Abstract
Currently there are several dozen human polymorphisms that have been loosely associated with cancer risk. Correlating such variants with cancer risk has been challenging, primarily due to factors such as genetic heterogeneity, contributions of diet and environmental factors, and the difficulty in obtaining large sample sizes for analysis. Such difficulties can be circumvented with the establishment of mouse models for human variants. Recently, several groups have modeled human cancer susceptibility polymorphisms in the mouse. Remarkably, in each case these mouse models have accurately reflected human phenotypes, and clarified the contribution of these variants to cancer risk. We recently reported on a mouse model for the codon 72 polymorphism in p53, and found that this polymorphism regulates the ability to cooperate with NFκB and induce apoptosis. Here-in we present evidence that this polymorphism impacts the apoptotic function of p53 in a tissue-specific manner; such tissue-specific effects of polymorphic variants represent an added challenge to human cancer risk association studies. The data presented here support the premise that modeling human polymorphisms in the mouse represents a powerful tool to assess the impact of these variants on cancer risk, progression and therapy.
Key words: p53, polymorphism, apoptosis, codon 72, NFκB
Introduction
The p53 protein is central to the ability of an organism to suppress tumor development. In response to oncogenic or genotoxic stress, p53 becomes activated as a transcription factor and transactivates genes including p21/waf1, which functions in growth arrest and senescence. p53 also transactivates genes such as Puma, Noxa and Bax, which play roles in apoptosis. In addition to its transcriptional role, p53 has a direct apoptotic role at the mitochondria.1–3 The central role of p53 in tumor suppression is best epitomized by the fact that this gene is mutated with remarkably high frequency (>50%) in human tumors. Other members of the p53 pathway are also mutated in human cancer. For example, the negative regulator of p53, MDM2, is amplified and overexpressed in certain cancers.4 Additionally, the negative regulator of MDM2, ARF, is mutationally inactivated in a significant percentage of human tumors.5 While the role of p53 as a tumor suppressor gene garners the majority of attention, it is important to note that p53 also plays an integral role in a diversity of biological processes, including reproduction, metabolism and innate immunity.
A distinctive feature of genes in the p53 pathway is that several functionally significant polymorphisms have been identified in these genes that affect the ability of p53 to induce apoptosis or senescence. For example, in the p53 gene itself there is an intensively investigated polymorphism at codon 72, which encodes either proline (P72) or arginine (R72). Experimental studies in cell lines suggest that the P72 variant possesses increased ability to induce growth arrest and senescence due to increased ability to transactivate p21/waf1.6–8 Conversely, the R72 variant possesses increased ability to induce apoptosis, due to increased mitochondrial localization, and in some cases superior ability to transactivate a subset of pro-apoptotic genes.9–11 At codon 47, p53 encodes either proline (wt) or serine (S47). The S47 variant has reduced phosphorylation of serine 46, along with decreased transcriptional and apoptotic function.12 Another common polymorphism lies in the promoter of the Mdm2 gene. SNP309 (single nucleotide polymorphism at base pair 309) in the Mdm2 promoter encodes either “G” or “T” alleles; the “G” allele produces an Sp1 site that increases the level of MDM2, thereby reducing the overall level of p53 protein.13 Studies on the association of these polymorphic variants with cancer risk have been controversial. Correlations between the codon 72 polymorphism and cancer incidence have produced conflicting results in different tumor types.14 Correlations between SNP309 of the MDM2 gene have held true only in certain tumor types, and in certain ethnic groups.15 Finally, the low frequency of the S47 polymorphism (1–5% of African Americans and Brazilians) precludes the ability to associate this polymorphism with cancer risk in a statistically significant way.16,17 These facts constitute a compelling argument for modeling these polymorphisms in the mouse.
Mouse Models of Human Polymorphic Variants Mimic Human Phenotypes
Recently Johnson et al. described two mouse models for the codon 72 polymorphism of p53; because amino acid 72 in mouse encodes alanine, this group opted to use homologous recombination to knock in exon 4 of the human p53 gene into the mouse. In the second model they generated transgenic mouse for the bacterial artificial chromosome (BAC) containing the human p53 gene, and crossed this mouse to the p53 knockout mouse. Consistent with the data on P72 and R72 accumulated in human cell lines, this group found increased apoptotic potential associated with the R72 variant in the two cell types they analyzed, the skin epidermis and the small intestine. Somewhat surprisingly, these apoptotic differences did not lead to differences in skin cancer incidence in a UV-induced model, suggesting that, at least in this cell type, apoptosis might not be the prevailing contributor to p53-mediated tumor suppression.11
Lozano and colleagues recently created a model for the SNP309 polymorphism in mouse. Like the codon 72 polymorphism, the region of the Mdm2 promoter containing SNP309 is not conserved in the mouse. Therefore, this group knocked the human intron 3, containing either allele of SNP309 (“G” or “T”), into the mouse Mdm2 locus. Remarkably, this SNP functioned the same in mouse as in human, and in the thymus the “G” allele of SNP309 was associated with 4-fold increased Mdm2 mRNA levels. Interestingly, in other tissues this polymorphism had significantly less effect on Mdm2 mRNA level, suggesting possible tissue-specific impact. Unlike the case for the codon 72 polymorphism mouse model, the impact of SNP309 on cancer was unequivocal: the SNP309 “G” allele had increased incidence of spontaneous cancer (p = 0.015), and in a mouse model of Li Fraumeni syndrome, decreased survival (p = 0.0005). Even more remarkable was the impact of this SNP on tumor type: mice with the SNP309 “G” allele developed mammary adenocarcinomas; this is an unusual tumor type in mice, and one not seen for “T” mice.18
The Codon 72 Polymorphism of p53 Regulates Interaction with NFκB
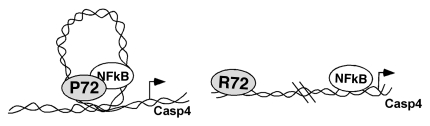
In our investigations on the biology of the p53 codon 72 polymorphism, we made use of the humanized knock-in model for p53 (Humanized p53 knock-in or Hupki). In this model, the murine p53 sequences from exon 4 through exon 9 have been replaced with the corresponding human p53 segment. There are 2 Hupki strains that have wild-type human p53 sequences, one with the codon 72 (in exon 4) encoding arginine and one encoding proline.19,20 In both strains the chimeric human-mouse p53 protein is functional and tumor suppressive.8,19–21 In analyzing the DNA damage response of inbred Hupki P72 and R72 mice, we noted that the P72 allele was consistently able to induce 2.5-fold increased apoptosis in the thymus, compared to R72. Micro-array analyses revealed that the overwhelming majority of p53-induced genes were identically regulated in P72 and R72 thymuses. However, we found that a small subset of 12 genes were upregulated to greater extent in thymocytes from P72 mice. The majority of these genes in this subset were NFκB target genes, and many played roles in innate immunity. One of these genes, Caspase 4 (also called Caspase 11 in mouse), possesses canonical binding sites for both p53 and NFκB; we used chromatin immunoprecipitation to prove that both p53 and NFκB bind to this promoter, and that silencing of the p65 subunit of NFκB rendered p53 unable to bind. Similar cooperativity between p53 and NFκB has been seen in the regulation of the Skp2 promoter.22 These studies led to our finding that the P72 variant of p53 possesses significantly increased ability to interact with p65, compared to R72, in both murine and human cells. The combined data favor a model whereby the codon 72 polymorphism regulates interaction and cooperation with NFκB (Fig. 1).
Figure 1.
Model for the differential cooperation between P72 and R72 variants of p53 with NFκB. In the model on the left, p53 and NFκB bind to the promoter of Caspase 4, and the increased ability of these two proteins to interact causes a looping out of chromatin, potentially favorable for recruitment of other transcription factors or the basal transcription machinery. On the right, the decreased ability of the R72 variant to bind to NFκB limits the potential for these two transcription factors to cooperate in transcription.
Tissue-Specific Effects of the Codon 72 Polymorphism
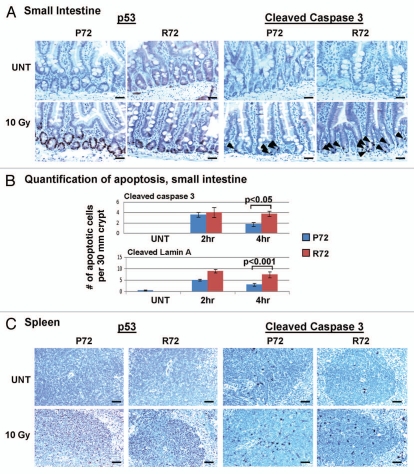
One of the compelling reasons for modeling the codon 72 polymorphism in mice was the potential for analyzing the impact of these variants in normal tissues, and the possibility that tissue-specific differences might exist in the activity of these variants. Whereas our studies indicated that gamma radiation led to a 2.5-fold increase in programmed cell death in the P72 thymus, the opposite correlation held true in the small intestine (Fig. 2A and B). In the small intestine, consistent with the findings by Johnson, we found that the R72 variant induces approximately two- to three-fold increased apoptosis.11 This finding was consistent in five independent experiments, and was statistically significant (p < 0.001). Interestingly, in the spleen, where p53-dependent apoptosis also occurs, there was no difference in the induction of apoptosis between P72 and R72 variants (Fig. 2C).
Figure 2.
Tissue-specific effects of the codon 72 polymorphism on apoptosis. (A) Immunohistochemistry using antisera for p53 and cleaved caspase 3 was performed on the small intestine of P72 and R72 mice that were untreated (UNT) or 4 h after 10 Gy gamma radiation. Increased levels of cleaved caspase 3 staining are visible in the crypt of R72 mice 4 hours after gamma radiation while the levels of p53 staining are comparable between P72 and R72 mice. The data depicted are representative of five independent experiments. Arrowheads: apoptotic cells. Scale bar: 50 um. (B) Quantification of cells positive for cleaved caspase 3 and cleaved lamin A in the small intestine of P72 and R72 mice 4 h after 10 Gy. The number of positive cells per 30 mm of crypt was counted and tabulated; the results shown are averaged from three independent experiments, and standard deviation bars are shown. p values were calculated using the Student's two-tailed t-test. Scale bar: 50 um. (C) Immunohistochemistry using antisera for p53 and cleaved caspase 3 was performed on the spleens of untreated (UNT) P72 and R72 mice or 4 hours after 10 Gy gamma radiation. The data depicted are representative of five independent experiments.
In terms of cancer risk, we found no difference in the tumor suppressive capabilities of P72 and R72 in the background of P/- and R/- mice. However, in the background of the Eu-myc model of Burkitt's lymphoma, where p53-mediated senescence is known to play a pivotal role,23 we found that the P72 variant, which possesses superior ability to induce senescence in human and murine cells,6 conferred increased survival in this model.8 The combined data suggest that the codon 72 polymorphism is likely to influence apoptosis and tumor suppression in a tissue- and/or tumor-specific manner; this may explain the controversial findings in human studies. These data also suggest that the pathways through which p53 induces apoptosis may be different in different cell types, a concept not fully appreciated until now; for example in the thymus p53 may functional primarily through the transcriptional pathway, via cooperation with NFκB, while in the small intestine other pathways may predominate. Therefore, mouse models of these polymorphic variants in the p53 pathway have the potential to increase our understanding for how p53 induces apoptosis in different cell types.
Concluding Remarks
It is nothing short of remarkable that two human-specific polymorphisms in the p53 pathway have been modeled successfully in the mouse, with phenotypes relevant to humans as the result. Interestingly, recently two other human-specific polymorphisms have been modeled successfully in the mouse. A common SNP in the gene encoding brain-derived neurotropic factor (Val66Met) was recently modeled in the mouse, and in both mouse and human this SNP was found to regulate learning.24 Similarly, a common SNP in the FGFR4 gene, Arg385, which was loosely associated with breast cancer in humans, was found to significantly promote breast tumor progression in mice.25 Taken together, these findings indicate that, while these polymorphisms may not be conserved in mouse, the relevant pathways and protein-protein interactions that they affect are conserved. These mouse models also serve as discovery tools with the potential to reveal other disease types where these polymorphisms can have impact. For example, the codon 72 polymorphism has been associated with other diseases, including ulcerative colitis and diabetes;26–28 studies in these mouse models should clarify the contribution of these polymorphic variants to disease.
Acknowledgments
This work was supported by funding from the National Institutes of Health. This project was also funded in part under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 2.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 3.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 4.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ, Bertwistle D, DENB W, Kuo ML, Sugimoto M, Tago K, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 6.Salvioli S, Bonafe M, Barbi C, Storci G, Trapassi C, Tocco F, et al. p53 codon 72 alleles influence the response to anticancer drugs in cells from aged people by regulating the cell cycle inhibitor p21WAF1. Cell Cycle. 2005;4:1264–1271. doi: 10.4161/cc.4.9.1978. [DOI] [PubMed] [Google Scholar]
- 7.Orsted DD, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival and risk of cancer in the general population. J Exp Med. 2007;204:1295–1301. doi: 10.1084/jem.20062476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank AK, Leu JI, Zhou Y, Devarajan K, Nedelko T, Klein-Szanto A, et al. The Codon 72 Polymorphism of p53 Regulates Interaction with NF{kappa}B and Transactivation of Genes Involved in Immunity and Inflammation. Mol Cell Biol. 2011;31:1201–1213. doi: 10.1128/MCB.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 10.Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhu F, Dolle ME, Berton TR, Kuiper RV, Capps C, Espejo A, et al. Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Res. 2010;70:5851–5859. doi: 10.1158/0008-5472.CAN-09-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280:24245–24251. doi: 10.1074/jbc.M414637200. [DOI] [PubMed] [Google Scholar]
- 13.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 15.Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 16.Almeida LO, Custodio AC, Pinto GR, Santos MJ, Almeida JR, Clara CA, et al. Polymorphisms and DNA methylation of gene Tp53 associated with extraaxial brain tumors. Genet Mol Res. 2009;8:8–18. doi: 10.4238/vol8-1gmr518. [DOI] [PubMed] [Google Scholar]
- 17.Pinto GR, Yoshioka FK, Silva RL, Clara CA, Santos MJ, Almeida JR, et al. Prognostic value of Tp53 Pro47Ser and Arg72Pro single nucleotide polymorphisms and the susceptibility to gliomas in individuals from Southeast Brazil. Genet Mol Res. 2008;7:207–216. doi: 10.4238/vol7-1gmr415. [DOI] [PubMed] [Google Scholar]
- 18.Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinbold M, Luo JL, Nedelko T, Jerchow B, Murphy ME, Whibley C, et al. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene. 2008;27:2788–2794. doi: 10.1038/sj.onc.1210932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- 21.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 22.Barre B, Perkins ND. The Skp2 promoter integrates signaling through the NFkappaB, p53 and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol Cell. 2010;38:524–538. doi: 10.1016/j.molcel.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene. 2010;29:1260–1269. doi: 10.1038/onc.2009.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, et al. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seitzer N, Mayr T, Streit S, Ullrich A. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res. 2010;70:802–812. doi: 10.1158/0008-5472.CAN-09-3239. [DOI] [PubMed] [Google Scholar]
- 26.Gaulton KJ, Willer CJ, Li Y, Scott LJ, Conneely KN, Jackson AU, et al. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes. 2008;57:3136–3144. doi: 10.2337/db07-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaji S, Salehi Z, Aminian K. Association of p53 codon 72 genetic polymorphism with the risk of ulcerative colitis in northern Iran. Int J Colorectal Dis. 2011;26:235–238. doi: 10.1007/s00384-010-1021-7. [DOI] [PubMed] [Google Scholar]
- 28.Vietri MT, Riegler G, Ursillo A, Caserta L, Cioffi M, Molinari AM. p53 codon 72 polymorphism in patients affected with ulcerative colitis. J Gastroenterol. 2007;42:456–460. doi: 10.1007/s00535-007-2026-z. [DOI] [PubMed] [Google Scholar]