Abstract
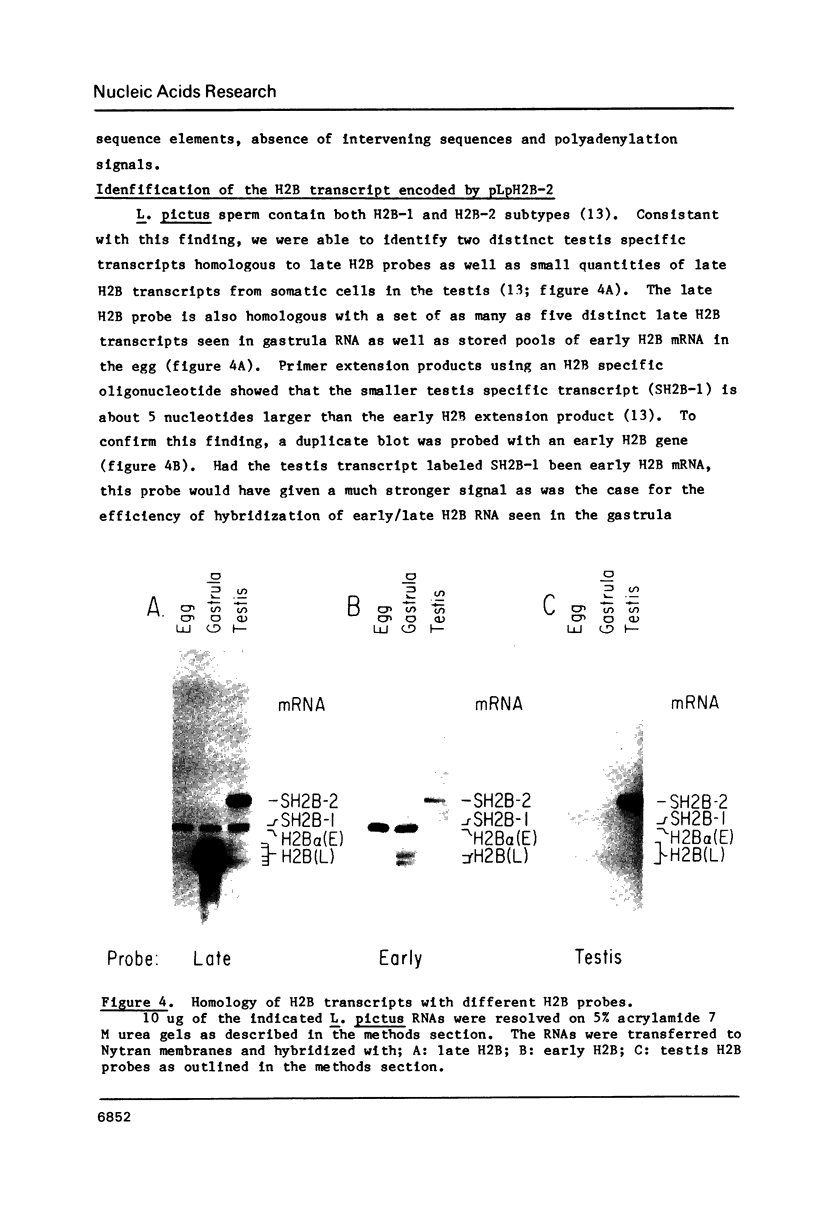
We have cloned and characterized the gene encoding a sperm specific H2B-2 histone subtype from the sea urchin L. pictus. The gene is not clustered with any other histone genes. However, it resembles other histone genes in many respects. The gene contains no intervening sequences or polyadenylation signals. Like other histone genes it contains the conserved 3' hairpin loop sequence and CAAGAAAGA box necessary for the processing of the 3' end of the histone transcript. Upstream of the gene in addition to the TATA box, and two copies of inverted CCAAT Boxes is a conserved sequence element found in many H2B genes. Unlike most histone genes, the H2B-2 gene is expressed in a sex and tissue specific manner only in testis. The sperm H2B-2 subtype encoded by this gene is 142 amino acids in length. The larger size of this H2B protein is accounted for by residues in the N-terminus of the protein consisting of a series of pentapeptide repeats typical of this histone subtype.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceci R. J., Gross P. R. Sea urchin sperm chromatin structure as probed by pancreatic DNase I: evidence for a noval cutting periodicity. Dev Biol. 1980 Nov;80(1):210–224. doi: 10.1016/0012-1606(80)90509-6. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Cetta A., Davidson E. H. The single-copy DNA sequence polymorphism of the sea urchin Strongylocentrotus purpuratus. Cell. 1978 Dec;15(4):1175–1186. doi: 10.1016/0092-8674(78)90044-2. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Barberis A. Synthesis of sperm and late histone cDNAs of the sea urchin with a primer complementary to the conserved 3' terminal palindrome: evidence for tissue-specific and more general histone gene variants. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5676–5680. doi: 10.1073/pnas.82.17.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Childs G., Nocente-McGrath C., Lieber T., Holt C., Knowles J. A. Sea urchin (lytechinus pictus) late-stage histone H3 and H4 genes: characterization and mapping of a clustered but nontandemly linked multigene family. Cell. 1982 Dec;31(2 Pt 1):383–393. doi: 10.1016/0092-8674(82)90132-5. [DOI] [PubMed] [Google Scholar]
- Cohn R. H., Lowry J. C., Kedes L. H. Histone genes of the sea urchin (S. purpuratus) cloned in E coli: order, polarity, and strandedness of the five histone-coding and spacer regions. Cell. 1976 Sep;9(1):147–161. doi: 10.1016/0092-8674(76)90060-x. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- DeLeon D. V., Cox K. H., Angerer L. M., Angerer R. C. Most early-variant histone mRNA is contained in the pronucleus of sea urchin eggs. Dev Biol. 1983 Nov;100(1):197–206. doi: 10.1016/0012-1606(83)90211-7. [DOI] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984 Jul 5;310(5972):71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Phosphorylation of sea urchin sperm H1 and H2B histones precedes chromatin decondensation and H1 exchange during pronuclear formation. Dev Biol. 1985 Mar;108(1):235–245. doi: 10.1016/0012-1606(85)90026-0. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Lund T., Murray E. J., Mellor A. L., Dahl H. H., Flavell R. A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982 Nov 11;10(21):6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. P., Robins A. J., Wells J. R. Independently evolving chicken histone H2B genes: identification of a ubiquitous H2B-specific 5' element. Nucleic Acids Res. 1982 Dec 11;10(23):7851–7863. doi: 10.1093/nar/10.23.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Childs G., Kedes L. Distinct organizations and patterns of expression of early and late histone gene sets in the sea urchin. Nature. 1983 Jan 13;301(5896):120–125. doi: 10.1038/301120a0. [DOI] [PubMed] [Google Scholar]
- Mous J., Stunnenberg H., Georgiev O., Birnstiel M. L. Stimulation of sea urchin H2B histone gene transcription by a chromatin-associated protein fraction depends on gene sequences downstream of the transcription start site. Mol Cell Biol. 1985 Oct;5(10):2764–2769. doi: 10.1128/mcb.5.10.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M., Thomsen G. H., Roeder R. G. Genomic organization and nucleotide sequence of two distinct histone gene clusters from Xenopus laevis. Identification of novel conserved upstream sequence elements. J Mol Biol. 1985 Oct 5;185(3):479–499. doi: 10.1016/0022-2836(85)90065-8. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Gross P. R. Histones and histone synthesis in sea urchin development. Dev Biol. 1974 Feb;36(2):286–298. doi: 10.1016/0012-1606(74)90052-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showman R. M., Wells D. E., Anstrom J., Hursh D. A., Raff R. A. Message-specific sequestration of maternal histone mRNA in the sea urchin egg. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5944–5947. doi: 10.1073/pnas.79.19.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Bergman L. W. Structure of sea urchin sperm chromatin core particle. J Biol Chem. 1980 Nov 25;255(22):10702–10709. [PubMed] [Google Scholar]
- Simpson R. T. Modulation of nucleosome structure by histone subtypes in sea urchin embryos. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6803–6807. doi: 10.1073/pnas.78.11.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoultchi A., Gross P. R. Maternal histone messenger RNA: detection by molecular hybridization. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2840–2844. doi: 10.1073/pnas.70.10.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., Von Holt C. The complete amino-acid sequence of histone H2B(1) from sperm of the sea urchin Parechinus angulosus. Eur J Biochem. 1977 Jul 15;77(2):263–275. doi: 10.1111/j.1432-1033.1977.tb11665.x. [DOI] [PubMed] [Google Scholar]
- Strickland W. N., Strickland M., Brandt W. F., Von Holt C., Lehmann A., Wittmann-Liebold B. The primary structure of histone H1 from sperm of the sea urchin Parechinus angulosus. 2. Sequence of the C-terminal CNBr peptide and the entire primary structure. Eur J Biochem. 1980 Mar;104(2):567–578. doi: 10.1111/j.1432-1033.1980.tb04460.x. [DOI] [PubMed] [Google Scholar]
- Strickland W. N., Strickland M., Brandt W. F., Von Holt C. The complete amino-acid sequence of histone H2B(2) from sperm of the sea urchin Parechinus angulosus. Eur J Biochem. 1977 Jul 15;77(2):277–286. doi: 10.1111/j.1432-1033.1977.tb11666.x. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Zalenskaya I. A., Pospelov V. A., Zalensky A. O., Vorob'ev V. I. Nucleosomal structure of sea urchin and starfish sperm chromatin. Histone H2B is possibly involved in determining the length of linker DNA. Nucleic Acids Res. 1981 Feb 11;9(3):473–487. doi: 10.1093/nar/9.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]