecular mechanism for apoptosis due to DEK depletion, western blotting was performed to analyze the expression of ten pro- and anti-apoptotic proteins. While striking changes in expression were not detected (Supplementary Figure 1A), the relative expression of Bcl-xL in DEKsh2 averaged approximately 70% of that observed in NTsh MCF7 and MDA-MB-486 cells. Additionally, our previous preliminary experiments suggested that Bcl-xL rescues DEK depletion-induced apoptosis (Wise-Draper et al., 2006). Indeed, exogenous Bcl-xL expression in MCF7 cells resulted in a significant reduction in the percentage of apoptotic DEKsh2 cells, as determined by sub-G1 DNA content (Supplementary Figure 1B). These data suggest that the observed slight Bcl-xL repression contributes to DEKsh-induced apoptosis, but by no means rule out a functional role for Bcl-xL-unrelated molecular mechanisms. Taken together, our data indicate that DEK promotes cell growth at least in part via the inhibition of apoptosis, regardless of estrogen receptor status, in breast cancer cells.
DEK enhances mammosphere formation
Since DEK expression modulated the growth rates of the culture population as a whole, we next asked if DEK may also contribute specifically to the growth of the breast cancer stem cell population. Cancer stem cells have been implicated in tumor recurrence and drug resistance (Dontu et al., 2005) and can be enriched in suspension cultures as mammospheres (Dontu et al., 2003). Additionally, this distinct sub-population of cancer cells can be prospectively isolated based on the ability to efflux Hoechst 33342 dye through ABC transporters to form the “side population” (Woodward et al., 2005). First, MCF7 cells were analyzed for Hoechst dye exclusion by flow cytometry. The over-expression of DEK nearly doubled the size of the side population whereas DEK depletion resulted in a decrease in the side population (Figure 3A). Transporter inhibition through the addition of verapamil completely eliminated the side population. We then used DEK deficient and proficient MCF7 cells cultured under non-adherent conditions in media formulated to foster mammosphere formation (Dontu et al., 2003). The over-expression of DEK resulted in increased mammosphere formation whereas the loss of DEK decreased mammosphere formation (Figure 3B). Lysates collected from mammospheres confirmed that DEK abundance was maintained (Figure 3C). Mammospheres were then passaged twice to assess cancer stem cell maintenance, with the second passage of mammospheres exhibiting the same degree of difference as observed in the first passage (Figure 3D). Finally, NTsh mammospheres were confirmed to be enriched for cells in the side population compared to DEKsh spheres (compare Figure 3A and 3E). These data indicate that DEK expression stimulates the growth of the cancer stem cell population in MCF7 cells.
Figure 3. DEK expression correlates with the proportion of cells in the cancer stem cell population.
(A) DEK expression regulates the side population (SP; highlighted in the box). Side population cells were identified based on the exclusion of Hoechst 33342 dye and analyzed by flow cytometry. Verapamil, which blocks dye efflux, was used as a negative control. (B) DEK over-expression in MCF7 cells increases the number of mammospheres while DEK depletion impairs mammosphere formation. The graph represents the quantification of the number of mammospheres generated per cell line after ten days in suspension culture. (C) Western blot analysis shows expression of exogenous DEK or DEK shRNA are maintained in mammospheres. (D) Mammospheres were grown as in (B) then dissociated with 0.05% trypsin to a single cell suspension. Cells were replated at one-fourth the original density and grown in suspension culture. The graph represents the relative number of mammospheres in passage one and passage two when DEK-modulated cells were compared with their respective controls. (E) Mammospheres are enriched for potential cancer stem cells in the side population and DEK depletion results in fewer side population cells in mammospheres. Mammospheres from NTsh or DEKsh2 MCF7 cells were dissociated and analyzed for the side population as in (A).
DEK promotes cellular motility and invasion
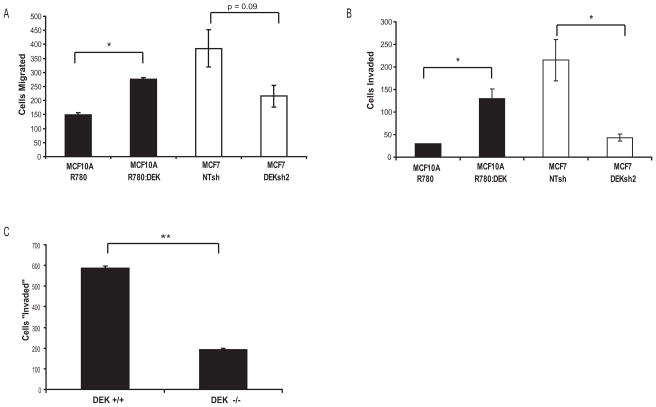
A hallmark of advanced breast cancer is distant metastases to the lungs, liver, and bone. DEK expression had been correlated with metastases in cDNA microarray studies and with cancer cell invasion in vitro in one lung cancer cell line (Shibata et al., 2010). Using transwell assays, we tested DEK proficient and deficient cells for motility and invasive potential in breast cells. MCF10A cells are non-transformed, and therefore harbor a low degree of motility when compared with transformed MCF7 cells ((Figures 4A and 4B) and (Privette et al., 2007)). DEK over-expression in MCF10A cells increased cellular motility (Figure 4A) and invasive potential (Figure 4B). Conversely, MCF7-DEKsh2 cells exhibited decreased cellular motility and invasion in transwell migration assays (Figures 4A and 4B). This significant decrease in invasive potential also was observed in MDA-MB-468 cells (Figure 7C). We next asked if the role of DEK in cellular motility was specific to cancer cells. Matrigel invasion assays showed that Dek knockout mouse embryonic fibroblasts (MEFs) had reduced chemotaxis-driven migration through collagen compared to Dek wild type MEFs (Figure 4C, DEK expression shown in Figure 6E). This data identifies a novel role for the DEK oncogene in regulating cellular invasion in both normal and breast cancer cells.
Figure 4. DEK promotes cellular motility and invasion in vitro.
(A) DEK over-expression increased cellular migration in non-tumorigenic MCF10A mammary epithelial cells (p < 0.001) while DEK depletion decreased cellular motility in MCF7 breast cancer cells ( p = 0.09). Transwell motility assays were performed using complete medium in the bottom chamber as a chemotactic incentive for migration. The total number of Giemsa-stained cells were counted and are shown graphically. (B) DEK over-expression increased the invasive potential of MCF10A cells (p = 0.06), whereas loss of DEK in MCF7 breast cancers dramatically decreased cellular invasive potential (p = 0.003) in vitro. Matrigel transwell invasion assays were performed using complete medium in the bottom chamber as a chemotactic incentive for migration.. The total number of Giemsa-stained cells were counted and are shown graphically. (C) Dek knockout MEFs have fewer migrating cells in a Matrigel transwell invasion assay, performed as described above, compared to wild type MEFs.
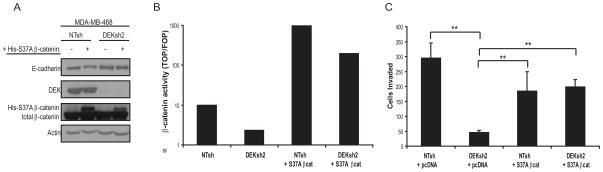
Figure 7. Constitutively active β-catenin rescues the loss of invasive potential in DEK deficient breast cancer cells.
(A) Expression of a transiently transfected, constitutively active His-tagged S37A β-catenin construct is confirmed by western blotting of total lysates from MDA-MB-468 cells. This construct was functional as indicated by the moderate down-regulation of E-cadherin. (B) S37A β-catenin partially restores transcriptional activity in MDA-MB-468 DEKsh2 cells as determined by TOP/FOP luciferase reporter assays, performed as described earlier. (C) S37A β-catenin expression in MDA-MB-468 DEKsh2 cells rescues the invasive potential through Matrigel transwell invasion assays to levels comparable to control cells. Matrigel transwell invasion assays were performed as described earlier and were plated 48 hours post-transfection. The statistically significant differences are indicated by two asterisks (p<0.001). All other comparisons, including NTsh + pcDNA versus NTsh + S37A β-catenin and NTsh + S37A β-catenin versus DEKsh2 + S37A β-catenin, are insignificant (p≥0.2).
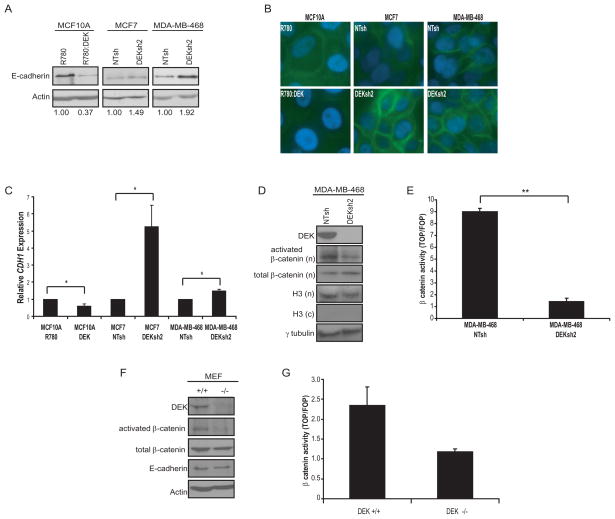
Figure 6. DEK regulates β-catenin transcriptional activity.
(A and B) DEK over-expression correlated with decreased E-cadherin levels while DEK depletion was associated with increased E-cadherin, as determined by western blotting (A) and immunofluorescence (B). Fold-change in E-cadherin expression, as determined by densitometry, is shown below each lane. (B) Cells were stained for E-cadherin (green) and nuclei were stained with DAPI (blue). (C) Transcription of CDH1 (E-cadherin) is altered following changes in DEK expression. Quantitative real-time RT-PCR for CDH1 expression indicated that DEK over-expression results in decreased CDH1 transcription while DEK depletion enhances CDH1 transcription. GAPDH was used as an interal control and relative expression was determined using the comparative Ct method. (D) MDA-MB-468 cells expressing DEK shRNA show a decrease in the amount of active, phosphorylated (Y142) β-catenin in the nucleus. Cells lysates were separated into nuclear (n) and cytoplasmic (c) fractions and probed by western blot analysis. γ-tubulin and histone H3 are used as loading and fractionation controls, respectively. (E) DEK deficient MDA-MB-468 cells have lower β-catenin transcriptional activity compared to control cells, as determined by TOP/FOP luciferase reporter assays. Cells were transfected with either pSUPER(8x)TOPFlash, which has eight TCF/LEF binding sites, or the negative control pSUPER(8x)FOPFlash with mutated TCF/LEF sites. Values are presented as the ratio of TOPFlash activity to FOPFlash activity. (F) Western blotting of whole cell lysates from wild type (+/+) and Dek knockout (-/-) MEFs depict decreased levels of active β-catenin (dephosphorylated S37 or T41) and decreased p63 expression in knockout MEFs. (G) TOP/FOP luciferase reporter assays indicate a decrease in β-catenin activity in knockout MEFs compared to wild type MEFs (p = 0.067 two-tailed, two-sample t-test, p = 0.03 one-tailed t-test).
DEK promotes tumor cell growth in vivo
We next tested the biological role of DEK in breast tumorigenesis and metastasis in vivo using an orthotopic model system. Control NTsh or DEKsh2 MDA-MB-468 cells were injected into the mammary fat pad of nulliparous female nude mice and then monitored for tumor size. Tumor initiation was comparable for both cell lines. However, beginning at six weeks, the tumors from MDA-MB-468 NTsh cells grew significantly larger than tumors from DEKsh2 cells (Figure 5A). Final tumor volumes after sacrifice also show that NTsh tumors were significantly larger than DEKsh2 tumors (Figure 5A bottom panel). The excised tumors were analyzed by immunohistochemistry for BrdU incorporation as a marker of proliferation and cleaved caspase 3 as a marker of apoptosis. Tumors were also analyzed for ΔNp63 since DEK over-expression resulted in increased ΔNp63 expression in primary human keratinocytes, and because p63 is essential for mammary gland development and is a hallmark of poorly differentiated breast carcinomas (Ribeiro-Silva et al., 2003, Wise-Draper et al., 2009b). Increased expression of the oncogenic isoform, ΔNp63, was observed in NTsh tumors compared to DEKsh2 tumors (Figure 5B). Expression of E-cadherin was increased in DEKsh2 derived tumors, supporting the noted decrease in cell invasion in vitro (Figure 5B). DEKsh2 derived tumors exhibited increased numbers of cleaved caspase-3 positive cells and decreased numbers of BrdU-positive cells (Figures 5C and 5D), demonstrating that DEK promotes cancer cell proliferation and survival in vivo. Finally, immunofluorescence studies of tumor sections derived from NTsh cells indicated co-expression of DEK and the proliferation marker PCNA within the same cells (Figure 5E), providing further evidence that DEK expression is associated with proliferating cancer cells.
Figure 5. DEK promotes breast tumor growth in vivo.
(A) (Top) Loss of DEK expression impairs tumor growth in nude mice. MDA-MB-468 breast cancer cells transduced with lentiviral non-targeting shRNA (NTsh) or DEK shRNA (DEKsh2) were injected into the mammary fat pad of female nude mice (N=10 per cell line) and tumor growth was monitored with calipers for ten weeks. (Bottom) At necropsy, tumors were removed from the mammary gland and measured in three dimensions with calipers to calculate the final tumor volume (volume = (π/6)*(LxWxH)). (B) (Top) H&E images of representative tumors. T = tumor and N = necrosis. Arrows indicate normal mammary ducts. Immunohistochemical staining of tumors showed a correlation between DEK (middle panel) and ΔNp63 (bottom panel) staining. The H&E images are at 100x magnification. (Right) Western blotting of tumor lysates show down-regulation of ΔNp63 and upregulation of E-cadherin in DEKsh2 tumors. (C) DEKsh2 tumors had a higher percentage of cleaved caspase-3 positive cells, indicative of apoptosis, compared to NTsh-derived tumors. (Right) Representative immunohistochemistry images for caspase 3 staining. Arrows indicate representative positive cells. (D) DEKsh2 tumors had fewer BrdU-positive cells compared to NTsh-derived tumors. (Right) Representative immunohistochemistry images for BrdU staining. (E) Tumor cells exhibit co-expression of DEK with PCNA. Sections from paraffin-embedded NTsh derived tumors were immunolabelled for DEK (red) and PCNA (green).
DEK promotes breast cancer cell invasion by inducing β-catenin activity
In order to define a potential mechanism for the role of DEK in cancer cell invasion, DEK proficient and deficient cells were analyzed for E-cadherin expression, which is typically downregulated in cells that have become more motile (Guarino et al., 2007). Although we did not observe an overt morphological change, DEK over-expression in the MCF10A cells caused decreased E-cadherin levels, while the loss of DEK in MCF7 and MDA-MB-468 cells resulted in a moderate increase in E-cadherin expression (Figures 6A and 6B). Furthermore, quantitative RT-PCR for E-cadherin (CDH1) confirmed these trends and indicated that this change in expression was due to transcriptional regulation (Figure 6C). CDH1 expression is negatively regulated by Wnt/β-catenin signaling, which is a major pathway driving cell motility and stem cell maintenance (Heuberger and Birchmeier, 2010, Li et al., 2003). We discovered that the down-regulation of DEK in MDA-MB-468 cells resulted in decreased levels of active β-catenin in the nucleus (Figure 6D). Similar results were observed in MCF7 DEKsh2 cells and increased levels of nuclear β-catenin were observed in DEK over-expressing cells (data not shown). Importantly, Dek knockout MEFs also exhibited decreased levels of activated β-catenin, indicating that DEK is important for β-catenin activity in multiple species and in both normal and cancerous cells (Figure 6F). This decrease in active β-catenin levels translated into repression of transcriptional activity, as detected by TOP/FOP luciferase reporter assays, in MDA-MB-468 and MEF cells (Figures 6E and 6G).
We attempted to rescue the decreased invasion phenotype by transiently expressing a constitutively active form of β-catenin, S37A β-catenin, in NTsh and DEKsh2 MDA-MB-468 cells (Figure 7A). Expression of S37A β-catenin partially rescued the defect in transcriptional activity relative to control cells (Figure 7B) and fully rescued the invasion defect in Matrigel transwell assays (Figure 7C). Therefore, the role of DEK in stimulating cellular motility and invasion is due, at least in part, to its ability to activate the β-catenin signaling pathway.
Discussion
Together, our findings are the first to report on the oncogenic activities of DEK in breast cancer and to define both functional and molecular mechanisms for DEK in breast cancer pathogenesis. DEK was highly expressed in breast cancer cell lines and was expressed in a higher percentage of tumor cells from high grade primary invasive ductal carcinomas compared to benign tissue and low grade carcinomas. We have found that DEK regulates cancer growth and survival in vitro and in vivo and can additionally contribute to breast cancer stem cell growth. We also report here the novel association of DEK with invasion through the β-catenin signaling pathway.
We demonstrate control of motility and invasion by DEK in breast cancer cells and in normal murine embryo fibroblasts, thus extending recent data published for neuroendocrine lung cancer cells (Shibata et al., 2010). Furthermore, we report the significant and novel finding of a molecular mechanism for this phenotype in that DEK can stimulateβ-catenin transcriptional activity. In addition, the decrease in invasive potential in DEK deficient cancer cells was rescued by the expression of a constitutively active β-catenin construct. Our identification of DEK as a regulator of β-catenin signaling may be supported by the recent finding, according to microarray data, that DEK can regulate Wnt10b expression (Shibata et al., 2010). Of importance, the regulation of β-catenin activity by DEK was observed in multiple species and in both normal fibroblasts and breast cancer cells, indicating that this is an evolutionarily conserved mechanism.
Cancer stem cells have been hypothesized to facilitate tumor recurrence due to their drug resistance and ability to reconstitute the entire tumor. The cancer stem cell hypothesis is particularly relevant to breast cancer following the discovery of breast cancer stem cells and the ability to culture them as mammospheres (Al-Hajj et al., 2003, Dontu et al., 2003). This is the first report to implicate DEK in the maintenance of the breast cancer stem cell-like population by showing that DEK over-expression in MCF7 breast cancer cells increased the number of mammospheres formed in culture and the percentage of cells in the side population. Wnt/β-catenin signaling is well known to contribute to cancer invasion and metastasis as well as cell migration during development, often through an epithelial-to-mesenchymal transition (EMT) (Heuberger and Birchmeier, 2010). However, β-catenin signaling is also crucial for the proliferation of breast cancer stem/progenitor cells in vivo (Li et al., 2003). Based on our data, we postulate that DEK up-regulation in primary high grade breast cancers may contribute to subsequent recurrence and metastasis through alterations in β-catenin signaling.
Our previous studies in human keratinocytes indicated that highly proliferative cancer cells are more susceptible to DEK depletion than less proliferative cells (Wise-Draper et al., 2009a). Targeting DEK expression may therefore be an efficient mechanism for inhibiting breast cancer growth and progression with minimal toxicity to neighboring normal cells with low proliferation indices. Interestingly, recent reports have indicated that DEK expression correlates with resistance to chemotherapeutic drugs like camptothecin, etoposide, neocarzinostatin, and doxorubicin, which is often used to treat breast cancer (Kappes et al., 2008, Khodadoust et al., 2009). The combination of conventional clastogenic agents with DEK-specific targeting approaches may therefore function synergistically in the treatment of patients with advanced breast cancer.
Methods
Cell Culture
Cell lines were obtained from the American Type Culture Collection. MCF7 cells were grown under ATCC recommended conditions. MDA-MB-468 cells were cultured in Modified Improved MEM (IMEM; Invitrogen, Carlsbad, CA) with 10% FBS and MCF10A cells were cultured and maintained as previously described (Soule et al., 1990). Transfections were performed using TransIT-LT1 reagent (Mirus Bio, Madison, WI) and cells were assayed after 48 hours.
Dek wild type and knockout mice (gift of Gerard Grosveld, St. Jude Children’s Hospital) were used to generate primary mouse embryonic fibroblasts (MEFs) (Wise-Draper et al., 2009a). MEFs were explanted from day 13.5 embryos and maintained in high glucose DMEM containing 10% FBS, 2mM L-glutamine, MEM non-essential amino acids, 0.055mM β-mercaptoethanol, and gentamycin. Immortalized MEFs were obtained by 3T3 serial passaging.
Western Blotting
Western blotting was performed as previously described using 40 μg of total protein and semi-dry transfer (Andreassen and Margolis, 1994, Wise-Draper et al., 2006). Membranes were probed with antibodies to DEK (1:1000, BD Biosciences, San Jose, CA), cyclin A (1:400, Santa Cruz, Santa Cruz Biotechnology, CA), E-cadherin (1:2500, BD Biosciences), p63 (1:200, Santa Cruz), total β-catenin (1:2000, BD Biosciences), activated phosphorylated (Y142) β-catenin (1:500, Abcam Inc, Cambridge, MA), activated β-catenin (1: 1000, clone 8E7 “anti-ABC”, Millipore, Billerica, MA), Histone H3 (1:2000, Abcam), γ-tubulin (1:1000, Sigma-Aldrich, St. Louis, MO) and Actin C4 (1:10,000, gift of James Lessard, Cincinnati Children’s Hospital). Fractionated lysates were prepared as previously described (Dignam et al., 1983). Protein from tumor tissue was isolated by pulverizing snap-frozen tissue with a mortar and pestle prior to the addition of lysis buffer. Band intensities were measured using Image J software.
Tissue Microarray
A breast tissue microarray (BRC961C_F, Pantomics, Inc., Richmond, CA) was stained by immunohistochemically stained for DEK as described below. The tissue microarray contained intact duplicate samples including normal breast (n=2), invasive adenocarcinomas (n=30, 28 ductal, one ductal papillary and one mucinous) and non-malignant breast disease including hyperplasia (n=3), fibrocystic changes (n=4) and fibroadenomas (n=3). All patients were female with the exception of the one case of ductal papillary adenocarcinoma. DEK staining was blindly scored as positive or negative based on the presence of any brown-stained nuclei.
Immunohistochemistry
Paraformaldehyde fixed and paraffin embedded tissues were sectioned, subjected to sodium citrate antigen retrieval, and stained using the M.O.M. Peroxidase kit (Vector Labs, Burlingame, CA) and 3,3′-Diaminobenzidine (DAB). The following antibodies were used: DEK (1:60, BD Biosciences), BrdU (1:100, Molecular Probes, Invitrogen), ΔNp63 (1:50, Santa Cruz), or cleaved caspase 3 (Asp175; 1:100, Cell Signaling Technology, Beverly, MA). Samples were counterstained with 0.1% Nuclear Fast Red (Poly Scientific, Bay Shore, NY) and preserved with Permount (Fisher Scientific, Pittsburgh, PA).
Retroviral and Lentiviral Transduction
Cells were transduced with retroviral constructs (R780 and R780:DEK) as described previously and sorted based on GFP expression (Wise-Draper et al., 2009a). Alternatively, cells were transduced with the lentiviral pLKO.1 constructs (Sigma Aldrich Mission shRNA library) and selected in 2 μg/ml puromycin. DEKsh2 represents construct pLKO.1_DEK832 and DEKsh5 represents construct pLKO.1_DEK523. NTsh is a non-targeting control. DEKsh2 functionality was published previously (Kappes et al., 2008). All cells were analyzed within four passages of selection.
Growth Curves
Cells were plated at equal density in six-well plates, trypsinized, and counted in triplicate using a hemacytometer.
Flow Cytometry
Apoptotic cells were labeled with the Caspase 3, Active Form, Apoptosis kit (BD Biosciences) according to manufacturer’s instructions. Samples were analyzed with a FACSCalibur flow cytometer (BD Bioscience). The side population was analyzed based on the exclusion of Hoecsht 33342 using an LSRII flow cytometer (BD Bioscience) as previously described (Goodell et al., 1996). Verapamil at a final concentration of 100 μM was used as a negative control to properly identify the population.
Mammosphere Culture
MCF7 cells were seeded at a density of 10,000 cells per ml in 6-well plates coated with 1% agarose and cultured in DMEM:F12 (1:1) media with 2% BSA, 10 ng/ml EGF, and 1.0 μg/ml each of hydrocortisone and insulin. The total number of mammospheres were counted after 10 days. Mammospheres were passaged as previously described and replated at a density of 2,500 cells per ml (Zhang et al., 2008). Images were obtained with a Leica DMIL microscope (Leica Microsystems, Bannockburn, IL) and SPOT imaging software (Diagnostic Instruments, Sterling Heights, MI).
Migration and Invasion Assays
Transwell assays, with or without Matrigel, were used to determine cell migration and invasive potential in vitro according to manufacturer’s instructions (BD Biosciences). Cells were seeded in supplement-free media and complete media was placed into the bottom chamber as a chemoattractant. Cells attached to the outer surface of the transwell chamber were alcohol-fixed and stained with Giemsa after 20 hours.
In vivo Tumorigenesis
Cells were orthotopically injected into the inguinal mammary fat pads of nulliparous, 12-week old female athymic nude mice as described (Allan et al., 2006). Tumors were measured with calipers and volume was calculated as [(π/6)xLxW2] (Euhus et al., 1986, Tomayko and Reynolds, 1989). Two hours prior to sacrifice, the mice were intraperitoneally injected with 600 μg of BrdU (200 μl of a 10 mM solution; GE Healthcare/Amersham, Piscataway, NJ). At necropsy, the mice were weighed and the tumors with any adjacent mammary gland and/or lymph nodes were excised, measured, weighed, fixed and embedded in paraffin. Lungs and livers were visually inspected for metastases. The usage and handling of mice were performed with the approval of the University of Cincinnati Institutional Animal Care and Use Committee.
Immunofluorescence
Tissue sections from paraffin-embedded MDA-MB-468 NTsh xenograft tumors from the mammary gland were deparaffinized, underdwent antigen retrieval with 10 mM sodium citrate, and were blocked with 5% normal donkey serum and immunostained with antibodies for DEK (1:50, Aviva Systems Biology, San Diego, CA) and PCNA (1:50, BD Biosciences). DEK and PCNA were visualized with rhodamine- and FITC-conjugated secondary antibodies, respetively (1:200, Jackson Immunoresearch, West Grove, PA) then counterstained with ProLong Gold antifade reagent with 4',6-diamidino-2-phenylindole (DAPI; Invitrogen Molecular Probes).
Cultured cells were seeded into 8-well chambered slides (BD Biosciences) and labeled as previously described (Prahalad et al., 2004) except 0.1% normal goat serum was used for blocking. Cells were labeled with anti-E-cadherin antibody (1:50, BD Biosciences) and FITC-conjugated anti-mouse secondary antibody (1:50, Jackson Immunoresearch, West Grove, PA), then counterstained with Vectashield plus DAPI (Vector Labs). All immunofluorescnce images were obtained with a Leica DMI 6000b microscope (Leica Microsystems) and OpenLab v5.5 imaging software (Perkin Elmer, Waltham, MA)
TOP/FOP Luciferase Reporter Assay
MDA-MB-468 cells were transfected as described above with pRL-TK (Renilla), and either pSUPER(8x)TOPFlash or pSUPER(8x)FOPFlash then assayed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Rescue of β-catenin signaling was performed by transfecting cells with the constitutively active S37A β-catenin construct cloned into the pcDNA6.V5-His plasmid. All plasmids were a gift from Aaron Zorn (Cincinnati Children’s Hospital). Data is presented as the ratio of relative light units (RLUs) of TOPFlash to FOPFlash.
Quantitative RT-PCR
mRNA was isolated from sub-confluent cells using Trizol reagent (Invitrogen, Carlsbad, CA) and one microgram of total mRNA was used to produce cDNA using the QuantiTect Reverse Transcription kit (Qiagen, Valencia CA). Expression was tested using SYBR Green detection and an ABI7300 Real Time PCR system (Applied Biosystems, Carlsbad, CA). Data was analyzed using the comparative Ct method (Schmittgen and Livak, 2008). The following primers were used: (1) E-cadherin Forward: 5-CAGAAAGTTTTCCACCAAAG-3 (2) E-cadherin Reverse: 5- AAATGTGAGCAATTCTGCTT -3 (3) GAPDH Forward: 5-GGTCTCCTCTGACTTCAACA-3 and (4) GAPDH Reverse: 5-ATACCAGGAAATGAGCTTGA-3.
Statistics
Statistical significance was assayed using two-tailed Student’s t-test or Chi Square test. All in vitro experiments represent the average of triplicate experiments and errors bars depict standard error. In the figures, one asterisk (*) indicates p<0.05, and two asterisks (**) indicates p<0.001.
Supplementary Material
Acknowledgments
We thank Yi Zheng for critical evaluation of this manuscript. We also thank James Lessard, Aaron Zorn, Jose Cancelas, James Mulloy, and James Wells for reagents and discussion, Gerard Grosveld for Dek knockout mice, Gina Kavanaugh for the immortalized Dek wild type and knockout mouse embryonic fibroblasts and the Viral Vector Core at at CCHMC. We would like to acknowledge the assistance of the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, supported in part by NIH AR-47363. This research was supported by Kirschstein National Research Service Awards (NRSA) F32CA139931 and T32HL091805 from the National Cancer Institute (L.M.P.V.), and Public Health Service grants CA116316 (S.I.W.), CA100002 (S.E.W.), and HL079193 (K. W-B.)
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest
References
- Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, et al. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5:881–90. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACS. Breast Cancer Facts & Figures 2009–2010. Atlanta: American Cancer Society, Inc; 2009. [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14 :1308–12. [PMC free article] [PubMed] [Google Scholar]
- Allan AL, George R, Vantyghem SA, Lee MW, Hodgson NC, Engel CJ, et al. Role of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancer. Am J Pathol. 2006;169:233–46. doi: 10.2353/ajpath.2006.051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Margolis RL. Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J Cell Biol. 1994;127:789–802. doi: 10.1083/jcb.127.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle. 2007;6:667–71. doi: 10.4161/cc.6.6.3988. [DOI] [PubMed] [Google Scholar]
- Bowles E, Corson TW, Bayani J, Squire JA, Wong N, Lai PB, et al. Profiling genomic copy number changes in retinoblastoma beyond loss of RB1. Genes Chromosomes Cancer. 2007;46:118–29. doi: 10.1002/gcc.20383. [DOI] [PubMed] [Google Scholar]
- Campillos M, Garcia MA, Valdivieso F, Vazquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–5. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5 :1202–7. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11 :1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Liu S, Wicha MS. Stem cells in mammary development and carcinogenesis: implications for prevention and treatment. Stem Cell Rev. 2005;1:207–13. doi: 10.1385/SCR:1:3:207. [DOI] [PubMed] [Google Scholar]
- Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–34. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- Evans AJ, Gallie BL, Jewett MA, Pond GR, Vandezande K, Underwood J, et al. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am J Pathol. 2004;164:285–93. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–18. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–16. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–23. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, et al. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–57. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–13. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–6. [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. Embo J. 2001;20:4987–97. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–8. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZL, Luo DZ, Wen JM. Expression and significance of tumor-related genes in HCC. World J Gastroenterol. 2005;11:3850–4. doi: 10.3748/wjg.v11.i25.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–20. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahalad P, Calvo I, Waechter H, Matthews JB, Zuk A, Matlin KS. Regulation of MDCK cell-substratum adhesion by RhoA and myosin light chain kinase after ATP depletion. Am J Physiol Cell Physiol. 2004;286:C693–707. doi: 10.1152/ajpcell.00124.2003. [DOI] [PubMed] [Google Scholar]
- Privette LM, Gonzalez ME, Ding L, Kleer CG, Petty EM. Altered expression of the early mitotic checkpoint protein, CHFR, in breast cancers: implications for tumor suppression. Cancer Res. 2007;67:6064–74. doi: 10.1158/0008-5472.CAN-06-4109. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Silva A, Zambelli Ramalho LN, Britto Garcia S, Zucoloto S. The relationship between p63 and p53 expression in normal and neoplastic breast tissue. Arch Pathol Lab Med. 2003;127:336–40. doi: 10.5858/2003-127-0336-TRBPAP. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–12. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Miyamoto M, Hosoda F, Gotoh M, Tsuta K, et al. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 2010 doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–5. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–54. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- Tubiana M, Koscielny S. The rationale for early diagnosis of cancer--the example of breast cancer. Acta Oncol. 1999;38:295–303. doi: 10.1080/028418699431357. [DOI] [PubMed] [Google Scholar]
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–10. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–94. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–19. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–17. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009a;69:1792–9. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper TM, Morreale RJ, Morris TA, Mintz-Cole RA, Hoskins EE, Balsitis SJ, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am J Pathol. 2009b;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–94. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–82. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.