Abstract
Background
Metastatic colorectal cancer is frequently treated with irinotecan, a topoisomerase-I inhibitor. The UGT1A1 gene encodes for an enzyme that metabolizes irinotecan and its genetic variants were shown to be associated with increased drug toxicity. We evaluated clinical outcomes associated with the UGT1A1*28 variant.
Methods
Included were 329 colorectal cancer patients, participants in the Israeli population-based MECC study, treated with a chemotherapy regimen that included irinotecan. Cases with metastases or disease recurrence were followed up for a median period of 2.0 years after the appearance of the event. Study end points were appearance of grade 3–4 hematological and gastroenterological toxicity, hospitalization due to toxic events, mostly neutropenia, fever, diarrhea or vomiting, and length of hospitalization and overall survival. UGT1A1*28 was genotyped from peripheral blood DNA by fragment analysis and reported as number of TATA sequence repeats in the promoter of the gene.
Results
The 7/7 variant of UGT1A1*28 was detected in 11.9% of the 329 participants. Grade 3–4 hematological toxicity was significantly higher in 7/7 carriers compared to 6/7 and 6/6 carriers (48.0%,10.2%,7.7% correspondingly, p<0.001), and so was the risk of toxicity related hospitalization (45.8%,25.3%,14.4% correspondingly, p=0.001). Both, short-term death within 2 months of treatment onset(12.8%, 5.2% and 2.9% correspondingly) and median overall survival(1.6,2.0,2.4 years correspondingly, log-rank p=0.01) were significantly worse in the 7/7 carriers. The age-stage-adjusted hazard ratio (HR) for cases with the 7/7 genotype compared to 6/6 was 1.7 (1.1–2.5).
Conclusions
UGT1A1*28 7/7 genotype is strongly associated with severe hematologic toxicity and higher hospitalization rate and predicts lower survival of colorectal cancer in users of Irinotecan.
Keywords: Irinotecan, Colorectal cancer, UGT1A1, Toxicity, Survival
Introduction
Metastatic colorectal cancer is frequently treated with Irinotecan (CPT-11), a topoisomerase-I inhibitor (1). The gene UGT1A1 encodes an enzyme that catalyzes the glucuronidation of the active irinotecan metabolite SN-38, which is eliminated in the liver by metabolic alteration to an inactive form SN-38G (2). Irinotecan toxicity is relatively commonly observed and myelo-suppression (neutropenia) and diarrhea are the most commonly reported dose-limiting toxicities (3,4). Homozygosity for UGT1A1*28 allele (7/7) have previously been shown to be associated with irinotecan-related hematological (5–13) or gastrointestinal (11, 14–16) toxicity. These findings led the Food and Drug Administration in 2005 to require that gene related information be added to the drug product label (17). It is as yet unclear if carriers of the different genetic variants also have a different prognosis (7, 8, 18, 19) or differ in number or length of hospitalizations (11). We retrospectively evaluated the association between UGT1A1 genetic variation, prevalence of severe toxicity and survival of irinotecan-treated colorectal cancer patients in a large cohort of consecutive patients.
Materials and Methods
Participants
Patients diagnosed with colorectal cancer within the framework of the Molecular Epidemiology of Colorectal Cancer (MECC) Study and who were treated with Irinotecan-based chemotherapy (FOLFIRI, IFL, TEGAFIRI, XELIRI) for metastatic disease were included in this sub-analysis. The MECC study is a population-based case-control study of all newly diagnosed colorectal cancer cases in Northern Israel. 2,135 cases from phase I of the study, which took place in 1998–2004, served as the source for the cases participating in the current analysis. The study was approved by the IRB committees of Carmel Medical Center (Haifa, Israel) and the University of Michigan, Ann Arbor.
The methods of the MECC Study were formerly described (20). In brief, newly diagnosed cases of colorectal cancer and their population-based, randomly selected, age-sex-residence matched controls were identified and after signing a consent form were interviewed with an extensive questionnaire and gave a venous blood sample, further separated into DNA, serum and lymphocytes.
Baseline clinical information included data extracted from pathology reports of all participating cases ( stage at diagnosis, grade, tumor location in the colon, histological type )
Follow-up
The medical records of 2,135 CRC cases recruited in phase I of the MECC study were sought. Of them, 2,039 (95.5%) were retrieved with either full follow up data (1762 cases) or partial follow up data (277 cases). 96 records could not be found, either because cases were detected in very early stage and did not enter oncology surveillance or because the patient died shortly after diagnosis. Medical records were extracted to provide information about events of tumor recurrence, appearance and location of metastases, appearance of second primary tumors in or outside the colon, as well as detailed treatment information. Date of death and cause of death were available from the Israeli population register.
Among the 2,039 MECC cases, 387 cases were identified as having been treated with irinotecan-based chemotherapy and serve as the study group for this report. Blood samples were available for genetic analysis for 329 of them (85.0%). Time of first evidence of event (metastasis or local recurrence) was recorded or imputed as time of start of treatment if time of event was missing in record (in 23 of 387, 5.9%). 214 of the 329 (63.5%) cases with genetic analyses could be retrieved and studied for evaluation of events of hematological or non-hematological toxicity.
Laboratory assays
Genomic DNA extracted from blood was used for genotyping either by simple determination of TATA box sequence in patient DNA or by implementation of fragment analysis. Isolation of UGT promoter region which carries the *28 mutation was performed with the following primers: Forward 5′-TTC CAG CCA GTT CAA CTG TTG -3′, Reverse 5′-GCC TTT GCT CCT GCC AG – 3′. PCR products underwent a sequencing PCR reaction using BigDye@Terminator reagents (Applied Biosystems). Forward primer was used for reaction initiation. Products were purified with BigDYE®XTerminator™ purification kit (Applied Biosystems) and were sequenced with the 3130xl Genetic Analyzer (Applied Biosystems). Fragment analysis was performed by isolating the UGT promoter region via PCR reaction with the following FAM labeled primers: Forward 5 -FAM′-AAA TTC CAG CCA GTT CAA CTG TTG TT -3′, Reverse 5′-GCC TTT GCT CCT GCC AG – 3′. PCR products were diluted 1:20 and 1μl was added to a reaction solution containing 8.5μl Formamide and 0.5μl GeneScan™ –500 ROX™ Size Standard (Applied Biosystems). Fragment analysis was performed with the 3130xl Genetic Analyzer (Applied Biosystems). The polymorphism UGT1A1*28 is characterized by the presence of an additional TA repeat in the TATA sequence of the UGT1A1 promoter, ((TA)7TAA, instead of (TA)6TAA). Fragment analysis determines fragment size with 211 nucleotide fragment for the 6 TA repeats and 213 for 7 TA repeats. Genotype data was available for 329 of the patients
Statistical Methods
The statistical analysis included an analysis of the risk of severe side effects in irinotecan-treated colorectal cancer cases and an analysis of the survival of irinotecan-treated cases. Participants in the side-effects analysis were 214 cases with full treatment information and genotype who were also among the 329 cases in the survival analysis genotype data.
The association between UGT1A1*28 genotypes and demographic variables were tested using Chi-Square tests (exact test when appropriate).
The association between UGT1A1*28 genotype and the incidence of Grade 3–4 toxicities were tested using Armitage test for trend. Similarly, the association with toxicity related hospitalization mortality (death within two months from treatment initiation) was assessed. Overall survival was calculated form the date of first metastases (or recurrence) diagnosis to death date or last date on study. Overall survival was presented with the use of Kaplan-Meier curves and was compared with the use of Log-Rank test. The Cox proportional hazards model was used to estimate hazard ratio, with adjustment to age and existence of metastasis at initial diagnosis and when relevant also to ethnicity.
Results
UGT1A1*28 genotype-specific toxicity of irinotecan-based treatments was studied in 214 colorectal cancer cases and genotype-specific overall survival was studied in 329 irinotecan-treated colorectal cancer cases.
No significant differences in the demographics of cases and in their clinical data (stage at diagnosis or overall survival) were noted among the overall series, those with genotype-specific survival data, and all cases with irinotecan treatment and toxicity data (Table 1).
Table 1.
Demographic and clinical comparison of study sub-groups
| Overall series | Genotyped cases | Cases with toxicity data | |
|---|---|---|---|
| n=387 | n=329 | n=214 | |
| Gender | |||
| Males | 48.7% | 48% | 46.3% |
| Females | 51.3% | 52% | 53.7% |
| Ethnicity | |||
| Jews | 83.9% | 82.4% | 85% |
| Non-Jews | 16.1% | 17.6% | 15% |
| Mean age (± SD) at diagnosis | 63.1±11.4 | 62.9±11.6 | 63.1±10.9 |
| Evidence for metastasis at diagnosis | 53.3% | 52.9% | 50% |
| Median (IR) of Time from Diagnosis to Treatment (wk) | 48 13–108 |
48 13–109 |
44 11–99 |
| Overall Survival (OS) | |||
| Number of deaths (%) | 338 (87.3%) | 286 (86.9%) | 185 (88.5%) |
| Median OS (Years from Initial Diagnosis) | 2.8 | 2.9 | 2.8 |
IR=Interquartile Range
Genotype frequency
UGT1A1*28 genotypes 6/6, 6/7, 7/7 were detected in 41.6%, 46.5%, and 11.9% of the patients respectively (Table 2). Genotype distribution was not significantly different between Jews (11.1% with 7/7 variant) and non-Jews (15.5% with 7/7 variant) and between males (9.5% with 7/7) and females (14.0% with 7/7). No difference was noted between the genetic subtypes, in age at diagnosis or stage at presentation (Table 2). Although, the 6/6 genotype tended to be diagnosed with lower proportion of stage IV.
Table 2.
Demographic and clinical characteristics of cases by UGT1A1*28 genotype
| 6/6 | 6/7 | 7/7 | ||
|---|---|---|---|---|
| Total population n= 329 | 137 (41.6%) | 153 (46.5%) | 39 (11.9%) | |
| Gender (p=0.135*) | ||||
| Males | n= 158 (100%) | 38.6% | 51.9% | 9.5% |
| Females | n= 171 (100%) | 44.4% | 41.3% | 14% |
| Ethnicity (p=0.207*) | ||||
| Jews | n= 271 (100%) | 40.2% | 48.7% | 11.1% |
| Non-Jews | n= 58 (100%) | 48.3% | 36.2% | 15.5% |
| Mean age (± SD) at diagnosis& | 62±11.9 | 63.6±11.6 | 63.1±10.6 | |
| Stage at diagnosis (p*=0.077) | ||||
| 1 | 4.6% | 2.1% | 2.8% | |
| 2 | 26.9% | 12.6% | 19.4% | |
| 3 | 27.7% | 31.5% | 30.6% | |
| 4 | 40.8% | 53.8% | 47.2% | |
| Unknown (n) | 7 | 10 | 3 | |
chi-Square test
Anova test, p=0.487
Treatment characteristics
Treatment protocols in our series included FOLFIRI (38% of treatments), IFL (41%) and other regimens (XELIRI, TEGAFIRI - 21%). No difference in treatment protocols was noted between the groups with different genotypes Table 3). In the overall irinotecan-treated series, treatment was initiated at a median time of 48 weeks after initial diagnosis, and 12 weeks from diagnosis of metastases or recurrence with a wide time range. Patients with 6/6 genotype had a significantly longer time to treatment onset than patients with 6/7 or 7/7 genotype (62 weeks versus 39 and 42, respectively) (Table 3). This longer time was a reflection of a significantly longer time to development of metastases/recurrence and not due to differences in time of treatment initiation after metastases were detected.
Table 3.
Differences, by UGT1A1*28 genotype, in time from diagnosis, metastatic disease and treatment
| 6/6 | 6/7 | 7/7 | ALL (n=329) | |
|---|---|---|---|---|
| n=137 | n=153 | n=39 | ||
| Time (in weeks) from initial diagnosis and treatment start (p=0.014)& | ||||
| Median | 62 | 39 | 42 | 48 |
| Inter-quartile Range | 22–122 | 11–86 | 11–97 | 13–109 |
| Time (in weeks) from 1st metastasis/recurrence to treatment start (p=0.13)& | ||||
| Median | 13 | 11 | 13 | 12 |
| Inter-quartile Range | 6–38 | 5–23 | 8–38 | 6–30 |
| Treatment protocol^ (p=0.69) | ||||
| FOLFIRI | 42% | 37% | 28% | 38% |
| IFL | 37% | 44% | 44% | 41% |
| Other irinotecan-based (TEGAFIRI, XELIRI) | 21% | 19% | 28% | 21% |
chi-Square test
Kruskal-Wallis test
In patients with genotype and full oncological follow-up
Drug toxicity
Among the 214 patients who had detailed treatment and toxicity information, a total of 164 grade 3–4 toxicity events were reported in 98 patients (46.9%). The most common toxicity events were: diarrhea (22.4%), vomiting (9.8%), leucopenia (9.3%), neutropenia (8.9%), infection (5.1%), neutropenic fever (3.7%) and mucositis (1.4%) (Table 4).
Table 4.
Grade 3–4 toxicity in irinotecan treated colorectal cancer cases, by UGT1A1*28 genotype
| 6/6 | 6/7 | 7/7 | ALL | P for trend | |
|---|---|---|---|---|---|
| n=91 | n=98 | n=25 | n=214 | ||
| Grade 3–4 Toxicity | |||||
| Hematological n (%) | 7 (7.7%) | 10 (10.2%) | 12 (48.0%) | 29 (13.6%) | P<=0.001 |
| Leucopenia % | 6.6% | 6.1% | 32% | 9.3% | P=0.005 |
| Neutropenia % | 5.5% | 8. 2% | 24.0% | 8.9% | P=0.019 |
| Neutropenic fever % | 0 | 2.0% | 24.0% | 3.7% | P<=0.001 |
| Non –hematological n (%) | 31 (34.1%) | 29 (29.6%) | 7 (28%) | 67 (31.3%) | P=0.46 |
| Diarrhea % | 27.5% | 18..4% | 20% | 22.4% | P=0.20 |
| Vomiting % | 4.4% | 12.2% | 20% | 9.8% | P=0.015 |
| Infection % | 4.4% | 7.1% | 0 | 5.1% | P=0.82 |
| Mucositis % | 2.2% | 1% | 0 | 1.4% | P=0.73 |
| Time to toxicity (weeks) Median (IR) | 5.9(2.1–8.6) | 3.2(2.1–5.9) | 2.1(1.2–3.1) | 3.1 (2–6.9) | P=0.016 |
| Hospitalization due to toxicity | |||||
| Yes n (%) | 13 (14.4) | 24 (25.3%) | 11 (45.8%) | 48 (23%) | P=0.001 |
| unknown | 1 | 3 | 1 | ||
| Short-term death*(%) | 2.9% | 5.2% | 12.8% | 5.2% | P=0.027 |
within two months from treatment onset, based on all treated patients
The median (Inter-quartile Range) time from start of treatment to appearance of 1st event of grade 3–4 toxicity was 5.9 (2.1–8.6) weeks for UGT1A1*28 6/6, 3.1 (2.1–5.9) weeks for 6/7 and shortest, 2.1 (1.2–3.1) weeks for cases with the 7/7 genotype, p=0.016.
Hematological toxicity was significantly more common in patients with the 7/7 genotype, 48% as compared with 10.2% and 7.7% in the 6/7 and 6/6 genotypes (p for trend<0.001). A similar elevated event rate was observed for leucopenia, neutropenia and neutropenic fever (Table 4).
There were no significant differences between cases with the genetic sub-types in the rate of non-hematological grade 3–4 toxicities although vomiting was found to be significantly increased with an additional copy of a 7 allele.
Hospitalization due to toxicity was recorded in 48 patients (23%). The frequency of hospitalization was significantly higher among the 7/7 genotype group (45.8%) compared to the 6/6 group (14.4%) and 6/7 group (25.3%) (p for trend=0.01). The median number of hospitalization days, for those who were hospitalized, was similar across genotype group (6, 5 and 5 for the 6/6, 6/7 and 7/7, respectively).
The pattern of increased toxicity and increased hospitalization rate among the cases with the 7/7 genotype was noted in all treatment protocol types (data not shown).
When toxicity analysis was stratified by ethnicity, the apparent association of the 7 allele with increased hematological grade 3–4 toxicity was observed in both Jewish and non-Jewish patients (data not shown).
Seventeen deaths were observed in the 2-month period following the start of irinotecan-based treatment. Death rates significantly increased with an additional copy of the 7 allele: 2.9%, 5.2% and 12.8%, for the 6/6, 6/7 and 7/7, respectively (p for trend=0.027).
Overall Survival
Among the 329 cases available for this analysis, a total of 286 deaths of any cause (86.9%) occurred during the follow-up period which started at the date of first metastasis or local recurrence event. The median follow-up period (from event) was of 2.0 years (and 6.0 years for the patients who were alive at end of follow-up period).
The median overall survival of the three genotype groups was significantly different: 2.4, 2.0 and 1.6 years, for the 6/6, 6/7 and 7/7 respectively (p=0.008, log-rank test) (Figure 1). The hazard ratio (HR), adjusted for age and stage at diagnosis (IV vs. other), comparing cases with the 7/7 genotype and the 6/6 genotype was 1.7 (95% CI=1.1 to 2.5). When analysis was stratified according to ethnicity, the adjusted HR comparing the 7/7 genotype to the 6/6 genotype was 1.6 (95% CI=1.0 to 2.5) among Jewish cases and 2.2 (95% CI =0.99 to 5.0) for non-Jewish cases. The age- stage-and ethnicity-adjusted HR for death of cases with 7/7 compared to 6/6 was 1.7 (95% CI= 1.1 to 2.3, p=0.010).
Figure 1.
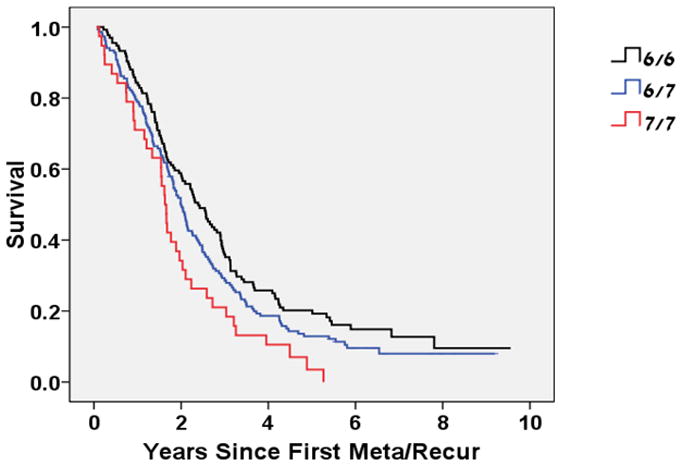
Kaplan-Meier curve of Overall Survival (measured from 1st metastasis or recurrence event) by UGT1A1*28 Genotype
p=0.008 (log-rank test)
Age-stage-ethnicity-adjusted HR (7/7 vs. 6/6) = 1.7 (1.1–2.3), p=0.01
Analysis of survival from time of initial diagnosis provided similar results with worse survival for the 7/7 genotype (median survival time of 3.5, 2.8 and 2.1 for the 6/6, 6/7 and 7/7 genotypes respectively (p=0.003 log-rank test, Figure 2) and an elevated age- stage- ethnicity-adjusted HR for death the 7/7 genotype of 1.6 (95% CI= 1.1 to 2.4, p=0.015).
Figure 2.
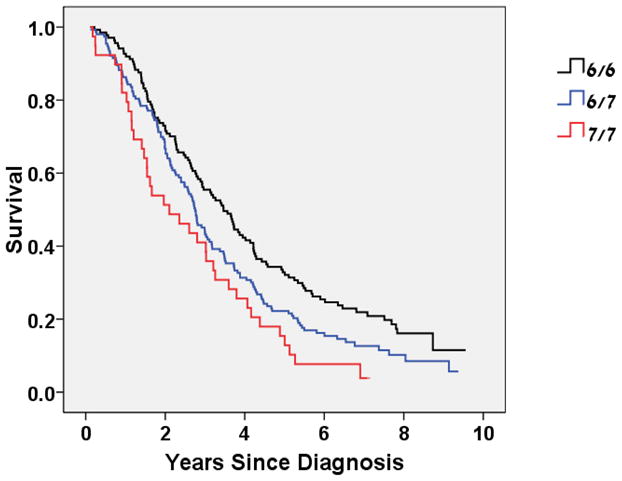
Kaplan-Meier curve of Overall Survival (measured from time of diagnosis) by UGT1A1*28 Genotype
P=0.003 (log-rank test)
Age-stage-ethnicity-adjusted HR (7/7 vs. 6/6) = 1.6 (1.1–2.4), p=0.015
Discussion
Our data suggests significantly higher rate of complications and worse survival among irinotecan-treated patients with advanced colorectal cancer who carry the 7/7 variant of UGT1A1*28.
Our finding of a higher rate of grade 3–4 toxicity in irinotecan-treated cases who were carriers of the 7/7 variant, is in line with findings in most other studies with different treatment protocols (5–16). A meta-analysis (11) suggested that such higher toxicity of at least hematological side effects is only seen when treatments with doses > 150 mg/m2 were employed. While we were unable to retrospectively calculate the exact doses given to our study participants, the treatment protocols used in our study suggest that most of our cases were likely to be in the commonly used therapeutic range of low doses. The many different irinotecan treatment regimens (IFL, Capecitabin/Irinotecan, Raltitrexed/Irinotecan, FOLFIRI, FOLFOXIRI, other) used in the different trials included in the meta-analysis could make the results difficult to directly compare to our results.
In our data no events of neutropenic fever were noticed among individuals with the 6/6 variant. This is in line with a recent UGT1A1 genotype-driven phase I study of irinotecan (18) which demonstrated the possibility of irinotecan dose escalation in patients with UGT1A1 wild type homozygous TA6/TA6 of up to 370 mg/m2/every 2 weeks. We see great importance in further investigation of the UGT 1A1 genotype dose-response relationship, evaluation of the clinical effectiveness and of the utility of UGT1A1 genotyping as related to true clinical benefit of different irinotecan protocols.
While most irinotecan pharmacogenetic trials focused on the predictive role of UGT1A1 variants on toxicity, only a few trials evaluated the prognostic value of these genetic markers (7, 8). Our study demonstrated a clear survival advantage to cases with wild-type variants among the cases that were treated with irinotecan of any regimen. A prospective study (7), with a comparable number of patients, which also reported limited higher hematological toxicity in 22 FOLFIRI treated carriers of the 7/7 variant found them to also have a higher response rate but no significant survival difference. However their median follow-up time was of only 15 months in which only 52% of the participants died. A second retrospective smaller study (8) in a Chinese population did not detect any association between the genetic status and progression-free or overall survival but had only 6 cases with the 7/7 variant. Our much longer follow-up time, with a median time for mortality events at 24 months and more, together with our larger number of participants, and especially of 7/7 variant carriers, could explain the difference between our findings and the findings in the other studies. Possible explanations for a survival disadvantage in 7/7 carriers of the UGT1A1 gene could lie either in the possibility that these patients are receiving suboptimal treatment due to the severity of side effects, or even drop out of treatment altogether. In addition, the mutation which influences glucoronidation processes in the liver could be of wider importance as the liver is not only the leading site of metastases development in colorectal cancer patients, but also the metabolic site of many other chemotherapeutic agents. If these are of importance, we would expect to see a survival disadvantage of 7/7 carriers also in cases that have not been treated with irinotecan-based regimen. Such data have not been reported yet, to the best of our knowledge.
The source of data for these analyses was a comprehensive population-based series of colorectal cancer cases diagnosed in Israel during the recent decade. In this sense, these data represent a real life experience of patients not involved in clinical trials, but rather being treated according to commonly used protocols. Our retrospective data, thus, might suffer from a certain level of selection bias as decisions about treatment initiation and type of treatment could differ between certain doctors and certain patients. This is a possible explanation for different irinotecan administration protocols in our study population. Another possible weakness of studies performed outside a trial set-up could be a difference in quality of collected data. We had to rely on data that appeared in the medical records, but were also able to rely on electronic records which have a close to absolute validity for events such as death, hospitalization and length of hospitalization and more. Nevertheless, even if selection bias and information bias are of concern, there is no reason to expect them to differentially influence our results as the genetic status of the involved cases was not known at time of treatment, not to the treating physicians and not to the study researchers. In addition, the prevalence of the 7/7 variant in our series was similar to the prevalence of the variant reported in other Caucasian populations. Genotyping for UGT1A1 was not incorporated into clinical practice or treatment choices for the patients included in our population-based study, and thus the total proportion of patients with the 7/7 variant treated with irinotecan based regiments is similar to the overall prevalence of the variant. Chemotherapy regimens and dosages were not influenced by genotype at the time these patients were treated.
UGT1A1*28 7/7 genotype is strongly associated with severe toxicity and hospitalizations and with lower overall survival in patients with advanced disease treated with irinotecan. These data support the FDA recommendation and product labeling to tailor treatment plans for colorectal cancer patients.
Acknowledgments
Funding for this study was provided by grant (1R01CA81488) from the National Cancer Institute.
Bibliography
- 1.Pizzolato JF, Saltz LB. The Camptothecins. Lancet. 2003;361:2235–42. doi: 10.1016/S0140-6736(03)13780-4. [DOI] [PubMed] [Google Scholar]
- 2.Innocenti F, Kroetz DL, Schuetz E, et al. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J Clin Oncol. 2009;27:2604–14. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armand JP, Extra YM, Catimel G, Abigerges D, Marty M, Clavel M. Rationale for the dosage and schedule of CPT-11 (irinotecan) selected for phase II studies, as determined by European phase I studies. Ann Oncol. 1996;7:837–42. doi: 10.1093/oxfordjournals.annonc.a010763. [DOI] [PubMed] [Google Scholar]
- 4.Rouits E, Charasson V, Petain A, et al. Pharmacokinetic and pharmacogenetic determinants of the activity and toxicity of irinotecan in metastatic colorectal cancer patients. Br J Cancer. 2008;99:1239–45. doi: 10.1038/sj.bjc.6604673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innocenti F, Ratain MJ. “Irinogenetics” and UGT1A: from genotypes to haplotypes. Clin Pharmacol Ther. 2004;75:495–500. doi: 10.1016/j.clpt.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Cecchin E, Innocenti F, D’Andrea M, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457–65. doi: 10.1200/JCO.2008.19.0314. [DOI] [PubMed] [Google Scholar]
- 7.Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061–8. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- 8.Liu CY, Chen PM, Chiou TJ, et al. UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer. 2008;112:1932–40. doi: 10.1002/cncr.23370. [DOI] [PubMed] [Google Scholar]
- 9.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–7. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 10.Cote JF, Kirzin S, Kramar A, et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin Cancer Res. 2007;13:3269–75. doi: 10.1158/1078-0432.CCR-06-2290. [DOI] [PubMed] [Google Scholar]
- 11.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–5. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 12.Biason P, Masier S, Toffoli G. UGT1A1*28 and other UGT1A polymorphisms as determinants of irinotecan toxicity. J Chemother. 2008;20:158–65. doi: 10.1179/joc.2008.20.2.158. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes KE, Zhang W, Yang D, et al. ABCB1, SLCO1B1 and UGT1A1 gene polymorphisms are associated with toxicity in metastatic colorectal cancer patients treated with first-line irinotecan. Drug Metab Lett. 2007;1:23–30. doi: 10.2174/187231207779814328. [DOI] [PubMed] [Google Scholar]
- 14.Ferraldeschi R, Minchell LJ, Roberts SA, et al. UGT1A1*28 genotype predicts gastrointestinal toxicity in patients treated with intermediate-dose irinotecan. Pharmacogenomics. 2009;10:733–9. doi: 10.2217/pgs.09.20. [DOI] [PubMed] [Google Scholar]
- 15.Massacesi C, Terrazzino S, Marcucci F, et al. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106:1007–16. doi: 10.1002/cncr.21722. [DOI] [PubMed] [Google Scholar]
- 16.de Jong FA, Kehrer DF, Mathijssen RH, et al. Prophylaxis of irinotecan-induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: a double-blind, randomized, placebo-controlled study. Oncologist. 2006;11:944–54. doi: 10.1634/theoncologist.11-8-944. [DOI] [PubMed] [Google Scholar]
- 17.FDA. http://www.accessdata.fda.gov/drugsatfda_docs/appletter/2005/020571s026ltr.pdf.
- 18.Toffoli G, Cecchin E, Gasparini G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:866–71. doi: 10.1200/JCO.2009.23.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palomaki GE, Bradley LA, Douglas MP, Kolor K, Dotson WD. Can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? An evidence-based review. Genet Med. 2009;11:21–34. doi: 10.1097/GIM.0b013e31818efd77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poynter JN, Gruber SB, Higgins PD, et al. Statins and the risk of colorectal cancer. N Engl J Med. 2005;352:2184–92. doi: 10.1056/NEJMoa043792. [DOI] [PubMed] [Google Scholar]


